Abstract
The MIRAGE (minimum information required for a glycomics experiment) initiative was founded in Seattle, WA, in November 2011 in order to develop guidelines for reporting the qualitative and quantitative results obtained by diverse types of glycomics analyses, including the conditions and techniques that were applied to prepare the glycans for analysis and generate the primary data along with the tools and parameters that were used to process and annotate this data. These guidelines must address a broad range of issues, as glycomics data are inherently complex and are generated using diverse methods, including mass spectrometry (MS), chromatography, glycan array-binding assays, nuclear magnetic resonance (NMR) and other rapidly developing technologies. The acceptance of these guidelines by scientists conducting research on biological systems in which glycans have a significant role will facilitate the evaluation and reproduction of glycomics experiments and data that is reported in scientific journals and uploaded to glycomics databases. As a first step, MIRAGE guidelines for glycan analysis by MS have been recently published (Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J., et al. 2013. The minimum information required for a glycomics experiment (MIRAGE) project – Improving the standards for reporting mass spectrometry-based glycoanalytic data. Mol. Cell Proteomics. 12:991–995), allowing them to be implemented and evaluated in the context of real-world glycobiology research. In this paper, we set out the historical context, organization structure and overarching objectives of the MIRAGE initiative.
Background
Advances in our ability to identify and quantify complex glycans and glycoconjugates has led to an increasing awareness of the key roles that these molecules play in a wide range of physiological and pathological processes, including cell adherence, cell–cell interactions, molecular trafficking, biosynthetic quality control, signal transduction and host–pathogen recognition. Various types of glycans and glycoconjugates are thus becoming recognized as essential participants in almost all biological processes. Structural analysis of glycoconjugates is technically challenging, requiring sophisticated analytical and computational techniques applied at the interface of biology and chemistry. Although recent technical advances in this area have led to the emergence of glycomics as a distinct discipline, progress is slowed by the unavailability of robust, generally applicable software tools required to process, annotate, archive and mine the data now being generated in this domain.
The complexity of glycans and the diversity of their structures and molecular contexts have necessitated the development of a wide range of experimental techniques and instrumentation for their analysis. Although mass spectrometry (MS) is the most frequently applied methodology for glycan analysis, array-based ligand-binding assays, high-performance liquid chromatography, capillary electrophoresis (CE), nuclear magnetic resonance (NMR) and several other techniques are now being routinely used for this purpose. Recent advances in analytical methodology and instrumentation make it possible to produce glycomics data with increased depth, speed and efficiency, resulting in the generation of extremely large and diverse datasets. However, the reporting and/or distribution of information obtained during a glycomics experiment pose unique challenges to the analyst. This includes the identification and presentation of relevant metadata that allows the results to be objectively evaluated and interpreted in a biological context and reproduced in the laboratories of other scientists.
It is important to note that glycomics cannot be viewed as a straightforward extension of proteomics. Glycomics and proteomics experiments share the same basic goal, i.e. the identification and quantification (where possible) of specific molecular structures in a particular biological context. However, the complex and often branched structures of glycans, in combination with the non-template-driven mechanisms leading to their biosynthesis, have made the emergence of glycomics as a discipline dependent on the ongoing development of new analytical approaches and computational tools that are not required for proteomics. The quality and information content of the annotated data generated by such tools can vary considerably, depending on the exact experimental conditions used to generate primary data, the suitability and configuration of the computational tools used to process this data, the quality of any databases that are invoked during the data processing and the validity of any assumptions that are made when assigning glycan structures in the presence of incomplete analytical data.
The validity of glycan structure assignments can be assessed only if the relevant experimental parameters, computational methods and underlying assumptions used to make the assignments are described. In addition glycan analysis is often performed not just using one method or technique but by utilizing several orthogonal methods including array-based ligand-binding assays, liquid chromatography (LC), CE, NMR, various types of MS such as mass profiling and tandem MS or hyphenated analytical methods such as GC–MS or LC–MS. Therefore, any information derived from each technique used has to be reported to provide a comprehensive and meaningful overview on the structure assignment, since each technique will provide additional information consolidating the structural assignment or illustrating exclusion of alternative structures.
