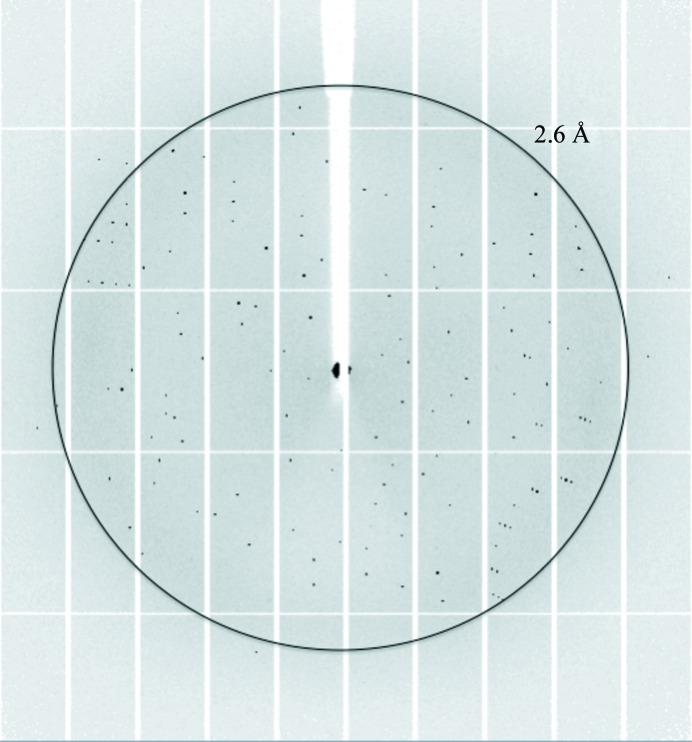
Manganese lipoxygenase of Geaumannomyces graminis was crystallized and X-ray diffraction data were collected to a resolution of 2.6 Å.
Keywords: manganese lipoxygenase, Gaeumannomyces graminis
Abstract
Lipoxygenases constitute a family of nonhaem metal enzymes with catalytic iron or, occasionally, catalytic manganese. Lipoxygenases oxidize polyunsaturated fatty acids with position specificity and stereospecificity to hydroperoxides, which contribute to inflammation and the development of cancer. Little is known about the structural differences between lipoxygenases with Fe or Mn and the metal-selection mechanism. A Pichia pastoris expression system was used for the production of the manganese lipoxygenase of the take-all fungus of wheat, Gaeumannomyces graminis. The active enzyme was treated with α-mannosidase, purified to apparent homogeneity and subjected to crystal screening and X-ray diffraction. The crystals diffracted to 2.6 Å resolution and belonged to space group C2, with unit-cell parameters a = 226.6, b = 50.6, c = 177.92 Å, β = 91.70°.
1. Introduction
Lipoxygenases are iron- or manganese-containing fatty-acid dioxygenases (Brash, 1999 ▶; Kuhn et al., 2005 ▶; Andreou & Feussner, 2009 ▶). Iron lipoxygenases (FeLOX) occur in plants, animals and a few fungi, whereas two manganese lipoxygenases (MnLOX), linoleate 13R-MnLOX and 9S-MnLOX, occur in wheat and rice pathogenic fungi of the Magnaporthaceae family, respectively (Su & Oliw, 1998 ▶; Wennman & Oliw, 2013 ▶). All lipoxygenases belong to the same gene family, which is characterized by sequence homology and by apparently conserved metal ligands (Brash, 1999 ▶; Cristea et al., 2005 ▶; Kuhn et al., 2005 ▶; Liavonchanka & Feussner, 2006 ▶). Lipoxygenases have important biological functions. They oxidize polyunsaturated fatty acids with 1Z,4Z-pentadiene units to hydroperoxides. This occurs by hydrogen abstraction at the bis-allylic carbon of the 1Z,4Z-pentadiene followed by oxygen insertion, which usually leads to the formation of cis–trans-conjugated hydroperoxy fatty acids (Brash, 1999 ▶). These hydroperoxides are precursors of signal molecules in animals, plants and fungi, and may take part in inflammation, cancer development and the chemical warfare between plants, fungi and other microorganisms (Brash, 1999 ▶; Oliw, 2002 ▶; Kuhn et al., 2005 ▶; Liavonchanka & Feussner, 2006 ▶; Andreou & Feussner, 2009 ▶).
