Abstract
Changes in gene dosage are a major driver of cancer, engineered from a finite, but increasingly well annotated, repertoire of mutational mechanisms1. This can potentially generate correlated copy number alterations across hundreds of linked genes, as exemplified by the 2% of childhood acute lymphoblastic leukemia (ALL) with recurrent amplification of megabase regions of chromosome 21 (iAMP21)2,3. We used genomic, cytogenetic and transcriptional analysis, coupled with novel bioinformatic approaches, to reconstruct the evolution of iAMP21 ALL. We find that individuals born with the rare constitutional Robertsonian translocation between chromosomes 15 and 21, rob(15;21)(q10;q10)c, have ~2700-fold increased risk of developing iAMP21 ALL compared to the general population. In such cases, amplification is initiated by a chromothripsis event involving both sister chromatids of the Robertsonian chromosome, a novel mechanism for cancer predisposition. In sporadic iAMP21, breakage-fusion-bridge cycles are typically the initiating event, often followed by chromothripsis. In both sporadic and rob(15;21)c-associated iAMP21, the final stages frequently involve duplications of the entire abnormal chromosome. The end-product is a derivative of chromosome 21 or the rob(15;21)c chromosome with gene dosage optimised for leukemic potential, showing constrained copy number levels over multiple linked genes. Thus, dicentric chromosomes may be an important precipitant of chromothripsis, as we show rob(15;21)c to be constitutionally dicentric and breakage-fusion-bridge cycles generate dicentric chromosomes somatically. Furthermore, our data illustrate that several cancer-specific mutational processes, applied sequentially, can co-ordinate to fashion copy number profiles over large genomic scales, incrementally refining the fitness benefits of aggregated gene dosage changes.
Acute lymphoblastic leukemia (ALL) is the most common childhood cancer, with an annual incidence of 35/million children aged 0-14 years4. Approximately 2% of these cases show intrachromosomal amplification of one copy of chromosome 21, iAMP21, which defines a distinct ALL subgroup3,5 with prognostic and therapeutic implications6,7.
In 95 patients with iAMP21 ALL enrolled in UK clinical trials, we found 3 (3.2%) with a constitutional Robertsonian translocation between chromosomes 15 and 21, rob(15;21)(q10;q10)c. In other ALL trials, we identified a further nine cases of iAMP21 associated with rob(15;21)c, at a similar fraction of cases. Robertsonian translocations are rearrangements between the short arms of acrocentric chromosomes (namely, 13-15, 21-22). They are found in ~1 in 1,000 newborns8,9, but rob(15;21)c accounts for only 0.5-1% of these. To confirm this, we interrogated cytogenetics databases. Only three patients among 93,000 referrals for haematological malignancies to the Munich Leukemia Laboratory and West Midlands Regional Genetics Laboratory carried rob(15;21)c. Similarly, only 16 cases were found among approximately 300,000 referrals to UK regional cytogenetics laboratories for investigation of infertility or previous Down syndrome birth.
From these data, we estimate the risk of iAMP21 ALL in carriers of rob(15;21)c to be ~2700–fold increased over the general population (Supplementary table 1). This association is remarkably specific. All patients in this study with rob(15;21)c had iAMP21 ALL, implying that they are not predisposed to other forms of ALL, nor other cancers, as far as we can ascertain. Furthermore, the only Robertsonian translocation associated with iAMP21 ALL was rob(15;21)c. For clarity, we use ‘rob(15;21)c’ to denote the germline configuration and ‘der(15;21)’ to describe the rearranged and amplified chromosome in these cases.
Using cytogenetics, fluorescence in situ hybridization (FISH) and copy number profiling, we studied 21 patients with sporadic iAMP21 ALL and 12 patients with ALL associated with rob(15;21)c. Five sporadic iAMP21 and four cases associated with rob(15;21)c were sequenced to identify genomic rearrangements10 (Supplementary table 2; Extended data figure 1; Supplementary figure 1). We applied deductive approaches, supported by confirmatory simulations, to reconstruct principles underlying the temporal evolution of iAMP21 ALL. This reasoning is explored in considerable detail, together with a sample-by-sample analysis, in Supplementary results, Extended data figures 3-8, Supplementary tables 3-6 and Supplementary figures 4-23.
