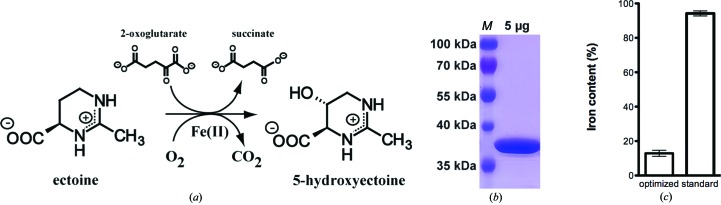
The ectoine hydroxylase from S. alaskensis was crystallized, yielding crystals diffracting to 2.1 Å resolution. The addition of iron prior to crystallization led to a different type of crystal, which may represent the iron-bound form of the enzyme.
Keywords: ectoine hydroxylase, Sphingopyxis alaskensis
Abstract
The ectoine hydroxylase (EctD) is a member of the non-haem-containing iron(II)- and 2-oxoglutarate-dependent dioxygenase superfamily. Its mononuclear iron centre is a prerequisite for the activity of this enzyme and promotes the O2-dependent oxidative decarboxylation of 2-oxoglutarate, which is coupled to a two-electron oxidation of the substrate ectoine to yield 5-hydroxyectoine. An expression and purification protocol for the EctD enzyme from Sphingopyxis alaskensis was developed and the protein was crystallized using the sitting-drop vapour-diffusion method. This resulted in two different crystal forms, representing the apo and iron-bound forms of the enzyme.
1. Introduction
Ectoine and its derivative 5-hydroxyectoine are well known members of the compatible solutes and are widely produced as protectants against osmotic stress by numerous microorganisms. Ectoine is synthesized from l-aspartate β-semialdehyde by the EctABC enzymes (Louis & Galinski, 1997 ▶; Ono et al., 1999 ▶). Ectoine is a superb stabilizer of macromolecules (Lippert & Galinski, 1992 ▶), an excellent cytoprotectant (Graf et al., 2008 ▶; Pastor et al., 2010 ▶) and is commercially used as sun protection agent in skincare products (Lentzen & Schwarz, 2006 ▶). A subgroup of ectoine producers convert ectoine to 5-hydroxyectoine, which possesses stress-protective and protein-function-preserving properties that are different from and often superior to those of ectoine (Borges et al., 2002 ▶; Bursy et al., 2008 ▶).
Hydroxylation of ectoine is catalyzed by the ectoine hydroxylase (EctD), a member of the non-haem-containing iron(II)- and 2-oxoglutarate-dependent dioxygenase superfamily (Prabhu et al., 2004 ▶; García-Estepa et al., 2006 ▶; Bursy et al., 2007 ▶). This hydroxylation reaction requires O2 and 2-oxoglutarate as co-substrates whereby CO2, succinate and 5-hydroxyectoine are formed (Bursy et al., 2007 ▶). The EctD-catalyzed reaction (Fig. 1a) is strictly dependent on a mononuclear iron centre promoting the O2-dependent oxidative decarboxylation of 2-oxoglutarate, which is coupled with a two-electron oxidation of the substrate ectoine (Widderich et al., 2014 ▶).
Here, we present the results of the purification, crystallization and preliminary X-ray crystallographic analyses from two different crystal types of the EctD protein from Sphingopyxis alaskensis, a microorganism that is well adapted to permanently cold marine environments (Ting et al., 2010 ▶). To solve its crystal structure, we plan to use the method of molecular replacement with the structure of the EctD protein from Virgibacillus salexigens (PDB entry 3emr; Reuter et al., 2010 ▶), which displays 50.8% amino-acid sequence identity to the S. alaskensis enzyme.