Three-dimensional structures are available of at least nine lipoxygenases from plants and animals, but three-dimensional structures of fungal lipoxygenases have not yet been reported (Boyington et al., 1993 ▶; Minor et al., 1996 ▶; Gillmor et al., 1997 ▶; Skrzypczak-Jankun et al., 1997 ▶; Oldham et al., 2005 ▶; Youn et al., 2006 ▶; Neau et al., 2009 ▶; Gilbert et al., 2011 ▶; Eek et al., 2012 ▶; Xu et al., 2012 ▶). Lipoxygenases consist of two domains: a small C2 PLAT domain with a β-barrel and a catalytic domain with α-helices and a mononuclear metal centre with Fe or Mn (Neau et al., 2009 ▶). The iron is bound to three His residues, the carboxyl O atom of the C-terminal amino acid, and usually to a distant Asn residue. Depending on the oxidative state of the lipoxygenase, a water molecule or OH− provides the third oxygen ligand to the metal.
The primary protein sequences of MnLOX and FeLOX can be aligned with regional similarities, and site-directed mutagenesis suggests that the primary iron ligands are conserved as metal ligands in 13R-MnLOX (Cristea et al., 2005 ▶). The second coordinating sphere may differ and contribute to the metal-selection process. The active sites of many FeLOX and MnLOX enzymes may bind their substrates in similar orientation. Alterations in the Sloane determinant of 9S-MnLOX and 13R-MnLOX changed the position specificity essentially as described for arachidonate 15-LOX (Wennman & Oliw, 2013 ▶). Replacement of a single amino acid (Phe337Ile) of 13R-MnLOX retained abstraction of the pro-S H atom at C-11 of linoleic acid but changed the oxygen insertion from suprafacial to mainly antarafacial (Wennman et al., 2012 ▶).
The redox potential of unbound Mn2+/Mn3+ is twice as large as for unbound Fe2+/Fe3+, and the difference is 0.7 V. The metal ligands of 13R-MnLOX may reduce the redox potential of the enzyme to close to 0.6 V, which is the redox potential of soybean LOX-1 (Nelson, 1988 ▶), to allow efficient oxidation of protein-bound Mn2+OH2 to Mn3+OH−. Superoxide dismutases with catalytic Mn or Fe bound to identical metal ligands also adjust the redox potentials of protein-bound metals to enable catalysis (Miller, 2012 ▶).
The crystal structure of 13R-MnLOX might provide detailed information on metal ligands, solvent molecules and the hydrogen-bond network of importance for the catalytic properties. We have therefore crystallized 13R-MnLOX and we report a preliminary crystallographic analysis.
2. Materials and methods
2.1. Expression of 13R-MnLOX
The 13R-MnLOX precursor consists of 618 amino acids (UniProt accession No. Q8X151), including a secretion signal of 16 amino acids which is cleaved off to generate the mature enzyme. 13R-MnLOX (602 amino acids) was secreted by Pichia pastoris using pPICZAα with the yeast α-secretion signal (pPICZAα_MnLOX_602; Cristea et al., 2005 ▶). P. pastoris was transformed as described in Cristea et al. (2005 ▶). Transformants were selected on yeast peptone dextrose agar plates with phleomycin (Zeocin; 100 µg ml−1) at 301 K (Cristea et al., 2005 ▶) and stored as glycerol stocks at 193 K.
