Abstract
Specification of endoderm is the prerequisite for gut formation in the embryogenesis of bilaterian organisms. Modern lineage labelling studies 1–3 have shown that in the sea urchin embryo model system, descendants of the veg1 and veg2 cell lineages produce the endoderm, and that the veg2 lineage also gives rise to mesodermal cell types. It is known that Wnt/β-catenin signalling is required for endoderm specification4–6 and Delta/Notch signalling is required for mesoderm specification7–9. Some direct cis-regulatory targets of these signals have been found10,11 and various phenomenological patterns of gene expression have been observed in the pre-gastrular endomesoderm. However, no comprehensive, causal explanation of endoderm specification has been conceived for sea urchins, nor for any other deuterostome. Here we propose a model, on the basis of the underlying genomic control system, that provides such an explanation, built at several levels of biological organization. The hardwired core of the control system consists of the cis-regulatory apparatus of endodermal regulatory genes, which determine the relationship between the inputs to which these genes are exposed and their outputs. The architecture of the network circuitry controlling the dynamic process of endoderm specification then explains, at the system level, a sequence of developmental logic operations, which generate the biological process. The control system initiates noninteracting endodermal and mesodermal gene regulatory networks in veg2-derived cells and extinguishes the endodermal gene regulatory network in mesodermal precursors. It also generates a cross-regulatory network that specifies future anterior endoderm in veg2 descendants and institutes a distinct network specifying posterior endoderm in veg1-derived cells. The network model provides an explanatory framework that relates endoderm specification to the genomic regulatory code.
Transcription factors, which are the products of regulatory genes, implement the genomic code for development by determining the set of expressed genes, and thus biological function. The spatially restricted expression of regulatory genes produces specific combinations of transcription factors, or regulatory states, in distinct spatial domains of the embryo. The complete set of regulatory interactions required for the formation and propagation of regulatory states explains the process of developmental specification, and this explanation is the ultimate goal of gene regulatory network (GRN) analysis.
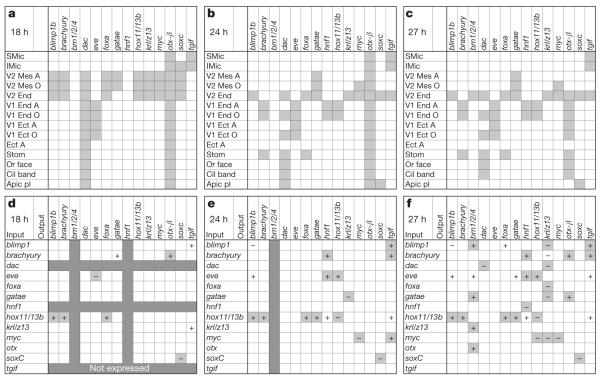
We have systematically analysed the GRN that determines the specification of the future endoderm in the embryo of the sea urchin Strongylocentrotus purpuratus, up to gastrulation. In this embryo, endoderm is derived from two cell lineages, which arise by a canonical and invariant cleavage process (Supplementary Fig. 1). The anterior compartments of the gut are formed by cells of the veg2 lineage, which is also the progenitor lineage of most mesodermal cell types. The posterior endoderm is formed by derivatives of the veg1 lineage. Comprehensive surveys12–16 of all predicted transcription factors in this genome showed that 14 regulatory genes are expressed specifically in endoderm-precursor cells before the beginning of gastrulation (30 h post-fertilization). Spatial expression patterns for these genes are summarized in Fig. 1a–c on the basis of evidence from double-fluorescent in situ hybridization (DFISH; Supplementary Figs 2 and 3) and earlier reports. We also provide a comprehensive digital summary of expression patterns at 3-h intervals for these and many additional genes in Supplementary Fig. 4.
