3-Hydroxybutyryl-CoA dehydrogenase from C. butyricum, an enzyme involved in n-butanol production, has been crystallized and X-ray diffraction data from the crystal have been collected and analyzed.
Keywords: (S)-3-hydroxybutyryl-CoA dehydrogenase, Clostridium butyricum, n-butanol
Abstract
(S)-3-Hydroxybutyryl-CoA dehydrogenase from Clostridium butyricum (CbHBD) is an enzyme that catalyzes the second step in the biosynthesis of n-butanol from acetyl-CoA by the reduction of acetoacetyl-CoA to 3-hydroxybutyryl-CoA. The CbHBD protein was crystallized using the hanging-drop vapour-diffusion method in the presence of 2 M ammonium sulfate, 0.1 M CAPS pH 10.5, 0.2 M lithium sulfate at 295 K. X-ray diffraction data were collected to a maximum resolution of 2.3 Å on a synchrotron beamline. The crystal belonged to space group R3, with unit-cell parameters a = b = 148.5, c = 201.6 Å. With four molecules per asymmetric unit, the crystal volume per unit protein weight (V M) is 3.52 Å3 Da−1, which corresponds to a solvent content of approximately 65.04%. The structure was solved by the molecular-replacement method and refinement of the structure is in progress.
1. Introduction
n-Butanol is one of the most promising biofuel sources, and the widely known anaerobic bacterial strain Clostridium acetobutylicum efficiently produces n-butanol through a carbohydrate catabolic pathway (Mitchell, 1998 ▶; Inui et al., 2008 ▶). Compared with bioethanol, n-butanol has several advantages as a biofuel source such as high energy content, lower corrosiveness, lower water solubility and ease of blending with motor-vehicle fuels (Dürre, 2007 ▶, 2008 ▶; Lee, Park et al., 2008 ▶). The n-butanol synthetic pathway consists of six tightly regulated steps catalyzed by independent proteins (Jones & Woods, 1986 ▶). Despite intensive studies to improve n-butanol production, n-butanol titres in Clostridium strains are usually less than 20 g l−1 (Mitchell, 1998 ▶), which prohibits their utilization in industrialized processes. Recently, the production of n-butanol using industrial hosts such as Escherichia coli, Pseudomonas putida and Bacillus subtilis has been attempted because their genetic and physiological characteristics are well defined and there are various genetic tools available to support their modification (Inui et al., 2008 ▶). However, the final titre of n-butanol was even worse than that of Clostridium strains, and did not exceed 1 g l−1 (Inui et al., 2008 ▶; Dürre, 2007 ▶, 2008 ▶; Lee, Park et al., 2008 ▶).
Engineering non-solventogenic microbes to produce a large amount of n-butanol has also been performed for the following reasons. Firstly, n-butanol may be toxic in bacterial cells and inhibits the growth of E. coli (Jones & Woods, 1986 ▶; Ezeji et al., 2010 ▶). Secondly, n-butanol synthesis disrupts the balance of energy carriers such as NADH/NAD+, which in turn results in a decrease in n-butanol production (Atsumi et al., 2008 ▶; Felnagle et al., 2012 ▶; Shen & Liao, 2008 ▶). Thirdly, the activities of the heterologous enzymes for n-butanol synthesis are host-cell specific, so each enzyme of the pathway should be optimized depending on the heterologous host. For example, about a fivefold higher titre of n-butanol is obtained in E. coli by a chimeric pathway using proteins from three different species (Steen et al., 2008 ▶; Nicolaou et al., 2010 ▶). These indicate that understanding the detailed enzymatic reactions and regulatory mechanisms of the key enzymes involved in the n-butanol biosynthetic pathway is inevitable for increased n-butanol production (Lee, Chou et al., 2008 ▶; Felnagle et al., 2012 ▶).
(S)-3-Hydroxybutyryl-CoA dehydrogenase from C. butyricum (CbHBD) is an enzyme that catalyzes the second step in the biosynthesis of n-butanol from acetyl-CoA by reducing acetoacetyl-CoA to 3-hydroxybutyryl-CoA (Fig. 1 ▶; Jones & Woods, 1986 ▶). A BLAST search using the PDB revealed that CbHBD has 43% amino-acid sequence homology to human heart short-chain l-3-hydroxyacyl-CoA dehydrogenase (HuHAD), an enzyme that catalyzes the oxidation of the hydroxyl group of l-3-hydroxyacyl-CoA to a keto group in the β-oxidation pathway (Barycki et al., 1999 ▶). As a step towards elucidating the structural and substrate-binding properties of CbHBD, we cloned the CbHBD coding gene and purified the recombinant CbHBD protein. Crystals of CbHBD with diffraction quality were obtained by the hanging-drop method. The crystal diffracted well and data were collected to a resolution of 2.3 Å; the structure was determined by the molecular-replacement method. Here, we describe the cloning, expression, purification, crystallization and X-ray crystallographic analysis of the CbHBD protein.