Transformed P. pastoris was first grown to generate biomass in buffered minimal glycerol complex medium in baffled flasks (301 K, 200 rev min−1, 48 h). Fermentation was carried out in a 10 l bioreactor (301 K) with 6 l buffered minimal methanol medium inoculated with washed cells to obtain an OD600 of 1. The conditions of the bioreactor were: aeration rate 6 l min−1; pressure 0.2 bar (1 bar = 100 kPa); dissolved oxygen 30% saturation; pH 5.0. The Ziegler–Nichols method was used to find the optimal proportional integral derivative feedback control system; the settings were adjusted if necessary during the fermentation. Methanol supplemented with 12 ml trace additives (6 g l−1 CuSO4.5H2O, 0.08 g l−1 KI, 3 g l−1 MnSO4.H2O, 0.2 g l−1 Na2MoO4, 0.02 g l−1 H3BO3, 0.5 g l−1 CoCl2, 20 g l−1 ZnCl2, 65 g l−1 FeSO4.7H2O, 0.2 g l−1 biotin and 5 ml H2SO4) per litre was added with an external pump. Polypropylene glycol was added to reduce foaming. The culture was harvested after 4–5 d when the OD600 reached at least 170. Yeast cells were precipitated by centrifugation at 5000g for 10 min. Ammonium sulfate (1 mol per litre of supernatant) was gradually added, the pH was adjusted to 7.0 with 10 M potassium hydroxide and the enzyme solution was stored at 193 K.
2.2. Purification of 13R-MnLOX
The protein solution was thawed and then centrifuged at 18 000g, filtered and loaded onto a column for hydrophobic interaction chromatography (70 ml Phenyl Sepharose CL-4B), which was eluted with an ÄKTA FPLC (Pharmacia) or a peristaltic pump (P-3; Pharmacia) at 294 K. The column was equilibrated with 25 mM potassium hydrogen phosphate pH 7.0, 1 M ammonium sulfate. Protein bound to the column was eluted in one step with 25 mM potassium hydrogen phosphate pH 7.0 and the fractions were assayed for 13R-MnLOX activity by UV spectroscopy (Wennman & Oliw, 2013 ▶).
The fractions with 13R-MnLOX activity were pooled. The volume was reduced and the buffer was changed to 0.1 M sodium acetate pH 5.0, 20 mM ZnCl2 by diafiltration (Ultracel 10K, Millipore). The protein concentration was estimated by UV analysis (NanoDrop). α-Mannosidase, which requires Zn2+ as a cofactor, was added at a protein ratio of 1:100 and the mixture was incubated at 294 K overnight. The protein solution was loaded onto a gel-filtration column (Superdex 200 HiLoad 16/60) with 25 mM Tris–HCl pH 7.5, 150 mM NaCl as the running buffer and eluted at 0.5 ml min−1 (ÄKTA FPLC) at room temperature. Fractions corresponding to 13R-MnLOX were pooled and analyzed by SDS–PAGE. The protein was washed with 25 mM Tris–HCl pH 7.5 and concentrated to ∼20 mg ml−1 by diafiltration. To assess the homogeneity and oligomerization state, we performed dynamic light-scattering analysis (Avid Nano W130i dynamic light-scattering system) at a concentration of 1 mg ml−1 in 25 mM Tris–HCl pH 7.5.
2.3. Crystallization
Crystals of 13R-MnLOX were grown using the hanging-drop vapour-diffusion method at 293 K. Initial crystallization trials were carried out using a Mosquito crystallization robot (TTP Labtech, Cambridge, England). The following commercially available crystallization screens were utilized to identify the best crystallization condition: PEG/Ion, Crystal Screen, Crystal Screen 2 (Hampton Research, USA), JCSG+ (Qiagen, Germany) and Morpheus protein crystallization screen (Molecular Dimensions). Using a 96-well plate, drops (0.3 µl) were prepared by mixing protein solution (∼20 mg ml−1 protein) with an equal amount of well solution. Crystals of 13R-MnLOX grew in many different conditions, but the best hits that gave crystals of approximately 20 × 30 ×50 µm in size were obtained from the Morpheus protein crystallization screen [e.g. solution A3, consisting of 0.1 M MES–imidazole pH 6.5, 20%(v/v) glycerol, 10%(w/v) PEG 4000, 0.03 M magnesium chloride, 0.03 M calcium chloride; Gorrec, 2009 ▶]. Crystals were taken directly from the 96-well plate and cooled in liquid N2.