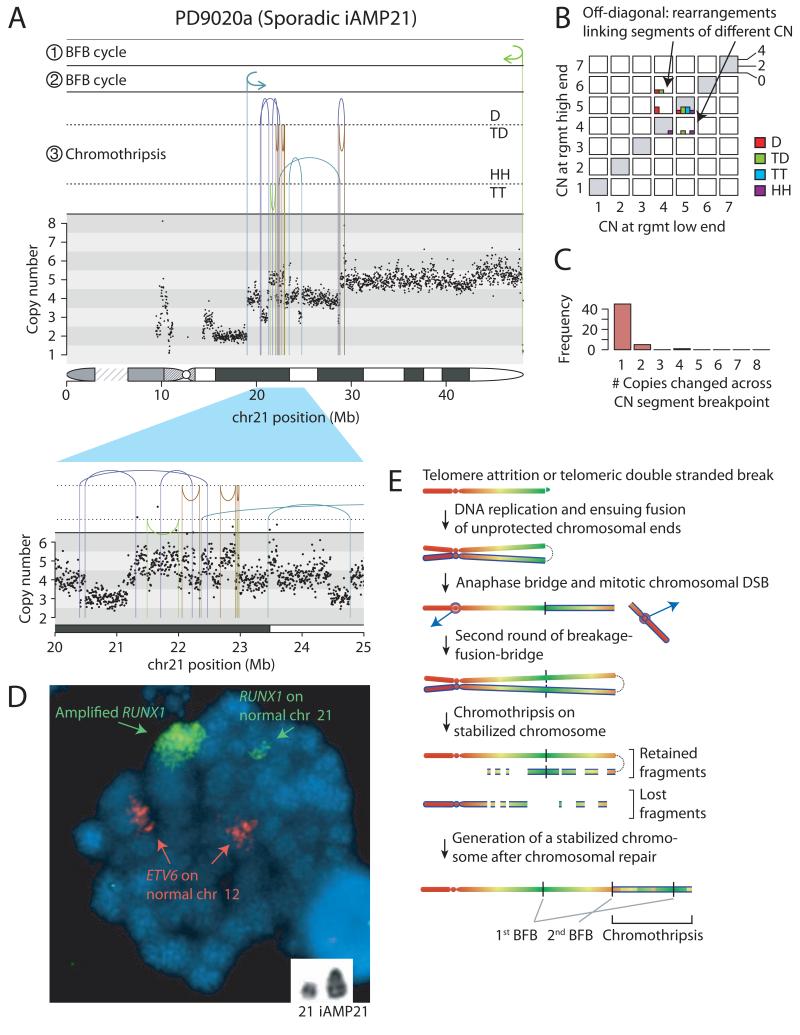
The broad themes are illustrated by two representative cases (Figures 1-2). In PD9020a, a patient with sporadic iAMP21, the boundaries of the amplified region are demarcated by fold-back inversion rearrangements (Figure 1A). These indicate breakage-fusion-bridge (BFB) repair11, previously proposed to trigger iAMP212,12,13. Breakage-fusion-bridge repair is a mutational process initiated by a telomeric double strand (ds) DNA break that is replicated in S phase. In G2, the two copies of the dsDNA break are fused by non-homologous end-joining (marked ① in figure 1A). This creates a dicentric chromosome in which the two centromeres are pulled to opposite poles during mitosis, forming an anaphase bridge. With cytokinesis, the bridge breaks, and the process can repeat in the next cell cycle (marked ② in figure 1A). In the region between the fold-back inversions in PD9020a, we also found a cluster of back-and-forth rearrangements (marked ③) of all four possible orientations, associated with copy number profiles that oscillate among three states (Figure 1A, zoomed-in panel). These clusters bear the hallmarks of chromothripsis14 (Extended data table 1; Extended data figure 2; Supplementary results), a mutational process in which a one-off catastrophic event shatters one or a few chromosomal regions leading to large numbers of localized genomic rearrangements15,16.
Figure 1. Rearrangements of chromosome 21 in patient PD9020a.
A: Rearrangement and copy number pattern. The temporal order of the three major rearrangement events are marked ①, ② and ③. Rearrangements are separated based on their orientation: D, deletion-type; TD, tandem duplication-type; HH, head-to-head inverted; TT, tail-to-tail inverted. B: Copy number jump distribution, showing the copy number at each end of each rearrangement. C: Copy number step distribution, showing the distribution in magnitude of copy number change at copy number segmentation breakpoints. D: Metaphase showing multiple signals for RUNX1 clustered on a single chromosome (large green signal) compared to the normal chromosome 21 (small paired green signals). The red signals indicate two normal copies of ETV6 on the chromosomes 12. Inset shows a partial G-banded karyotype of chromosomes 21. E: Model for the evolution of the iAMP21 chromosome. At each stage, newly synthesized sister chromatids are distinguished by a blue outline.