Figure 1.
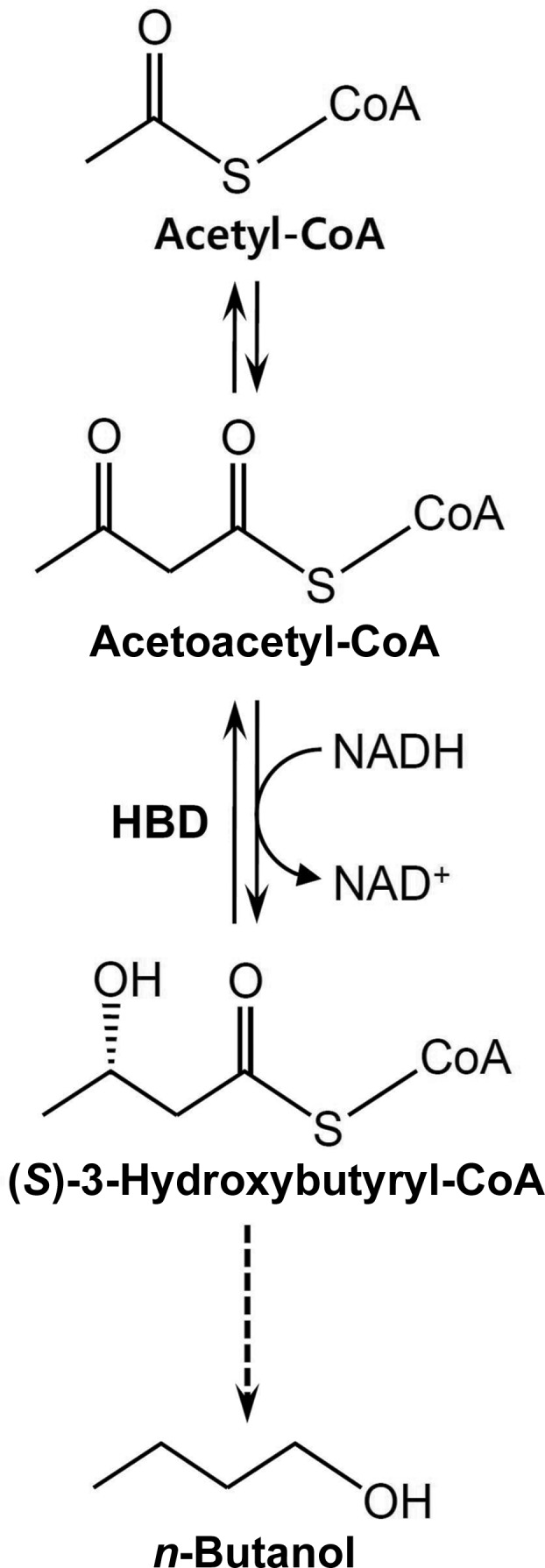
Enzymatic reaction of CbHBD. CbHBD is an enzyme involved in n-butanol biosynthesis by converting acetoacetyl-CoA to 3-hydroxybutyryl-CoA.