Figure 1. Endodermal gene expression and perturbation matrix.
a–c, Spatial expression (grey cells) of 14 endodermal regulatory genes at three time points post-fertilization. A, aboral; Apic pl, apical plate; Cil band, ciliated band; Ect, ectoderm; End, endoderm; LMic, large micromeres; Mes, mesoderm; O, oral; Or face, oral face; SMic, small micromeres; Stom, stomodeum; V1, veg1; V2, veg2. d–f Interactions among regulatory genes at three time points post-fertilization (data from ref. 17 and Supplementary Fig. 5). The change in output-gene expression after injection of a morpholino oligonucleotide targeting the input gene is denoted ‘−’ if expression is significantly increased (that is, the input gene represses the output gene) and ‘+’ if expression is significantly decreased (the input gene activates the output gene). Changes incorporated as regulatory linkages in the endoderm GRN are indicated by light grey cells (Supplementary Table 1); white cells with ‘+’ or ‘−’ denote significant effects that are not considered to be direct (Supplementary Tables 2 and 3); dark grey cells denote genes that are not expressed at the time point shown.
To establish a causal explanation for the dynamic process of regulatory-state separation in the respective spatial fate domains of the veg2 and veg1 lineages, we carried out a system-wide perturbation analysis (more than 6,500 data points; Supplementary Fig. 5). The expression of each transcription factor was downregulated by treating embryos with specific morpholino antisense oligonucleotides (MASO), and the effects on all other regulatory genes, as well as on many representative genes expressed in non-endodermal domains, were measured quantitatively and often assessed spatially as well. These perturbation results were interpreted using the logic and evidence detailed in Supplementary Tables 1–3. The probable direct interactions, some of which have already been confirmed by cis-regulatory analysis, are represented in Fig. 1d–f. The spatial regulatory-state matrix in Fig. 1a–c can be considered as the output of the gene interaction matrix in Fig. 1d–f. Perturbation results were combined with previous cis-regulatory evidence to formulate the GRN model.
By 15 h post-fertilization, endoderm progenitor cells constitute two distinct, concentrically arranged regulatory states (Supplementary Fig. 1)17. In the more vegetal tiers of cells, encompassing the veg2 endoderm precursor cells, eight regulatory genes are rapidly turned on. Most of these are under the spatial control of cis-regulatory Tcf sites11,17–19 , which bind the factor mediating Wnt signal transduction, and there are a few additional regulatory interactions among them17. By contrast, only one regulatory gene, even skipped (eve), is expressed in the peripheral veg1 endoderm precursors, with no detectable impact on any other regulatory gene at this stage17. Before 18 h after fertilization, the two endoderm regulatory states are expressed in most or all of the cells in the veg1 and veg2 lineages.
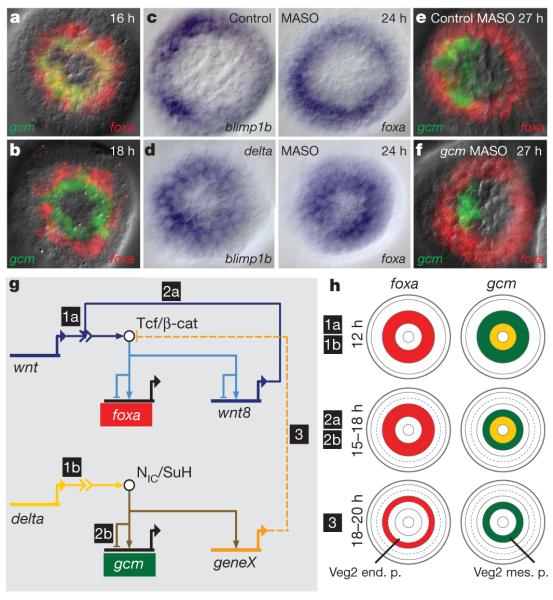
At this stage, the veg2 lineage consists of two concentric rings of cells, the inner ring destined to become mesoderm and the outer ring, anterior endoderm (Supplementary Fig. 1). The future mesoderm expresses both endoderm and mesoderm GRNs, whereas the future endoderm expresses only an endoderm regulatory state17. Thus, DFISH using probes that detect the endoderm regulatory gene forkhead box A (foxa) and the mesoderm regulatory gene glial cells missing (gcm) shows that there is overlapping expression of both genes in an inner ring of veg2-derived cells, whereas the peripheral cells of the veg2 lineage (the presumptive endoderm) express foxa alone (Fig. 2a). The gcm gene is at the top of the early mesoderm GRN hierarchy and is activated by signalling through the Delta/Notch pathway, via its cis-regulatory Suppressor of Hairless (Su(H)) target sites10. The expression of gcm is therefore restricted to the inner ring of veg2-derived cells, which are exposed to the Delta-presenting skeletogenic cells at the vegetal pole (Supplementary Fig. 1). Cis-regulatory modules that respond to Tcf or Su(H) act as ‘X,1–X’ spatial information processors20 in that they stimulate or permit gene expression in cells (‘X’) with an activated signal transduction pathway, but repress the same target genes in all other cells (‘1–X’). Thus, Su(H) and Tcf cis-regulatory interactions account for the spatial specificity of the initial, co-existing GRNs in the veg2 lineage.