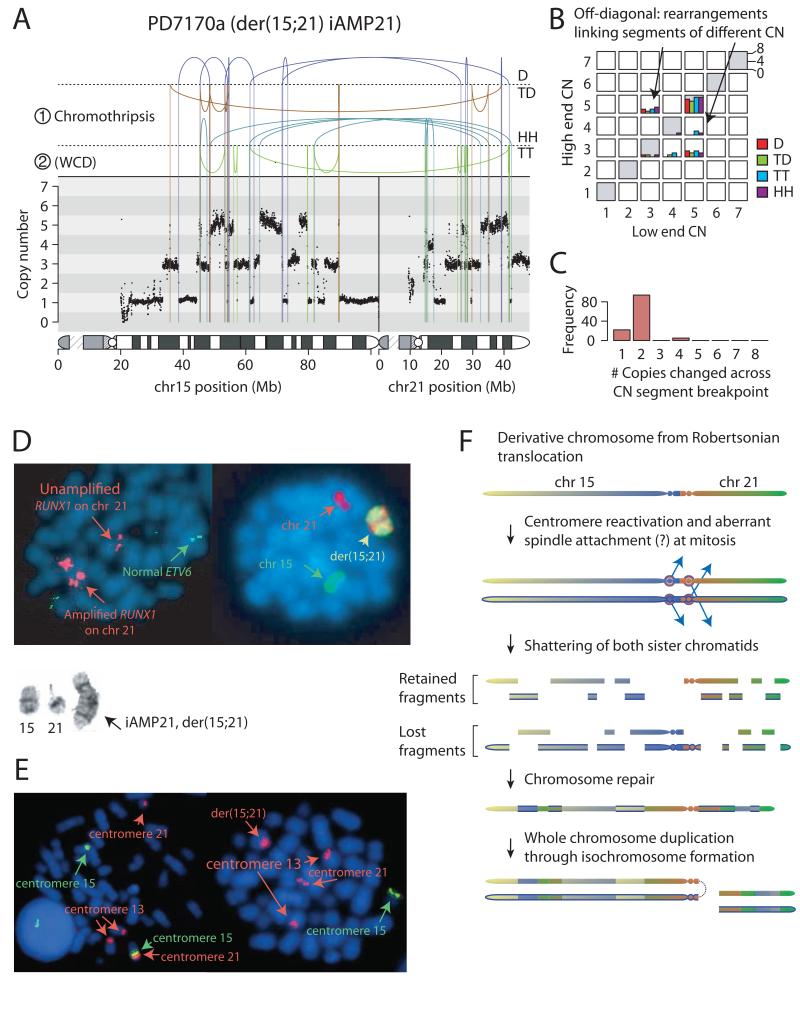
Figure 2. Rearrangements of der(15;21) in patient PD7170a.
A: Rearrangement and copy number pattern. The temporal order of the two major rearrangement events are marked ① and ②. Rearrangements are separated based on their orientation: D, deletion-type; TD, tandem duplication-type; HH, head-to-head inverted; TT, tail-to-tail inverted. B: Copy number jump distribution, showing the copy number at each end of each rearrangement. C: copy number step distribution, showing the distribution in magnitude of copy number change at copy number segmentation breakpoints. D: Representative metaphases: Left-hand cell shows multiple signals for RUNX1 (large red signals) clustered on two regions on the abnormal chromosome. And normal copies of ETV6 on chromosome 12 (green). Right-hand cell has been painted for chromosomes 15 (green) and 21 (red). Inset shows partial G-banded karyotype: normal chromosome 15, normal chromosome 21 and isochromosome der(15;21). E: Left-hand cell shows representative metaphase from a non-leukaemic cell in patient PD10009a with rob(15;21)c, hybridized with centromere-specific probes for chromosomes 15 (green) and 13 and 21 (red), confirming that the Robertsonian chromosome is dicentric. Right-hand cell shows a leukaemia metaphase in which der(15;21) iAMP21 chromosome retains the chromosome 21 centromere (red), but not the chromosome 15 centromere (green). F: Model for evolution of iAMP21 in rob(15;21)c. Newly synthesized sister chromatids are indicated by a blue outline.