2.4. X-ray diffraction data collection and preliminary analysis
Data were collected with a Pilatus detector at a wavelength of 0.976 Å at 100 K on beamline ID29 at the European Synchrotron Radiation Facility, Grenoble, France. The data were processed using XDS (Kabsch, 2010 ▶). The integrated data were then scaled using the scaling program SCALA (Winn et al., 2011 ▶). A set of 5% of the reflections was set aside and used to calculate the quality factor R free (Brünger, 1992 ▶).
Details of data collection and processing are presented in Table 1 ▶. The crystal of MnLOX protein belonged to space group C2, with unit-cell parameters a = 226.6, b = 50.6, c = 177.92 Å, β = 91.70°.
Table 1. Data-collection and processing statistics.
| Beamline | ID29, ESRF |
| Detector | Pilatus 6M |
| Wavelength (Å) | 0.976 |
| Oscillation range (°) | 0.15 |
| No. of images | 1127 |
| Space group | C121 |
| Unit-cell parameters (Å, °) | a = 226.6, b = 50.6, c = 177.92, β = 91.7 |
| Resolution range (Å) | 49.4–2.6 |
| No. of observed reflections | 160569 |
| No. of unique reflections | 55058 |
| Average multiplicity | 2.9 |
| Completeness (%) | 97.8 (98.7) |
| R merge † | 0.095 (0.367) |
| R meas ‡ | 0.134 (0.517) |
| 〈I/σ(I)〉 | 7.6 (2.4) |
R
merge = 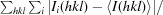
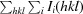 .
.
R
meas = R
r.i.m. = 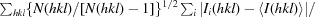
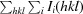 .
.
The native Patterson and self-rotation function were calculated using the programs MOLREP and FFT (Winn et al., 2011 ▶). For the self-rotation function an integration radius of 25 Å was used for the data between 20 and 3 Å resolution.
3. Results and discussion
13R-MnLOX was expressed in P. pastoris and secreted into the growth medium. The yield of secreted enzyme varied with the cell density, but the bioreactor culture yielded at least ten times more enzyme than in a bench-top shaker and at least 6 mg enzyme per litre as judged from the oxidation of linoleic acid 18:2n -6 by small aliquots of the crude growth medium and UV analysis.
13R-MnLOX was purified as described by Su & Oliw (1998 ▶) and the homogeneity of purified recombinant 13R-MnLOX protein was confirmed by SDS–PAGE and size-exclusion chromatography. Dynamic light-scattering experiments were carried out for both nondeglycosylated and deglycosylated (α-mannosidase-treated) 13R-MnLOX. The molecular weight and polydispersity were estimated to be 110 kDa and 0.08% for the glycosylated and 74 kDa and 0.12% for the nondeglycosylated protein, respectively. The homogeneity was estimated to be approximately 100% in both cases.
Many different crystallization screens were used to identify the best crystallization condition for 13R-MnLOX. Crystals with variations in morphology appeared initially in many conditions of the Morpheus protein crystallization screen (Fig. 1 ▶). In some conditions these crystals grew larger with regular faces and sharper edges within 3–7 d. In an attempt to improve the size of the crystals, a grid screen was performed around some of the successful conditions obtained from the Morpheus crystallization screen. Unfortunately, the crystals obtained from the grid screens did not grow any larger than the original crystals obtained from the screen. 13R-MnLOX is a highly stable protein (Su & Oliw, 1998 ▶; Cristea et al., 2005 ▶) and we confirmed that crystals contained active enzyme.