2. Expression and purification of the recombinant CbHBD
The forward and reverse primers were designed as 5′-GCGCGCATATGAAAAAAGTATTTGTACTTGGTGCAG-3′ and 5′-GCGCGCTCGAGTTTAGAATAATCGTAGAATCCTTTTC-3′ to introduce NdeI and XhoI cleavage sites, respectively. The CbHBD coding gene (Met1–Lys282, molecular mass 30.6 kDa) was amplified by polymerase chain reaction (PCR) using C. butyricum chromosomal DNA as a template. The PCR product was then subcloned into pET-30a (Invitrogen) with a 6×His tag at the C-terminus. The resulting expression vector pET30a:CbHBD was transformed into an Escherichia coli B834 strain, which was grown in 1 l LB medium containing kanamycin (50 mg ml−1) at 310 K. At an OD600 of 0.8, CbHBD protein expression was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After 20 h at 291 K, the cells were harvested by centrifugation at 4000g for 20 min at 277 K. The cell pellet was resuspended in buffer A (40 mM Tris–HCl pH 8.0, 5 mM β-mercaptoethanol) and disrupted by ultrasonication. The cell debris was removed by centrifugation at 13 500g for 30 min and the lysate was applied onto an Ni–NTA agarose column (Qiagen). After washing with buffer A containing 20 mM imidazole, the bound proteins were eluted with 300 mM imidazole in buffer A (Fig. 2 ▶). Finally, the trace amount of contaminants was removed by size-exclusion chromatography using a Superdex 200 prep-grade column (320 ml, GE Healthcare) equilibrated with buffer A containing 1 mM dithiothreitol (DTT). The protein eluted at a molecular mass of ∼60 kDa, indicating that the CbHBD protein with molecular weight of 30 kDa forms a dimeric structure. All purification experiments were performed at 277 K. The purity of the final protein was assessed by SDS–PAGE. The purified protein was concentrated to 30 mg ml−1 in 40 mM Tris–HCl pH 8.0, 1 mM DTT.
Figure 2.
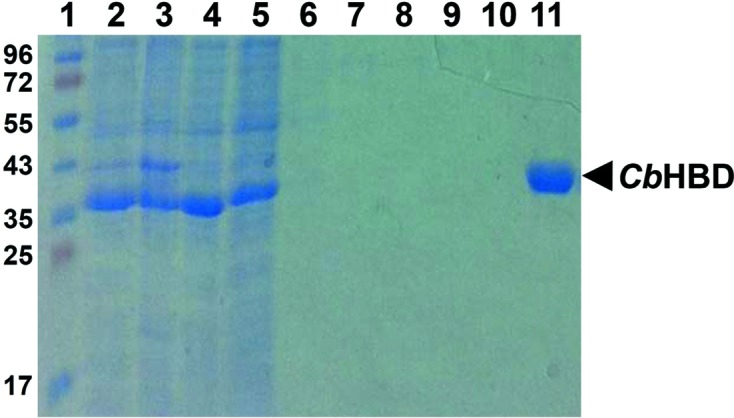
SDS–PAGE of purification of recombinant CbHBD protein. Lane 1 shows molecular-weight markers (labelled in kDa). Lanes 2–11 show the purification procedure of CbHBD using Ni–NTA chromatography. Lane 2, whole cell extract; lanes 3 and 4, pellet fraction and supernatant after centrifugation of the whole cell extract, respectively; lane 5, flowthrough from Ni–NTA column; lanes 6–10, wash with 0, 2.5, 5, 7.5 and 10 mM imidazole, respectively; lane 11, elution with 300 mM imidazole. Eluted CbHBD protein with a 6×His tag at the C-terminus is indicated on the right side of the gel.
3. Crystallization
Crystallization of the purified CbHBD protein was initially performed with commercially available sparse-matrix screens from Hampton Research and Emerald Bio using the hanging-drop vapour-diffusion method at 295 K. Each experiment consisted of mixing 1.5 µl protein solution (20 mg ml−1 in 40 mM Tris–HCl pH 8.0, 1 mM DTT) with 1.5 µl reservoir solution and then equilibrating this drop against 0.5 ml reservoir solution. CbHBD crystals were observed from several crystallization screening conditions. After several steps of improvement using the hanging-drop vapour-diffusion method, crystals of the best quality appeared in 7 d and reached maximum dimensions of 0.2 × 0.2 × 0.2 mm using 2 M ammonium sulfate, 0.1 M CAPS pH 10.5, 0.2 M lithium sulfate as the reservoir solution (Fig. 3 ▶).
Figure 3.