Figure 2. Separation of endoderm and mesoderm regulatory states.
a, b, DFISHs detecting expression of foxa and gcm at the indicated times post-fertilization. c, d, Demonstration that delta morpholino blocks mesodermal clearance of foxa and blimp1b; WMISH using blimp1b and foxa probes on control (c) and delta MASO-treated (d) embryos. e, f, DFISHs showing that the foxa expression pattern is independent of Gcm expression. g, Model of GRN interactions that determine the segregation of endodermal and mesodermal cell fates. Chronology is indicated by numbers 1–3. Wnt and Delta signals emanate from skeletogenic micromeres; geneX mediates interference with β-catenin activity. NIC/SuH, Su(H)–intracellular Notch domain complex; Tcf/β-cat, Tcf/β-catenin complex. h, Schematic showing spatial patterns of gene expression in the developing embryo, with chronology as in g. Note the additional ring of veg2 descendants after 15 h, arising by radial cleavage; end. p., endoderm precursors; mes. p., mesoderm precursors.
Within a few hours, the sea urchin embryo accomplishes one of the most important regulatory transitions in embryonic development, the permanent separation of endodermal fate from mesodermal fate in sister cells descendant from the same endomesodermal precursors. All but one of the eight endodermal regulatory genes cease to be expressed in mesodermal precursors by 24 h post-fertilization, the exception being the myc gene (Supplementary Fig. 2). Thus, the expression domains of foxa and gcm, which are partially overlapping at 16 h, become exclusive after 18 h, as shown by DFISH (Fig. 2a, b). The genomic mechanism of regulatory-state exclusion is elegant: the same Tcf sites that are used to initiate the endoderm GRN in the veg2 lineage are used again to extinguish it in mesoderm precursors. The mechanism depends on Delta/Notch signalling, which is also the inducer of mesoderm gene expression. In embryos with perturbed expression of either Delta or Notch, the endodermal regulatory genes foxa, blimp1b and dachshund (dac) continue to be expressed in the presumptive mesodermal domain at 24 h (Fig. 2c, d, Supplementary Fig. 6 and data not shown). A similar result was reported for foxa in another sea urchin species21. The exclusion of the endoderm GRN is independent of mesoderm specification per se, as perturbation of gcm expression does not lead to ectopic foxa expression in mesoderm progenitors (Fig. 2e, f). A foxa cis-regulatory study has demonstrated directly that Tcf target sites are required for transcriptional repression of this gene in mesoderm precursor cells11. A possible explanation is that in cells receiving Notch signalling, the availability of nuclear β-catenin is reduced, leading to Tcf/Groucho-mediated repression. This repression specifically affects veg2 endoderm regulatory genes. Thus, hox11/13b and eve, which are both expressed at this stage in veg1 endoderm progenitors, are not affected by interference with Delta/Notch signalling (Supplementary Fig. 6). The expression of the endoderm GRN in endoderm precursors is, in general, completely independent of Delta/Notch signalling (Supplementary Fig. 5).