Two features of this genomic architecture allow reconstruction of the temporal evolution of the iAMP21 chromosome. First, the rearrangements frequently link together genomic segments of different copy number (off-diagonal histograms, Figure 1B). Secondly, as we traverse chromosome 21 from first to last base-pair, the copy number segments on either side of each breakpoint position typically differ in copy number by one (Figure 1C). FISH confirms widespread RUNX1 signals along the iAMP21 chromosome (Figure 1D). Based on reasoning outlined in detail in Supplementary results, these features indicate that chromothripsis occurred after two BFB cycles and was likely the final major event, stabilising the chromosome (Figure 1E).
In PD7170a, a der(15;21) iAMP21 derived from rob(15;21)c, the picture is dominated by a series of back-and-forth rearrangements spanning chromosomes 15 and 21 (Figure 2A-C). Cytogenetic and FISH studies confirmed that it was the Robertsonian chromosome undergoing rearrangement (Figure 2D-E). A sizable number of rearrangements link together segments of different copy number (off-diagonal histograms, Figure 2B), and copy number oscillates among three, rather than two, states. Together with occasional inverted rearrangements with no breakpoints between the two joined ends, this pattern argues that chromothripsis was the initiating event, and that the chromothripsis process involved both sister chromatids of the Robertsonian chromosome (Extended data figures 7-8; supplementary results; supplementary table 4). Importantly, the shattered sister chromatids are repaired into one derivative chromosome, thereby amplifying the copy number of some chromosomal regions (Figure 2F). The amplification was completed by whole chromosome duplication of the der(15;21) chromosome through isochromosome formation (Figure 2D).
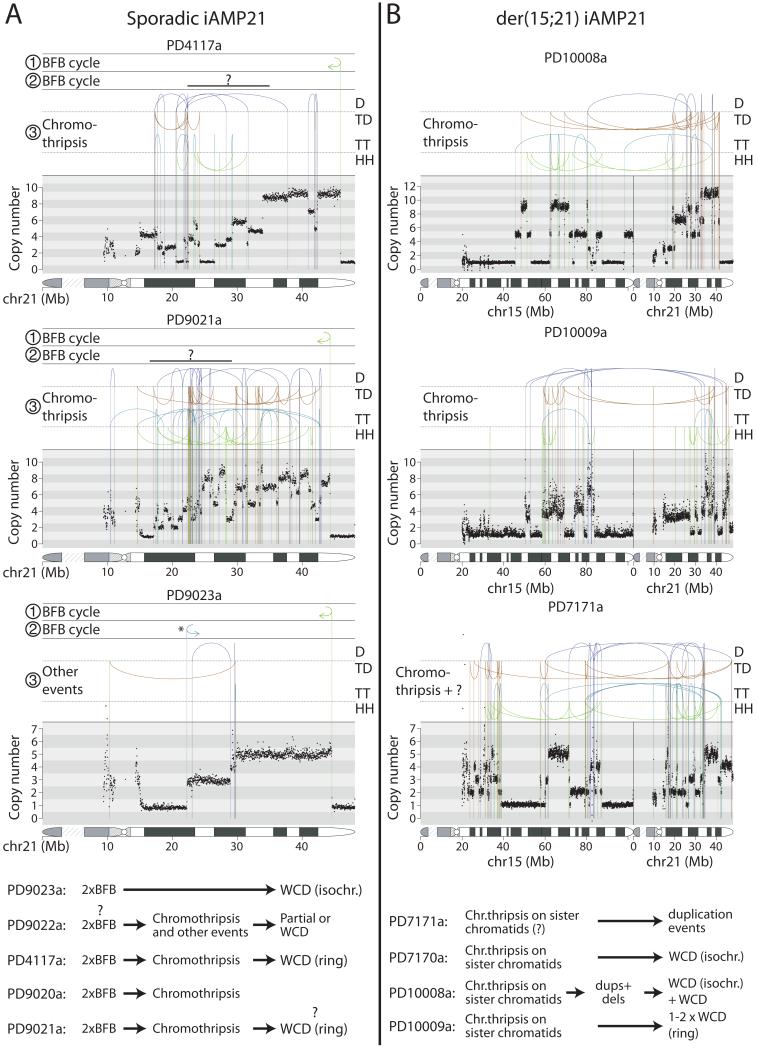
These broad temporal sequences of events are reflected in the other samples (Figure 3; Extended data figure 6; Supplementary results). In the other four sequenced cases of sporadic iAMP21, a telomeric fold-back inversion suggests at least one BFB cycle. In each, this was a critical early event, defining the break between the most amplified region of chromosome 21 and subtelomeric loss. Chromothripsis occurred after the BFB cycles in three cases. Finally, partial or whole chromosome duplications usually completed the evolution. In the other three sequenced der(15;21) iAMP21 cases, amplification was initiated by chromothripsis involving both sister chromatids of the rob(15;21)c chromosome. This appeared to be followed by further rearrangements in two cases and was completed by whole chromosome duplications.