2. Materials and methods
2.1. Overexpression and purification
Plasmid pMP40 (ectD +) was used for the overexpression of the S. alaskensis EctD protein (SaEctD, Accession No. YP_617990). The S. alaskensis ectD gene was amplified via PCR from chromosomal DNA using custom-synthesized DNA primers (ectD_Spha_fwd, ATGGTAGGTCTCAAATGCAAGACCTCTACCCCTCGCGC; ectD_Spha_rev, ATGGTAGGTCTCAGCGCTTGCCGGCACCGTTTCGACGAG). BsaI restriction sites were synthetically added to the ends of the DNA primers, allowing cloning of the amplified full-length ectD gene into plasmid pASK-IBA3 and thereby fusing it to a C-terminal Strep-tag II affinity peptide. The resulting plasmid pMP40 carries the S. alaskensis ectD gene under the control of the TetR-responsive and anhydrotetracycline (AHT)-inducible tet promoter carried by the pASK-IBA3 plasmid backbone. As a consequence, overexpression of the cloned S. alaskensis ectD gene can be induced by adding AHT (purchased from IBA GmbH) to the growth medium. The SaEctD-Strep-tag II hybrid protein was purified by affinity chromatography on Strep-Tactin Superflow material (purchased from IBA GmbH).
To provide the SaEctD enzyme for crystallization trials, Escherichia coli BL21 (pMP40) cells were grown at 310 K in Minimal Medium A (MMA) supplemented with ampicillin (100 µg ml−1) in a 2 l Erlenmeyer flask (filled with 1 l medium) in an aerial shaker set to 180 rev min−1. At an OD578 of 0.7 of the culture, overexpression was induced by the addition of AHT (final concentration of 0.2 mg ml−1); the temperature was than dropped to 303 K, the speed of the aerial shaker was reduced to 100 rev min−1 and the cells were propagated for an additional 2 h. The cells were then harvested by centrifugation (10 min at 4800g in a Hettich Rotana speed centrifuge at 277 K). The pelleted cells were resuspended in buffer A (20 mM TES pH 8, 100 mM KCl) and they were then disrupted by passing them three times in the cold (277 K) through a French Pressure Cell Press (SLM Aminco) at 1000 psi (1 psi = 6.895 kPa). Cellular debris was removed by ultracentrifugation (60 min at 100 000g and 277 K) and the cleared supernatant was loaded onto a Strep-Tactin Superflow column that had been equilibrated with five bed volumes of buffer A. The column was then washed with ten column volumes of buffer A. The EctD-Strep-tag II protein was eluted from the affinity chromatography material with three column volumes of buffer A containing 2.5 mM desthiobiotin. The eluted EctD-Strep-tag II protein was concentrated with Vivaspin 6 columns (Sartorius Stedim Biotech GmbH, Göttingen, Germany) to a concentration of about 10 mg ml−1 before it was used for crystallization trials. 200–300 mg EctD-Strep-tag II protein per litre of cell culture were routinely obtained using this overproduction and purification scheme. The protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, Schwerte, Germany) and an extinction coefficient of 41 035 M −1 cm−1 at 280 nm and the molecular mass (35.29 kDa) of the full-length EctD including the Strep-tag II. The iron content of SaEctD was determined as described by Lovenberg et al. (1963 ▶). The purity of the SaEctD protein was assessed by SDS–PAGE (12% polyacrylamide; Fig. 1 ▶ b). The SaEctD protein was shock-frozen in liquid nitrogen and stored at 193 K until use for crystallization.
Figure 1.
(a) Enzyme reaction mediated by ectoine hydroxylase (EctD). (b) SDS–PAGE analysis of SaEctD. Lane M, molecular-mass marker. (c) The iron content of protein samples was measured for the optimized SaEctD overproduction protocol for crystallography purposes and the standard SaEctD protein production protocol used for enzyme activity assays (Widderich et al., 2014 ▶).
2.2. Crystallization and preliminary X-ray analysis of SaEctD
For all crystallization trials the full-length SaEctD protein including the Step-tag II peptide was used.