In summary, the cis-regulatory Tcf responsiveness of early endodermal genes results first in the activation of an endodermal GRN and then, together with cleavage geometry, in the spatial separation of endodermal and mesodermal regulatory states and hence of biological fates (Fig. 2h). In contrast to many ‘binary’ cell-fate decisions that occur later22, this one involves no mutually acting repressors and no other bistable switch features. In fact, it is determinative, like much of early development, rather than bi-stable: there is no preceding intermediate state. Our results exclude an earlier model23 proposing that clearance of blimp1b expression from the mesodermal domain19,24 is responsible for clearance of wnt8 expression from this domain, on the assumption that Blimp1 is a necessary driver of wnt8 expression. This could ultimately lead to the downregulation of most endodermal regulatory genes, by removal of the Tcf/β-catenin signal that activates them. However, although mutation of Blimp1-binding sites reduces the activity of a small wnt8 cis-regulatory construct18, the same mutation does not affect expression of a bacterial artificial chromosome expression construct containing the whole genomic wnt8 cis-regulatory system (Supplementary Fig. 7). In any case, the expression of wnt8 begins in veg2-derived cells long before the onset of blimp1b expression in these cells18,24.
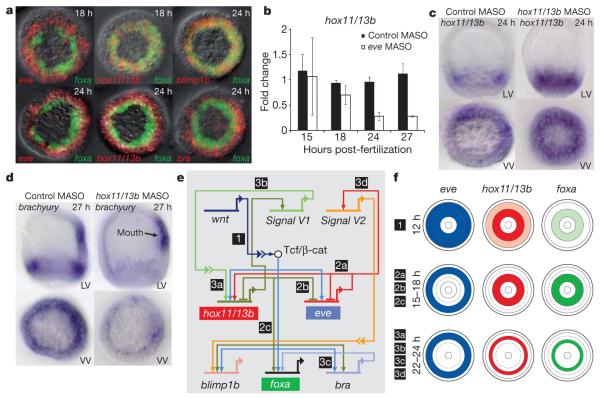
A few hours after the complete separation of endodermal and mesodermal cell fates, marked changes occur in the endodermal regulatory states. These result in the recruitment of two canonical hindgut regulatory genes into the veg1 endoderm GRN, which specifies future hindgut cell fate. By 24 h post-fertilization, hox11/13b and brachyury, which are both expressed in veg2 endoderm at 18 h, are being transcribed instead in veg1 endodermal progenitors, where eve also continues to be expressed (Fig. 3a). The dynamic changes in the spatial expression of hox11/13b and brachyury can be explained by the results of perturbation experiments (Figs 1 and 3 and Supplementary Fig. 5). Hox11/13b expression is activated by Eve in descendants of veg1 cells at 24 h, because injection of eve MASO reduces hox11/13b expression only after 24 h (Fig. 3b). As seen previously for blimp1b and eve, auto-repression is required for the change in the hox11/13b expression domain and this auto-repression results in its clearance from veg2 endoderm. Accordingly, injection of hox11/13b MASO interferes with the clearance of hox11/13b transcripts from veg2 endoderm (Fig. 3c). In both the early veg2 and the later veg1 endoderm GRNs, Hox11/13b functions as a driver of brachyury expression, as shown by the specific reduction of endodermal brachyury expression in embryos injected with hox11/13b MASO (Fig. 3d).
Figure 3. Separation of anterior and posterior endoderm regulatory states.
a, DFISHs showing the dynamic change in gene expression patterns. At 18 h, hox11/13b is co-expressed with foxa in veg2-derived cells, and eve is expressed in veg1 descendants exclusively with respect to foxa. At 24 h, hox11/13b and brachyury (bra) are expressed only in veg1-derived cells; blimp1b and foxa expression continue to overlap. b, Expression of hox11/13b in veg1 domain depends on eve expression. The histogram shows quantitative PCR measurements of hox11/13b ± s.d. (n = 3). c, Clearance of hox11/13b from veg2 descendants and its activation in veg1 descendants depend on Hox11/13b expression. LV, lateral view; VV, vegetal view. d, The expression of brachyury in veg1-derived cells depends on Hox11/13b expression. e, A chronological model of GRN interactions determining anterior versus posterior regulatory states. Chronology is indicated by numbers 1–3. f, Schematic showing spatial patterns of gene expression, with chronology as in e.