The MIRAGE guidelines provide a framework that allows this information to be identified and presented in a consistent manner in order to enhance the value of structural analyses that are disseminated by both scientific journals and databases. Scientific journals can use the MIRAGE guidelines as the foundation for developing their own checklists and guidelines for publishing glycomics data. In fact, the recently published MIRAGE guidelines for glycan analysis by MS (Kolarich et al. 2013) were evaluated by members of the glycobiology community (including representatives of several journals such as Glycobiology and Molecular and Cellular Proteomics (MCP)) at a workshop in Athens, Georgia in August 2012 before establishing guidelines for publishing mass spectral (MS) based glycomics data, later called “Athens Guidelines” (Wells and Hart 2013) (see also instructions to authors for Glycobiology). The MIRAGE guidelines are not strictly enforced by the editors of MCP or Glycobiology, as the MIRAGE guidelines are considerably more detailed than the Athens Guidelines. However, the two sets of guidelines are mutually consistent and complementary.
The MIRAGE guidelines provide reviewers with specific, technical descriptions of the metadata required to evaluate the glycomics analysis, and can thus be used by a qualified reviewer as a basis for making judgments regarding the validity of specific conclusions in the manuscript. However, they are not intended to substitute for the review process itself or to define acceptance criteria for submitted manuscripts.
The criteria for acceptance of glycomics data for inclusion into a database are likely to be somewhat different than those for acceptance in a scientific journal. For example, the curatorial process for inclusion of a dataset in a database may not include review by an expert. In such cases, rigorous guidelines for the inclusion of metadata are crucial, as these metadata are a necessary prerequisite for the (automated or manual) selection of relevant, trusted data from the database for comparison to new datasets or inclusion in data-mining investigations. We expect that the MIRAGE guidelines (developed by analysts, biologists and bioinformaticians with glycomics expertise) will serve as a convenient foundation to define the information that has to be stored in a database to provide comprehensive and reproducible datasets to its users.
MIRAGE
In 2006, participants at the Workshop on Analytical and Bioinformatic Glycomics, agreed that there is an urgent need to develop infrastructure, including standardized protocols for the exchange and reporting of structural glycan data and metadata to implement “a worldwide network of databases containing experimental and analytical data relevant to the structures and functions of glycans” (Packer et al. 2008). A subsequent NIH workshop organized by the Consortium for Functional Glycomics in 2009 extended and refined these requirements, emphasizing the need to “Define specific criteria that make it possible for experts and non-experts to rapidly assess the depth and quality of a structural characterization that is described in a publication or structural database entry.” (Workshop on Analytic and Bioinformatic Glycomics 2009). In July 2011, an international group of scientists attending the 2nd Beilstein Symposium on Glyco-Bioinformatics in Potsdam (Germany) established the MIRAGE initiative under the auspices of the Beilstein-Institut (http://www.beilstein-institut.de). MIRAGE (i.e. the “Minimum Information Required for A Glycomics Experiment”) integrates worldwide efforts to develop reporting guidelines for glycomics analytic data with the goal of facilitating the interpretation, evaluation and reproduction of these data. These guidelines are intended to improve the quality of glycomics datasets published in journals and stored in databases.
Glycomics analysis offers unique challenges that necessitate reporting and archiving standards that are distinct from those already established for proteomics (such as the MIAPE standard; Taylor et al. 2007, 2008). The MIRAGE standards thus serve as critical elements of the infrastructure required to integrate relevant scientific knowledge into a worldwide glycomics bioinformatics system capable of addressing diverse needs of the scientific community. In this context, success of the MIRAGE project represents a critical step toward fulfilling the recommendations of the NAS Committee on Assessing the Importance and Impact of Glycomics and Glycosciences (Walt et al. 2012), which include the development of a “well-documented glycan structure database that can be linked back to the original experimental data.”
Coordination and consulting
The MIRAGE project is steered by an international panel of scientists with expertise in diverse disciplines, including medicinal and developmental glycobiology, carbohydrate chemistry, glycoanalytics and glyco-bioinformatics, ensuring that the guidelines will encompass the most frequently used and relevant technologies.
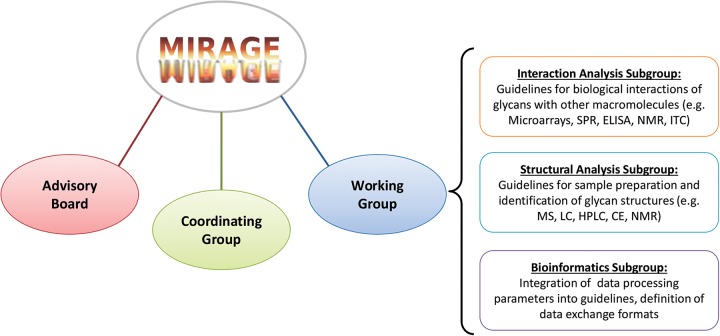
The MIRAGE project incorporates three organizational components: the working group, the coordinating group and the advisory board (Figure 1). The working group defines the tasks, makes general decisions and integrates detailed guidelines, which are developed by subgroups focusing on structural analysis, interaction analysis and bioinformatics.