2.2.1. Apo SaEctD
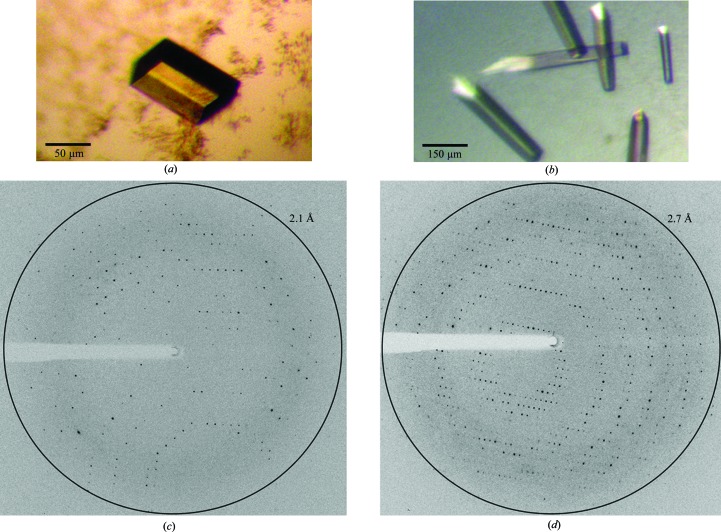
In order to find initial crystallization conditions we used standard screening kits from Qiagen, Hilden, Germany (Nextal JCSG Core Suites I–IV) and Molecular Dimensions, Suffolk, England (MemGold, MemGold 2, MIDAS) in Corning 3553 sitting-drop plates at 293 K. 0.5 µl of the homogeneous protein solution of EctD (10 mg ml−1 in 20 mM TES pH 7.5, 80 mM NaCl) was mixed with 0.5 µl reservoir solution. From the roughly 700 conditions tested, 12 conditions resulted in initial crystal hits. By optical inspection we chose four of them for optimization experiments in which we varied two parameters against each other (e.g. pH against PEG concentration) in sitting-drop trials at 293 K with drops composed of 1.5 µl protein solution and 1.5 µl reservoir solution. The largest and best diffracting crystals resulted from solutions consisting of 100 mM MES pH 6.0, 200 mM calcium acetate, 30%(w/v) PEG 400. To improve diffraction properties we tested this condition in combination with the additive and detergent screens from Hampton Research, Aliso Viejo, USA as described with 1/10 of the drop volume of the corresponding additive or detergent. The detergent n-dodecyl-N′,N-dimethylglycine yielded the crystals that diffracted the best (Fig. 2 ▶ a). Optimized crystallization trials were then performed using the sitting-drop vapour-diffusion method at 293 K. 1.5 µl of the homogeneous protein solution of EctD (10 mg ml−1 in 20 mM TES pH 7.5, 80 mM NaCl) was mixed with 1.5 µl reservoir solution consisting of 100 mM MES pH 6.0, 200 mM Ca acetate, 30%(w/v) PEG 400 and 1.5 mM n-dodecyl-N′,N-dimethylglycine and equilibrated over 300 µl reservoir solution. Crystals grew within 6–12 d to their final dimensions of around 30 × 30 × 50 µm. Crystals were cryoprotected by slowly and cautiously adding 1 µl 100% glycerol to the crystallization drop by stepwise pipetting before cooling the crystals in liquid nitrogen.
Figure 2.
Crystals of SaEctD without additional Fe (a) and (b) supplemented with FeIICl2 prior to crystallization. Diffraction images of SaEctD without (c) and with (d) the addition of Fe ions prior to crystallization [oscillation width 0.4° (c) and 0.2° (d)].
2.2.2. Fe-SaEctD
Starting from the optimized condition of the apo SaEctD crystals, we performed crystallization trials for Fe-SaEctD. The conditions were as described for the apo SaEctD protein, but the protein solution was premixed with 100 mM FeCl2 to a final concentration of 4 mM and incubated on ice for 10–15 min. EctD crystals were grown under the above-mentioned conditions but with 3.5 mM n-dodecyl-N′,N-dimethylglycine. They grew within 6–12 d at 293 K to final dimensions of around 40 × 40 × 180 µm (Fig. 2 ▶ b). The crystals were cryoprotected as described above.
Data sets were collected from a single crystal of either apo SaEctD or Fe-SaEctD on beamline ID23eh2 at the ERSF, Grenoble, France at 100 K. These data sets were processed using the XDS package (Kabsch, 2010a ▶) and scaled with XSCALE (Kabsch, 2010b ▶).
3. Results
The ectoine hydroxylase from S. alaskensis (SaEctD) was overexpressed in E. coli BL21 carrying plasmid pMP40 as a recombinant EctD-Strep-tag II protein and purified by affinity chromatography using a Strep-Tactin Superflow column. By slight variations of the initial expression protocol (Bursy et al., 2007 ▶; Reuter et al., 2010 ▶; Widderich et al., 2014 ▶) (see §2), the amount of purified SaEctD protein was increased tenfold to 200–300 mg l−1. Protein purity was validated by SDS–PAGE (Fig. 1 ▶ b). The purity of SaEctD was assumed to be at least 95% by optical inspection. The homogeneity of SaEctD was analytically checked by size-exclusion chromatography.