Figure 3. Rearrangement patterns of the iAMP21 chromosome in the remaining patients.
Rearrangement and copy number patterns for chromosome 21 of sporadic iAMP21 ALL patients (A) and der(15;21) rearrangements in der(15;21) iAMP21 ALL patients (B). The inferred temporal orders of the major rearrangement events are shown with symbols ①, ② and ③. In patients PD4117a and PD9021a, the fold-back rearrangement demarcating the second BFB repair breakpoint have probably been lost or obscured due to subsequent rearrangement events, and a ‘?’ symbol is used to denote the uncertainty of their location. Inferred evolution of the derivative iAMP21 chromosomes are shown in the bottom panel. WCD – whole-chromosome duplication, WC – whole chromosome. Events with incomplete understanding are labeled ‘?’.
These data provide insight into why there is such specific enrichment of iAMP21 ALL in carriers of rob(15;21)c. Universally, the amplification is initiated by chromothripsis that affects both sister chromatids of the Robertsonian chromosome. This suggests that rob(15;21)c has a structural abnormality that specifically predisposes it, after replication, to the catastrophic shattering of chromothripsis. Using FISH, we demonstrated that the rob(15;21)c chromosome has centromeres from both chromosomes 15 and 21, and is thus dicentric (Figure 2E, Extended data figure 1). Our hypothesis, therefore, is that the two centromeres of the Robertsonian chromosome can occasionally confound attachment of mitotic spindles to the sister kinetochores, such that each chromatid connects to spindles emanating from opposite poles (Figure 2F). During anaphase, this merotelic attachment would lead to lagging of both sister chromatids, rendering them jointly prone to chromothripsis. The der(15;21) iAMP21 chromosomes consistently lose the chromosome 15 centromere, shown by FISH (Figure 2E) and sequencing (Figures 2A, 3B), potentially enhancing stability of the derivative chromosome. In sporadic iAMP21 cases, chromothripsis frequently follows BFB cycles. While we cannot know whether chromothripsis is an immediate consequence, it is plausible that the dicentric chromosome created by BFB repair could trigger chromothripsis, analogous to that seen with the dicentric rob(15;21)c17,18.
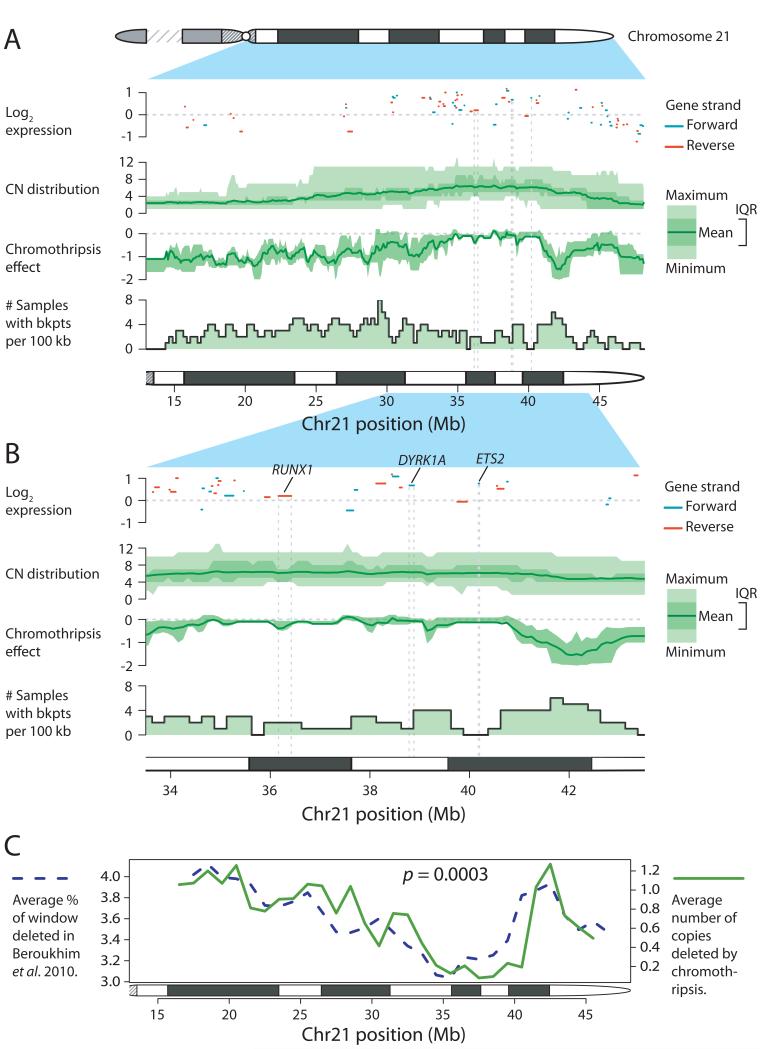
The preceding analysis provides insight into the mutational processes shaping chromosome 21, but unless the resulting chromosome profile confers a selective advantage on the clone, it will not expand. We combined copy number profiles and gene expression data3 from additional patients with iAMP21 ALL (Figure 4). A consensus copy number profile emerged in which regions from ~35.9-36.4Mb and ~38.0-40.0Mb of chromosome 21 were consistently the most highly amplified and over-expressed, including genes important in haematological malignancies such as RUNX1, DYRK1A and ETS22.
Figure 4. Chromothripsis alters the copy number landscape of chromosome 21 in a non-random fashion.