Fig. 1.
MIRAGE incorporates three organizational components: the working group, the coordinating group and the advisory board.
The efforts of the structural analysis subgroup have already led to the publication of guidelines for reporting glycoanalytic mass spectral (MS) data (Kolarich et al. 2013). This guideline has an organizational structure similar to that of the MIAPE-MS guideline for proteins (Taylor et al. 2007) but the content is significantly different, addressing the specific requirements of glycan analytics. Future work will encompass diverse sample preparation techniques and alternative techniques for the identification of glycan structures, such as LC, HPLC, CE or NMR.
The interaction analysis subgroup considers methods to define the biological interactions of glycans and glycoconjugates with other macromolecules, such as glycan-binding proteins, glycan-potentiated signal receptors or microbes such as bacteria and viruses. Initial efforts have focused on evaluating and describing glycan microarray analyses and the data generated by these experiments. Future work will encompass diverse technologies such as surface plasmon resonance, flow cytometry, enzyme-linked immuno-sorbent assays, saturation-transfer difference NMR and isothermal titration calorimetry.
The bioinformatics subgroup is charged with integrating data processing parameters into the guidelines for each laboratory method and defining separate guidelines for exchanging the information defined in the guidelines between different software systems and databases.
The second component of MIRAGE is the co-ordination group, which is concerned with the general organization of meeting, community participation and dissemination of the documents to both the members of MIRAGE and the broader scientific community. The third component of MIRAGE is the advisory board, which is composed of internationally recognized glycoscientists who support MIRAGE and have agreed to devote the time required to oversee the efforts of the first two groups.
Process of guideline development
The aim of reporting guidelines is to support the scientific community in publishing experimental data of high quality with respect to integrity, comprehensiveness and reproducibility by providing a framework to recommend details that should be reported along with the data. It is critical for the success of these guidelines that they do not appear to dictate how experiments and analysis should be designed or implemented. In order to gain general acceptance, MIRAGE thus shares several essential prerequisites with other guideline initiatives such as MIAPE (Taylor et al. 2007). These are:
Sufficiency: the guidelines should adequately describe information about the experimental data and the experimental conditions and methods used to generate the data to enable individuals to understand, critically evaluate, interpret and reproduce the data.
Practicability: the guidelines should be concise, understandable and limited to specific parameters that have a significant effect on the outcome of an experiment, facilitating compliance by scientists who use them.
Stability: the guidelines must be stable over a time period that is adequate to ensure consistency and comparability in data reporting. Nevertheless, the guidelines must accommodate technical and scientific advances that should be considered when a new technique is sufficiently mature and robust for widespread use.
Scientific journals will play a crucial role in encouraging acceptance of the guidelines by recommending that authors refer to the guidelines when submitting their data. In many cases, the status of the guidelines may change from a simple recommendation to a requirement that must be considered by all participants in the publication process (including authors, reviewers, curators and editors).
The development of a MIRAGE guideline is a multistep process. Maturation of the document through this process increases its visibility and encourages consensus among stakeholders, facilitating its acceptance by the community. Although this multistep development is time consuming and laborious, it has been proven to be an effective process for developing the MS guidelines (Kolarich et al. 2013), which have gained broad support from the MS and scientific publishing communities.
Transparency as a premise for a successful guideline proposition
The successful development and administration of any reporting standard involves balancing specifications that are sufficient to ensure the integrity and reproducibility of the reported data with the need for flexibility in designing and implementing experiments and the free dissemination of valuable experimental results (Klipp et al. 2007). Any restriction of scientific freedom regarding the generation or description of data should be avoided. Although scientific advances become valuable to the community only when new techniques and the data they generate are effectively communicated to others, innovation required for such advances must not be stifled by standards aimed at improving the information content of scientific reports.