Since the presence of a correctly complexed iron ligand is critical for EctD-mediated enzyme catalysis (Widderich et al., 2014 ▶), we determined the iron content of the purified protein and found between 0.12 and 0.14 mol of iron per mole of EctD (Fig. 1 ▶ c). This low iron content is likely to be due to the newly developed strong expression protocol, since the initial protocol (Widderich et al., 2014 ▶) yielded 0.9 mol iron per mol of EctD protein (Fig. 1 ▶ c).
Recently, it has been shown that the iron ligand can be added to the purified enzyme, leading to a restored enzyme activity of SaEctD (Widderich et al., 2014 ▶). For crystallization experiments we tested whether there is an effect on the crystallization behaviour and/or crystal quality when the SaEctD protein was supplemented with additional Fe ions prior to crystallization.
Diffracting crystals of apo SaEctD (i.e. no additional iron added) and Fe-SaEctD (supplemented with FeCl2) were obtained with 100 mM MES pH 6.0, 200 mM calcium acetate, 30%(w/v) PEG 400 and different concentrations of n-dodecyl-N′,N-dimethylglycine (1.5 and 3.5 mM) using the sitting-drop vapour-diffusion method (Figs. 2 ▶ a and 2 ▶ b). After adding glycerol as a cryoprotectant crystals were shock-frozen in liquid nitrogen. Native data sets for both protein crystal species were collected at 100 K. Crystals of apo SaEctD diffracted to a maximum resolution of 2.1 Å and those of Fe-SaEctD diffracted to a maximum resolution of 2.7 Å (Figs. 2 ▶ c and 2 ▶ d).
Preliminary data processing using the XDS package resulted in different unit-cell parameters and, more interestingly, the crystals displayed different space groups. Whereas apo SaEctD crystallized in space group C2221, Fe-SaEctD displayed a P212121 symmetry (see Table 1 ▶). The apo form contains one monomer per asymmetric unit whereas the Fe-supplemented form contains a dimer. V M values were calculated to be 2.4 Å3 Da−1 for the apo and 2.3 Å3 Da−1 for the Fe-SaEctD crystal with a solvent content of 50% and 47%, respectively (Matthews, 1968 ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Apo SaEctD | Fe-SaEctD | |
|---|---|---|
| Beamline | ID23eh2, ESRF | ID23eh2, ESRF |
| Detector | MAR225 | MAR225 |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 0.87260 | 0.87260 |
| Crystal-to-detector distance (mm) | 245 | 291 |
| Rotation range per image (°) | 0.4 | 0.2 |
| Total rotation range (°) | 100 | 120 |
| Exposure time per image (s) | 2.1 | 0.4 |
| Space group | C2221 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 83.48 | 78.16 |
| b (Å) | 86.51 | 87.52 |
| c (Å) | 95.34 | 96.05 |
| α = β = γ (°) | 90 | 90 |
| Resolution (Å) | 30–2.1 (2.2–2.1) | 30–2.7 (2.8–2.7) |
| No. of observed reflections | 85055 | 91395 |
| No. of unique reflections | 20251 | 18652 |
| Mean redundancy | 4.2 (4.1) | 4.9 (5.0) |
| Completeness (%) | 99.7 (99.8) | 99.6 (99.9) |
| 〈I/σ(I)〉 | 15.1 (2.8) | 19.8 (2.9) |
| Mosaicity (°) | 0.09 | 0.19 |
| R meas † | 6.2 (49.9) | 5.8 (58.9) |
| R pim ‡ | 3.6 (42.0) | 2.9 (47.3) |
| Overall B factor from Wilson plot (Å2) | 43.1 | 63.8 |
| Matthews coefficient V M (Å3 Da−1) | 2.41 | |
| Monomer | 2.44 | 4.65 |
| Dimer | 2.33 | |
| Solvent content (%) | 49.6 | 47.2 |
R
meas = 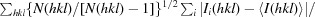
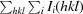 .