Chromosome arm-level (A) and zoomed-in view (B) of chromosome 21, showing gene expression, copy number (CN) distribution, chromothripsis effect and distribution of rearrangement breakpoints. In the gene expression panels, positive strand genes are shown in blue and negative strand genes are shown in red. C: Correlation between average rate of deletion in the Beroukhim et al.19 dataset and chromothripsis effect for chromosome 21. IQR – inter-quartile range.
The final stage of iAMP21 generation usually involves duplication of the whole derivative chromosome, through whole chromosome duplication, isochromosome or ring formation. All duplications occurred after chromothripsis, suggesting that chromothripsis might be remodeling chromosome 21 in a non-random fashion. We used the inferred temporal evolution of somatic rearrangements to extract and average copy number changes resulting from chromothripsis (Figure 4). As expected, chromothripsis spared the most amplified regions, whereas on average 1-2 copies were deleted from other parts of the chromosome. Analysis of der(15;21) iAMP21 suggested that regions of chromosome 15 were also consistently lost or retained, although sample numbers are small (Extended data figure 9).
Strikingly, the consensus chromothripsis landscape in iAMP21 closely mirrored the copy number profile of chromosome 21 averaged over thousands of cancer samples across different cancer types19,20 (p=0.0003; Figure 4C; Supplementary figures 27-30). This suggests that chromothripsis plays a critical role in optimising the copy number landscape of chromosome 21 to maximise the net selective advantage gained from subsequent rounds of whole chromosome duplication.
From a detailed dissection of the mutational forces causing one particular subtype of one particular cancer, findings with general significance have emerged. Carriers of constitutional rob(15;21)c chromosomes are specifically but massively predisposed to iAMP21 ALL. Usually, constitutional risk of cancer is mediated by variation in coding sequence or gene regulation, but here it appears to be transmitted through a propensity for the Robertsonian chromosome to undergo chromothripsis after replication. This may be because it is dicentric and prone to anaphase bridging, which would dovetail with the frequent occurrence of chromothripsis following BFB cycles in sporadic iAMP21 ALL. This hypothesis is consistent with the finding that lagging chromosomes during anaphase can become sequestered in micronuclei and subjected to chromosomal pulverisation before rejoining the main nucleus17,18. More generally, the study of iAMP21 ALL has illustrated how large-scale copy number changes can be optimised by spatially and temporally coordinated genomic instability taking several complementary forms. BFB cycles can generate rapid-fire, focal amplification; chromothripsis causes loss of multiple, non-contiguous chromosomal regions; and whole chromosome duplication gives expansive, low-amplitude amplification. Their combined activity, therefore, gives considerably more flexibility to shaping large-scale chromosomal copy number profiles than any one process alone. Of course, clones sample these mutational processes randomly, so only when the aggregate fitness of such changes is positive will the clone have the selective advantage to expand.