Figure 1.
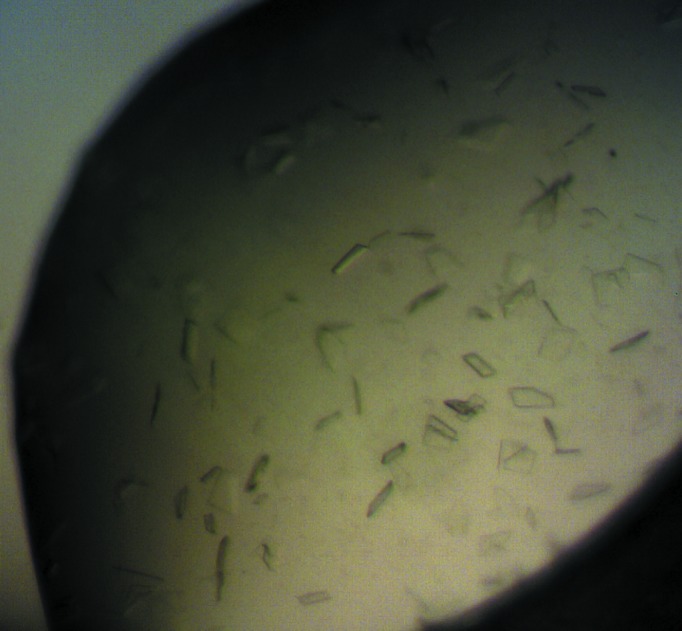
Image of crystals of 13R-MnLOX grown with the Morpheus protein crystallization screen.
One of the advantages of the Morpheus screen is that the buffer system and the precipitant mixes also act as cryoprotectants, which allowed us to flash-cool the crystals directly after removing them from the drop. Elimination of one extra step of moving the crystal into a cryosolution before cooling was likely to reduce crystal stress, an important factor for the diffraction power of these crystals since most of the crystals decayed or lost their diffraction power before a complete data set could be collected. A complete data set with more than 1100 consecutive images with an oscillation range of 0.15° and exposure time of 0.043 s was collected to 2.6 Å resolution from a single crystal (Fig. 2 ▶).
Figure 2.
X-ray diffraction pattern from 13R-MnLOX. The outer resolution ring indicates 2.6 Å resolution.
The data were processed with XDS and iMosflm (Battye et al., 2011 ▶) and both programs unambiguously identified the space group as C2, with approximate unit-cell parameters a = 226.6, b = 50.6, c = 177.92 Å, β = 91.7°. Preliminary analysis of the data using the Matthews probability calculator (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) suggested two or three molecules in the asymmetric unit, with a Matthews coefficient of 3.8 Å3 Da−1 and a solvent content of 68% for two molecules and a Matthews coefficient 2.6 Å3 Da−1 and a solvent content of 52% for three molecules in the asymmetric unit.
The self-rotation function confirmed the presence of a crystallographic twofold axis, but there were no peaks present corresponding to a noncrystallographic threefold axis. However, the self-rotation function shows the presence of two peaks, which suggests there may be a noncrystallographic twofold axis lying in a plane perpendicular to the crystallographic twofold axis. Analysis of the native Patterson showed no significant translation peaks. The largest non-origin peak had a height of only 14% of the Patterson origin peak, which indicates that there is no translation between the molecules in the asymmetric unit.
Several three-dimensional structures of lipoxygenases are available, but the sequence identity between 13R-MnLOX and known structures is less than 27%. Three-dimensional structures of fungal lipoxygenases have not yet been reported. Attempts to solve the structure by molecular replacement using known structures as search models, e.g. coral arachidonate 11R-LOX and 8R-LOX, animal arachidonate 15-LOX and 5-LOX, and plant sLOX-1, proved to be unsuccessful. Fortunately, 13R-MnLOX contains two cysteine and nine methionine residues in addition to the catalytic metal, which makes it a good candidate for future attempts to solve the structure by multiple or single anomalous dispersion using the Mn or S atoms as the source of anomalous scattering.