Eve expression defines the veg1 regulatory state from 15 h post-fertilization but its detectable regulatory functions begin only after 24 h. The assembly of the veg1 endoderm GRN, which is spatially activated by Eve, is temporally motivated by a predicted signal (V1) expressed under the control of the veg2 endoderm GRN. We note that hox11/13b expression remains restricted to veg2 endoderm precursors in embryos injected with hox11/13b MASO (Fig. 3c). As summarized in Fig. 3e and f, the signal called V1, which is probably Wnt16 (data not shown), is expressed under the control of Hox11/13b in the veg2 lineage to induce hox11/13b expression in veg1 endoderm progenitors. There may be a signal from veg1 to veg2 as well: blimp1b, brn1/2/4, gatae and tgif, which continue to be expressed in veg2 endoderm, are indirectly affected by the knockdown of eve expression in veg1 descendants (Fig. 1f). A second putative signal (V2) is expressed under the control of Eve and activates expression of blimp1b and gatae in veg2 endoderm precursors. Blimp1 then activates brn1/2/4 and tgif expression (Fig. 1f). As a possible consequence of signal V2, blimp1b expression becomes restricted to peripheral tiers of foxa-expressing cells just before gastrulation, when blimp1b transcripts accumulate in cells adjacent to the eve (and V2) expression domain (Supplementary Fig. 3). Blimp1b, gatae and tgif, as well as the tgif driver gene myc, are all expressed in the midgut at the late gastrula stage and we propose that a future midgut regulatory state might be initiated here.
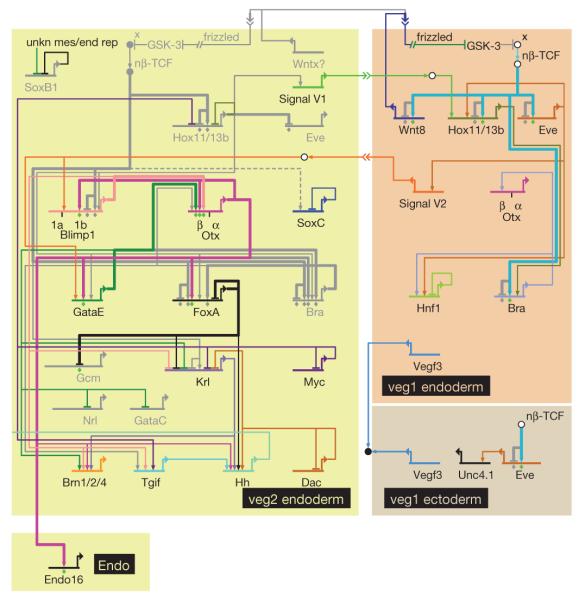
Figure 4 shows models of the ultimate anterior (veg2) and posterior (veg1) endoderm GRNs operating just before the onset of gastrulation. A few additional regulatory genes are activated in the final hours before gastrulation: brn1/2/4, gatae, dac and tgif are expressed in veg2-derived cells and hnf1 is expressed in the veg1 endoderm domain by 27 h after fertilization. In the same time period, the number of direct cross-regulatory linkages increases markedly, as indicated by the results of perturbations shown in Fig. 1d–f (from 6 linkages at 18 h to 26 at 27 h). The models proposed here include previously identified linkages such as the positive feedback circuit between blimp1b, otx and gatae. Almost immediately, with the inception of gastrulation, the anterior endoderm GRN will be required to direct gastrular invagination and accordingly, this GRN achieves autonomy by this time point with the cross-regulatory structure shown in Fig. 4. In contrast, although the posterior endoderm network is uniquely specified, its structure is much simpler at this time because hindgut invagination is still many hours in the future.