.
R
p.i.m. = 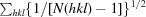
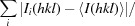
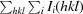 .
.
When one compares the data statistics of apo SaEctD with those of Fe-treated SaEctD, the slight differences in the unit-cell parameters in combination with the different space groups and the asymmetric unit content may be a hint that the apo crystals lack the iron catalyst whereas in the Fe-supplemented crystals the iron might be present. The structure determination of the SaEctD protein using both crystal forms via molecular replacement using the VsEctD structure (PDB entry 3emr; Reuter et al., 2010 ▶) as a template model is currently in progress.
Acknowledgments
We thank the staff of the P14 beamline at the EMBL, Hamburg, Germany, for kind support during crystal screening. We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities, especially Sean McSweeny from ID23eh2. We also gratefully acknowledge the ‘Fit For Excellence’ Fund of the Heinrich Heine University, the SFB 987, the IMPRS Marburg and the Emeritus group of R. K. Thauer for financial support. We thank Marco Pittelkow for the construction of the expression plasmid.
References
- Borges, N., Ramos, A., Raven, N. D., Sharp, R. J. & Santos, H. (2002). Extremophiles, 6, 209–216. [DOI] [PubMed]
- Bursy, J., Kuhlmann, A. U., Pittelkow, M., Hartmann, H., Jebbar, M., Pierik, A. J. & Bremer, E. (2008). Appl. Environ. Microbiol. 74, 7286–7296. [DOI] [PMC free article] [PubMed]
- Bursy, J., Pierik, A. J., Pica, N. & Bremer, E. (2007). J. Biol. Chem. 282, 31147–31155. [DOI] [PubMed]
- García-Estepa, R., Argandoña, M., Reina-Bueno, M., Capote, N., Iglesias-Guerra, F., Nieto, J. J. & Vargas, C. (2006). J. Bacteriol. 188, 3774–3784. [DOI] [PMC free article] [PubMed]
- Graf, R., Anzali, S., Buenger, J., Pfluecker, F. & Driller, H. (2008). Clin. Dermatol. 26, 326–333. [DOI] [PubMed]
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Lentzen, G. & Schwarz, T. (2006). Appl. Microbiol. Biotechnol. 72, 623–634. [DOI] [PubMed]
- Lippert, K. & Galinski, E. A. (1992). Appl. Microbiol. Biotechnol. 37, 61–65.
- Louis, P. & Galinski, E. A. (1997). Microbiology, 143, 1141–1149. [DOI] [PubMed]
- Lovenberg, W., Buchanan, B. B. & Rabinowitz, J. C. (1963). J. Biol. Chem. 238, 3899–3913. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Ono, H., Sawada, K., Khunajakr, N., Tao, T., Yamamoto, M., Hiramoto, M., Shinmyo, A., Takano, M. & Murooka, Y. (1999). J. Bacteriol. 181, 91–99. [DOI] [PMC free article] [PubMed]
- Pastor, J. M., Salvador, M., Argandoña, M., Bernal, V., Reina-Bueno, M., Csonka, L. N., Iborra, J. L., Vargas, C., Nieto, J. J. & Cánovas, M. (2010). Biotechnol. Adv. 28, 782–801. [DOI] [PubMed]
- Prabhu, J., Schauwecker, F., Grammel, N., Keller, U. & Bernhard, M. (2004). Appl. Environ. Microbiol. 70, 3130–3132. [DOI] [PMC free article] [PubMed]
- Reuter, K., Pittelkow, M., Bursy, J., Heine, A., Craan, T. & Bremer, E. (2010). PLoS One, 5, e10647. [DOI] [PMC free article] [PubMed]
- Ting, L., Williams, T. J., Cowley, M. J., Lauro, F. M., Guilhaus, M., Raftery, M. J. & Cavicchioli, R. (2010). Environ. Microbiol. 12, 2658–2676. [DOI] [PubMed]
- Widderich, N., Pittelkow, M., Höppner, A., Mulnaes, D., Buckel, W., Gohlke, H., Smits, S. H. J. & Bremer, E. (2014). J. Mol. Biol. 426, 586–600. [DOI] [PubMed]