With these human aspects in mind, it is clear that development of the MIRAGE guidelines should be a highly transparent process that respects the viewpoints of the many different stakeholders and thereby encourages maximum participation and continued support by the community. The parallel development of similar guidelines by different groups would lead to confusion and must be avoided. Therefore, once a new MIRAGE guideline has been reviewed by the advisory committee, it is made available to the general public via the MIRAGE project web site (http://glycomics.ccrc.uga.edu/MIRAGE/index.php). This site also provides information about the mission and history of the MIRAGE group, along with descriptions of the diverse projects and their progress. In addition, several hundred international researchers working on glycan analysis, structure and function are proactively approached via email for advice, for example, to obtain feedback on draft versions of the guidelines. The MIRAGE committee recognizes the need for broadly based input, and welcomes all comments, advice and help in developing the guidelines.
Conclusion
The MIRAGE project has been established to define data reporting guidelines for glycomics databases and publications. They describe the essential information necessary to understand and reproduce a glycomics experiment without dictating how the experiment itself should be performed. Adoption of these guidelines by scientific journals in the form of checklists or additions to the author instructions will increase the quality and reproducibility of the published results. In addition, adoption of the guidelines by glycomics databases will facilitate methods for processing and mining of the data produced by glycoanalytic experiments.
At the moment of writing this paper a guideline for glycomics MS experiments has been released and published (Kolarich et al. 2013). Further guidelines for other techniques used for glycomics experiments, such as glycan array experiments or LC separation in combination with or without MS, are in currently in preparation and will be finished in summer 2014.
Future activities of the MIRAGE project will be not just to create guidelines for other types of experiments but also to promote existing guidelines with scientific journals to accomplish the integration of these guidelines by the journals.
Funding
The Beilstein-Institut, a non-profit foundation established under civil law and located in Frankfurt am Main, Germany, has provided funding for advancement of the MIRAGE initiative. This work has also been supported by several other funding agencies: DK and ER are both supported by the Max Planck Society, and in addition, DK is funded by the Marie Curie grant PCIG09-GA-2011-293847. Both, ER and PMR have a grant from the European Union's Seventh Framework Programme (FP7-Health-F5-2011, grant agreement no 278535 “HighGlycan”); in addition, PMR has a grant from the European Union's Seventh Framework Programme (FP7/2007-2013, grant no. 259896); AD and SH are supported by the Biotechnology and Biological Sciences Research Council (grants BB/K016164/1 and BB/I017011/1); MC and NP are supported by the Australian Government project NeCTAR financed by the Education Investment Fund; JZ and CC are supported by the NIH/NIGMS grant P41GM104603; WY, RR, MT and LW are supported as part of the NIH/NIGMS funded National Center for Glycomics and Glycoproteomics (8P41GM103490). NK's contribution to this initiative is supported by The Swedish Foundation for International Cooperation in Research and Higher Education. TF and YL are supported by a Wellcome Trust Biomedical Resource grant WT099197MA. Funding to pay the Open Access publication charges for this article was provided by Beilstein-Institut.
References
- Klipp E, Liebermeister W, Helbig A, Kowald A, Schaber J. Nat. Biotech. 2007;25:390. doi: 10.1038/nbt0407-390. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, et al. The minimum information required for a glycomics experiment (MIRAGE) project – Improving the standards for reporting mass spectrometry-based glycoanalytic data. Mol. Cell Proteomics. 2013;12:991–995. doi: 10.1074/mcp.O112.026492. doi:10.1074/mcp.O112.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: Bioinformatics and biomarkers in disease. An NIH White Paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11–13, 2006) Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: The MIBBI project. Nat Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, et al. The minimum information about a proteomics experiment (MIAPE) Nat. Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Walt DA, Aoki-Kinoshita AF, Bendiak B, Bertozzi CR, Boons GJ, Darvill A, Hart G, Kiessling LL, Lowe J, Moon RJ, et al. Transforming glycoscience: A roadmap for the future. Washington, DC: The National Academies Press; 2012. http://www.ncbi.nlm.nih.gov/books/NBK109958/ [PubMed] [Google Scholar]
- Wells L, Hart GW. Glycomics: Building upon proteomics to advance glycosciences. Mol Cell Proteomics. 2013;12:833–835. doi: 10.1074/mcp.E113.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workshop on Analytic and Bioinformatic Glycomics, Bethesda, MA, USA, April 16–18, 2009 http://glycomics.scripps.edu/SubgroupWorkshop/WorkshopApril2009Rpt.pdf .