METHODS SUMMARY
Information was available from 21 iAMP21 patients and 12 with iAMP21 and rob(15;21)(q10;q10)c (Supplementary Table 2). Paired-end sequencing data were generated as 37-75bp paired reads from 400-500bp fragments as previously described10. The deductive reasoning for reconstructing temporal evolution of complex rearrangements followed principles formulated previously21. Confirmatory PCR across the breakpoints was performed for the vast majority of identified rearrangements (Supplementary results).
Supplementary Material
ACKNOWLEDGEMENTS
We thank member laboratories of the United Kingdom Cancer Cytogenetic Group (UKCCG) for providing cytogenetic data and material. Primary childhood leukemia samples used in this study were provided by the Leukaemia and Lymphoma Research Childhood Leukaemia Cell Bank working with the laboratory teams in the Bristol Genetics Laboratory, Southmead Hospital, Bristol: Molecular Biology Laboratory, Royal Hospital for Sick Children, Glasgow: Molecular Haematology Laboratory, Royal London Hospital, London: Molecular Genetics Service and Sheffield Children’s Hospital, Sheffield. We also thank all the members of the NCRI Childhood Cancer and Leukaemia Group (CCLG) Leukaemia Subgroup for access to material and data on clinical trial patients. This work was supported by the Wellcome Trust (077012/Z/05/Z); Leukaemia and Lymphoma Research Specialist Programme and European Research Council (249891). PJC has a Wellcome Trust Senior Clinical Research Fellowship (WT088340MA). PVL is supported by a postdoctoral research fellowship and PV is a Senior Clinical Investigator funded by the Research Foundation – Flanders (FWO). P.S. is funded by the European Research Council (grant #249891). Genome sequence data have been deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/, hosted by the EBI) with accession number EGAD00001000658.
REFERENCES
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rand V, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011;117:6848–6855. doi: 10.1182/blood-2011-01-329961. [DOI] [PubMed] [Google Scholar]
- 3.Strefford JC, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci U S A. 2006;103:8167–8172. doi: 10.1073/pnas.0602360103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stiller CA, Kroll ME, Boyle PJ, Feng Z. Population mixing, socioeconomic status and incidence of childhood acute lymphoblastic leukaemia in England and Wales: analysis by census ward. Br J Cancer. 2008;98:1006–1011. doi: 10.1038/sj.bjc.6604237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moorman AV, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109:2327–2330. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 6.Moorman AV, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol. 2013;31:3389–3396. doi: 10.1200/JCO.2013.48.9377. [DOI] [PubMed] [Google Scholar]
- 7.Heerema NA, et al. Intrachromosomal amplification of chromosome 21 is associated with inferior outcomes in children with acute lymphoblastic leukemia treated in contemporary standard-risk children’s oncology group studies: a report from the children’s oncology group. J Clin Oncol. 2013;31:3397–3402. doi: 10.1200/JCO.2013.49.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamerton JL, Canning N, Ray M, Smith S. A cytogenetic survey of 14,069 newborn infants. I. Incidence of chromosome abnormalities. Clin Genet. 1975;8:223–243. doi: 10.1111/j.1399-0004.1975.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29:103–108. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair PB, et al. Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy. Hum Mol Genet. 2011;20:2591–2602. doi: 10.1093/hmg/ddr159. [DOI] [PubMed] [Google Scholar]
- 13.Robinson HM, Harrison CJ, Moorman AV, Chudoba I, Strefford JC. Intrachromosomal amplification of chromosome 21 (iAMP21) may arise from a breakage-fusion-bridge cycle. Genes Chromosomes.Cancer. 2007;46:318–326. doi: 10.1002/gcc.20412. [DOI] [PubMed] [Google Scholar]
- 14.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TM, et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res. 2013;23:217–227. doi: 10.1101/gr.140301.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenman CD, et al. Estimation of rearrangement phylogeny for cancer genomes. Genome Res. 2012;22:346–361. doi: 10.1101/gr.118414.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.