Figure 4. Anterior and posterior GRNs just before gastrulation.
BioTapestry presentation as ‘view from the nucleus’ at 30 h (for an interactive version, including expression and perturbation results for each gene, as well as the temporal sequence of appearance or disappearance of linkages, see http://sugp.caltech.edu/endomes/#BioTapestryViewer). Linkages shown in grey were active at earlier time points but by 30 h are extinguished. Unkn mes/end rep, unknown mesodermal and endodermal repressor of soxb1. For a discussion of the circuitry, see text.
Specific regulatory states thus distinguish anterior and posterior endoderm progenitors. These regulatory states are the outputs of GRNs composed of distinct sets of genes and regulatory interactions. Here we show, for the first time, the primary mechanistic basis for the different contributions of veg1 and veg2 endoderm to the future gut. Rather than the progressive differentiation of a broadly initiated, common ‘endomesoderm’ or ‘endoderm’ GRN, cell-fate specification results from the parallel activation of distinct GRNs, long before functional and morphological differentiation.
Regulatory system analysis generates a causal framework that extends vertically from the individual regulatory transactions encoded by the genome to the architecture of the control systems and thence to their ultimate outputs, the phenomena of dynamic spatial specification. This illuminates developmental biology in many ways. For example, we can now see why the endodermal cell lineages have different fates and how they acquire them; why and how the patterns of gene expression change; how the parts of the future gut are encoded and how they are pre-specified in a stepwise manner by the operation of the genomic regulatory system.
METHODS
MASO injection and RNA isolation
MASOs were provided by GeneTools and sequences are given in Supplementary Table 5. MASOs were microinjected into fertilized eggs at 100–400 μM in 0.12 M KCl, as described in ref. 26 and 28. Control MASOs consisting of random sequences 25 nucleotides long were injected at the same or higher concentrations as gene-specific MASOs. Experiments were performed on 2–4 independent embryonic batches. Embryos were cultured at 15 °C and about 100–200 embryos per sample were lysed at 24 h or 27 h after fertilization. Total RNA was isolated using the RNeasy Micro Kit (Qiagen).
Quantitative PCR analysis
Complementary DNA was synthesized using the iScript cDNA synthesis kit (BioRad). Quantitative PCR was performed using iTaq SYBR green supermix (BioRad) on an amount of cDNA equivalent to 0.6 embryos in a 10 μl reaction with the gene-specific primers listed in Supplementary Table 4. Gene expression levels were normalized to levels of ubiquitin expression. Changes in expression levels were determined by comparing normalized expression levels in MASO-injected embryos to normalized expression levels in uninjected control embryos29. Changes were considered significant if gene expression levels decreased more than threefold or increased more than twofold in embryos injected with a gene-specific MASO. Target gene expression was considered to be affected by a regulatory factor at a given time point if the injection of a regulatory-gene-specific MASO, but not the injection of control MASOs, resulted in significant changes in target gene expression in the majority of experiments. Computational procedures included the automated reduction of perturbation data (used to generate Supplementary Fig. 5) and the application of the GRN platform BioTapestry27.
Whole-mount in situ hybridization
Probe templates were either derived from a cDNA library or generated by PCR amplification of cDNA synthesized from the RNA of 27 h embryos with gene-specific primers listed in Supplementary Table 6. Antisense RNA probes labelled with digoxigenin or fluorescein were generated using the corresponding RNA labelling mix (Roche). Whole-mount in situ hybridizations (ISHs) were performed according to standard methods25,30. Briefly, embryos were fixed in 4% paraformaldehyde, 32.5% sea water, 32.5 mM maleic acid (pH7) and 162.5 mM NaCl at 4 °C overnight. Hybridizations were performed overnight at 65 °C using a probe concentration of 1 ng μl−1. For single whole-mount ISH, probeswere detected using anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (1:1,000 dilution) and NBT/BCIP (nitro-blue tetrazolium chloride/ 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt). Hybridizations for double whole-mount ISH were performed according to a protocol described previously18. Probes were detected by horseradish-peroxidase-conjugated anti-digoxigenin or anti-fluorescein Fab fragments (1:1,000 dilution) using substrates provided in the TSA Plus Cyanine3/Fluorescein System (Perkin Elmer). Staining occurred at a substrate dilution of 1:400 in 1× Plus Amplification Diluent or in Tris-buffered saline with 0.005% H2O2.