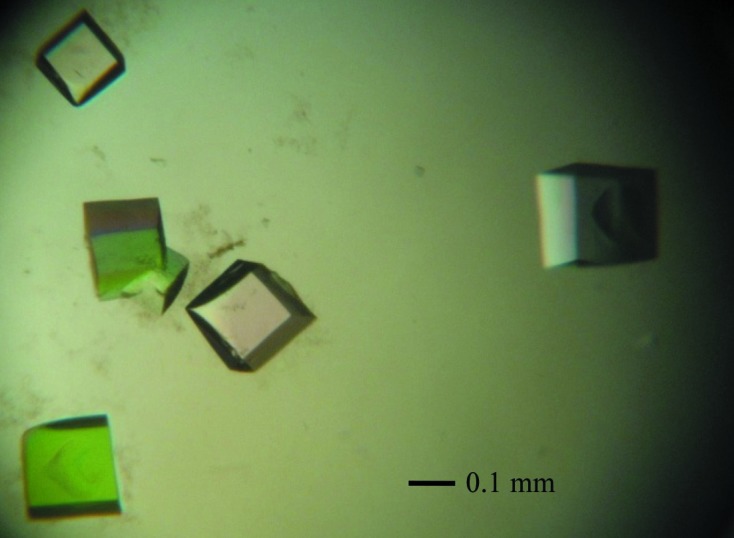
Trigonal crystals of CbHBD. Crystals of the best quality were crystallized at 2 M ammonium sulfate, 0.1 M CAPS pH 10.5, 0.2 M lithium sulfate.
4. X-ray analysis
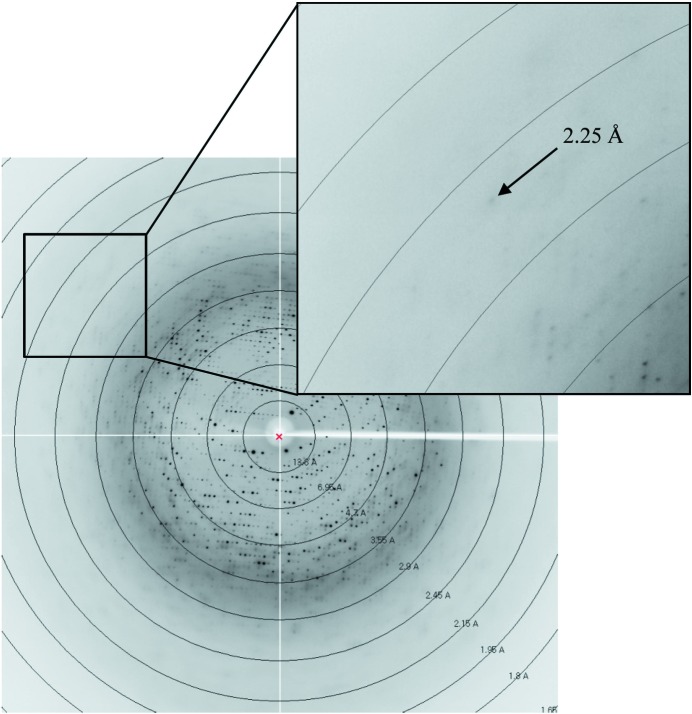
The crystals were transferred to cryoprotectant solution consisting of 0.2 M lithium sulfate, 0.1 M CAPS pH 10.5, 2 M ammonium sulfate, 30%(v/v) glycerol, fished out with a loop larger than the crystals and flash-coooled using a liquid-nitrogen cryostream at 100 K. The data were collected to a resolution of 2.3 Å on the 7A beamline (MXII) at the Pohang Accelerator Laboratory (PAL; Pohang, Republic of Korea) using a Quantum 270 CCD detector (ADSC, USA) (Fig. 4 ▶). The data were then indexed, integrated and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶). The crystals belonged to space group R3, with unit-cell parameters a = b = 148.5, c = 201.6 Å. With four molecules of CbHBD per asymmetric unit, the crystal volume per unit of protein mass was 3.52 Å3 Da−1 (Matthews, 1968 ▶), which corresponds to a solvent content of approximately 65.04%.
Figure 4.
Diffraction pattern of the CbHBD crystal. The crystal diffracted to a maximum resolution of 2.3 Å.
We attempted the molecular-replacement method of phase determination and a solution was found using human l-3-hydroxyacyl-CoA dehydrogenase (HuHAD; PDB entry 1f0y; Barycki et al., 2000 ▶) as a search model. CbHBD has an amino-acid sequence identity of 43% to HuHAD (Barycki et al., 2000 ▶). MOLREP (Vagin & Teplyakov, 2010 ▶) located four polypeptide-model molecules in the asymmetric unit. The resulting solution had a correlation coefficient and R factor of 0.505 and 57.6%, respectively. After rigid-body refinement using REFMAC5 (Murshudov et al., 2011 ▶) from the CCP4 suite (Winn et al., 2011 ▶) in the resolution range 50–2.3 Å, the R factor and R free were 54.4 and 55.7%, respectively, with an overall correlation coefficient of 0.662. The initial electron-density map, which was of good quality with backbones well defined by electron density, allowed us to build a three-dimensional model of CbHBD. After initial model building of 100 CbHBD amino-acid residues and restrained refinement, the R factor and R free were 33.5 and 35.6%, respectively, with an overall correlation coefficient of 0.812. Crystallographic model building and refinement of the structure to 2.3 Å resolution are in progress. The data statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics of CbHBD.