Acknowledgments
This work was supported by VR Medicine and by Uppsala University.
References
- Andreou, A. & Feussner, I. (2009). Phytochemistry, 70, 1504–1510. [DOI] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Boyington, J. C., Gaffney, B. J. & Amzel, L. M. (1993). Science, 260, 1482–1486. [DOI] [PubMed]
- Brash, A. R. (1999). J. Biol. Chem. 274, 23679–23682. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Cristea, M., Engström, K., Su, C., Hörnsten, L. & Oliw, E. H. (2005). Arch. Biochem. Biophys. 434, 201–211. [DOI] [PubMed]
- Eek, P., Järving, R., Järving, I., Gilbert, N. C., Newcomer, M. E. & Samel, N. (2012). J. Biol. Chem. 287, 22377–22386. [DOI] [PMC free article] [PubMed]
- Gilbert, N. C., Bartlett, S. G., Waight, M. T., Neau, D. B., Boeglin, W. E., Brash, A. R. & Newcomer, M. E. (2011). Science, 331, 217–219. [DOI] [PMC free article] [PubMed]
- Gillmor, S. A., Villaseñor, A., Fletterick, R., Sigal, E. & Browner, M. F. (1997). Nature Struct. Biol. 4, 1003–1009. [DOI] [PubMed]
- Gorrec, F. (2009). J. Appl. Cryst. 42, 1035–1042. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kuhn, H., Saam, J., Eibach, S., Holzhütter, H. G., Ivanov, I. & Walther, M. (2005). Biochem. Biophys. Res. Commun. 338, 93–101. [DOI] [PubMed]
- Liavonchanka, A. & Feussner, I. (2006). J. Plant Physiol. 163, 348–357. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Miller, A. F. (2012). FEBS Lett. 586, 585–595. [DOI] [PMC free article] [PubMed]
- Minor, W., Steczko, J., Stec, B., Otwinowski, Z., Bolin, J. T., Walter, R. & Axelrod, B. (1996). Biochemistry, 35, 10687–10701. [DOI] [PubMed]
- Neau, D. B., Gilbert, N. C., Bartlett, S. G., Boeglin, W., Brash, A. R. & Newcomer, M. E. (2009). Biochemistry, 48, 7906–7915. [DOI] [PMC free article] [PubMed]
- Nelson, M. J. (1988). Biochemistry, 27, 4273–4278.
- Oldham, M. L., Brash, A. R. & Newcomer, M. E. (2005). J. Biol. Chem. 280, 39545–39552. [DOI] [PubMed]
- Oliw, E. H. (2002). Prostaglandins Other Lipid Mediat. 68–69, 313–323. [DOI] [PubMed]
- Skrzypczak-Jankun, E., Amzel, L. M., Kroa, B. A. & Funk, M. O. (1997). Proteins, 29, 15–31. [PubMed]
- Su, C. & Oliw, E. H. (1998). J. Biol. Chem. 273, 13072–13079. [DOI] [PubMed]
- Wennman, A., Jernerén, F., Hamberg, M. & Oliw, E. H. (2012). J. Biol. Chem. 287, 31757–31765. [DOI] [PMC free article] [PubMed]
- Wennman, A. & Oliw, E. H. (2013). J. Lipid Res. 54, 762–775. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xu, S., Mueser, T. C., Marnett, L. J. & Funk, M. O. (2012). Structure, 20, 1490–1497. [DOI] [PMC free article] [PubMed]
- Youn, B., Sellhorn, G. E., Mirchel, R. J., Gaffney, B. J., Grimes, H. D. & Kang, C. (2006). Proteins, 65, 1008–1020. [DOI] [PMC free article] [PubMed]