Supplementary Material
Acknowledgements
We acknowledge technical assistance from J. Yun, who executed much of the perturbation analysis matrix, and from A. Puszynska and E. Erkenbrack, who contributed to whole-mount in situ hybridization results. We are grateful to E. Rothenberg for a detailed critique of the manuscript. I.S.P. was the recipient of a fellowship from the Swiss National Science Foundation in the initial stages of this work. The research was supported by National Institutes of Health grant HD37105 to E.H.D.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions This research was conceived by I.S.P. and E.H.D. and all experiments were designed and executed by I.S.P. with the assistance acknowledged above. The results were interpreted by I.S.P. and E.H.D., who also contributed jointly to the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Cameron RA, Fraser SE, Britten RJ, Davidson EH. Macromere cell fates during sea urchin development. Development. 1991;113:1085–1091. doi: 10.1242/dev.113.4.1085. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, McClay DR. The allocation ofearly blastomeres tothe ectoderm and endoderm is variable in the sea urchin embryo. Development. 1997;124:2213–2223. doi: 10.1242/dev.124.11.2213. [DOI] [PubMed] [Google Scholar]
- 3.Ransick A, Davidson EH. Late specification of Veg1 lineages to endodermal fate in the sea urchin embryo. Dev. Biol. 1998;195:38–48. doi: 10.1006/dbio.1997.8814. [DOI] [PubMed] [Google Scholar]
- 4.Byrum CA, Xu R, Bince JM, McClay DR, Wikramanayake AH. Blocking Dishevelled signaling in the noncanonical Wnt pathway in sea urchins disrupts endoderm formation and spiculogenesis, but not secondary mesoderm formation. Dev. Dyn. 2009;238:1649–1665. doi: 10.1002/dvdy.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 6.Wikramanayake AH, et al. Nuclear β-catenin-dependent Wnt8signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 7.Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- 8.Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in Notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126:5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- 9.Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 10.Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 11.de-Leon SB, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc. Natl Acad. Sci. USA. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 13.Howard-Ashby M, et al. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev. Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 14.Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev. Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev. Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev. Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minokawa T, Wikramanayake AH, Davidson EH. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev. Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 19.Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev. Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 2009;583:3948–3958. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137:83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 24.Livi CB, Davidson EH. Regulation of spblimp1/krox1a, an alternatively transcribed isoform expressed in midgut and hindgut of the sea urchin gastrula. Gene Expr. Patterns. 2007;7:1–7. doi: 10.1016/j.modgep.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Ransick A. Detection of mRNA by in situ hybridization and RT-PCR. Methods Cell Biol. 2004;74:601–620. doi: 10.1016/s0091-679x(04)74024-8. [DOI] [PubMed] [Google Scholar]
- 26.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl Acad. Sci. USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim. Biophys. Acta. 2009;1789:363–374. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rast JP, et al. Recovery of developmentally defined gene sets from high-density cDNA macroarrays. Dev. Biol. 2000;228:270–286. doi: 10.1006/dbio.2000.9941. [DOI] [PubMed] [Google Scholar]
- 29.Materna SC, Oliveri P. A protocol for unraveling gene regulatory networks. Nature Protocols. 2008;3:1876–1887. doi: 10.1038/nprot.2008.187. [DOI] [PubMed] [Google Scholar]
- 30.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl Acad. Sci. USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.