Values in parentheses are for the highest resolution shell.
| Beamline | 7A, PAL |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Oscillation (°) | 1.0 |
| Mosaicity (°) | 0.81 |
| Total rotation range (°) | 180 |
| Space group | R3 |
| Unit-cell parameters (Å, °) | a = b = 148.5, c = 201.6, α = β = 90.0, γ = 120.0 |
| Resolution (Å) | 50.0–2.30 (2.38–2.30) |
| Total reflections | 243475 |
| Unique reflections | 65804 |
| Completeness (%) | 93.5 (89.6) |
| R merge † (%) | 6.6 (29.6) |
| 〈I/σ(I)〉 | 29.9 (2.9) |
| Multiplicity | 3.7 (3.1) |
R
merge = 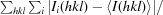
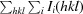 , where I
i(hkl) and 〈I(hkl)〉 are the observed individual and mean intensities of a reflection, respectively.
, where I
i(hkl) and 〈I(hkl)〉 are the observed individual and mean intensities of a reflection, respectively.  is the sum over the individual measurements of a reflection and
is the sum over the individual measurements of a reflection and 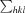 is the sum over all reflections.
is the sum over all reflections.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (NRF-2009-C1AAA001-2009-0093483) and by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by the MEST (ABC- 2012-053895).
References
- Atsumi, S., Hanai, T. & Liao, J. C. (2008). Nature (London), 451, 86–89. [DOI] [PubMed]
- Barycki, J. J., O’Brien, L. K., Bratt, J. M., Zhang, R. G., Sanishvili, R., Strauss, A. W. & Banaszak, L. J. (1999). Biochemistry, 38, 5786–5798. [DOI] [PubMed]
- Barycki, J. J., O’Brien, L. K., Strauss, A. W. & Banaszak, L. J. (2000). J. Biol. Chem. 275, 27186–27196. [DOI] [PubMed]
- Dürre, P. (2007). Biotechnol. J. 2, 1525–1534. [DOI] [PubMed]
- Dürre, P. (2008). Ann. N. Y. Acad. Sci. 1125, 353–362. [DOI] [PubMed]
- Ezeji, T., Milne, C., Price, N. D. & Blaschek, H. P. (2010). Appl. Microbiol. Biotechnol. 85, 1697–1712. [DOI] [PubMed]
- Felnagle, E. A., Chaubey, A., Noey, E. L., Houk, K. N. & Liao, J. C. (2012). Nature Chem. Biol. 8, 518–526. [DOI] [PubMed]
- Inui, M., Suda, M., Kimura, S., Yasuda, K., Suzuki, H., Toda, H., Yamamoto, S., Okino, S., Suzuki, N. & Yukawa, H. (2008). Appl. Microbiol. Biotechnol. 77, 1305–1316. [DOI] [PubMed]
- Jones, D. T. & Woods, D. R. (1986). Microbiol. Rev. 50, 484–524. [DOI] [PMC free article] [PubMed]
- Lee, S. K., Chou, H., Ham, T. S., Lee, T. S. & Keasling, J. D. (2008). Curr. Opin. Biotechnol. 19, 556–563. [DOI] [PubMed]
- Lee, S. Y., Park, J. H., Jang, S. H., Nielsen, L. K., Kim, J. & Jung, K. S. (2008). Biotechnol. Bioeng. 101, 209–228. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mitchell, W. J. (1998). Adv. Microb. Physiol. 39, 31–130. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nicolaou, S. A., Gaida, S. M. & Papoutsakis, E. T. (2010). Metab. Eng. 12, 307–331. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Method Enzymol, 276, 307–326. [DOI] [PubMed]
- Shen, C. R. & Liao, J. C. (2008). Metab. Eng. 10, 312–320. [DOI] [PubMed]
- Steen, E. J., Chan, R., Prasad, N., Myers, S., Petzold, C. J., Redding, A., Ouellet, M. & Keasling, J. D. (2008). Microb. Cell Fact. 7, 36. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.