Abstract
Social structure is proposed to influence the transmission of both directly and environmentally transmitted infectious agents. However in natural populations, many other factors also influence transmission, including variation in individual susceptibility and aspects of the environment that promote or inhibit exposure to infection. We used a population genetic approach to investigate the effects of social structure, environment, and host traits on the transmission of Escherichia coli infecting two populations of wild elephants: one in Amboseli National Park and another in Samburu National Reserve, Kenya. If E. coli transmission is strongly influenced by elephant social structure, E. coli infecting elephants from the same social group should be genetically more similar than E. coli sampled from members of different social groups. However, we found no support for this prediction. Instead, E. coli was panmictic across social groups, and transmission patterns were largely dominated by habitat and host traits. For instance, habitat overlap between elephant social groups predicted E. coli genetic similarity, but only in the relatively drier habitat of Samburu, and not in Amboseli, where the habitat contains large, permanent swamps. In terms of host traits, adult males were infected with more diverse haplotypes, and males were slightly more likely to harbor strains with higher pathogenic potential, as compared to adult females. In addition, elephants from similar birth cohorts were infected with genetically more similar E. coli than elephants more disparate in age. This age-structured transmission may be driven by temporal shifts in genetic structure of E. coli in the environment and the effects of age on bacterial colonization. Together, our results support the idea that, in elephants, social structure often will not exhibit strong effects on the transmission of generalist, fecal-oral transmitted bacteria. We discuss our results in the context of social, environmental, and host-related factors that influence transmission patterns.
Introduction
Social structure is thought to play a profound role in the transmission of both directly and environmentally transmitted infectious agents [1], [2], [3], [4]. This is because social structure–including patterns of affiliation, mating behavior, dispersal, and territoriality–determines contact and habitat use among members of a population. There is strong evidence that social structure can influence the transmission of directly transmitted organisms [5]–[7], but social structure can also be important for environmentally transmitted agents. For instance, for fecal-oral transmitted organisms, members of the same social unit tend to use the same foraging areas or water sources; hence, hosts might be most often exposed to fecal contamination from members of their own social group [8], [9], [10], [11]. Understanding when and how social structure influences transmission is important because it has implications for population management and the evolutionary costs and benefits of social behavior [3], [4], [5], [12], [13].
To date, most support for socially structured transmission of infectious agents comes from theoretical models [3], [12], [13], [14]. While theory predicts important effects of sociality on transmission, empirical evidence remains relatively scarce (but see [9], [11], [15]). Hence, for most host species and infectious agents, it is unclear how important social structure is compared to other factors that also influence transmission patterns. Here we consider four such factors that may obscure or enhance the signature of host social structure on transmission [4], [16], [17]. First, host specificity and the transmission mode of the infectious agent should determine how strongly its transmission is influenced by the social structure of a single host species. Infectious agents that are specific to a single host species and are transmitted by physical contact among hosts are more likely to reflect host social structure than generalist agents transmitted via environments or vectors. Second, host traits such as age or sex may influence differences in susceptibility and hence transmission. For instance, immune responses can change with age; hence, some age classes may dominate transmission more than others [18], [19]. In terms of host sex, males are less immunocompetent than females in many vertebrates [20], [21], [22], [23], [24], and sex-specific differences in behavior may also lead to sex-specific patterns of transmission [21], [22], [23]. Third, competitive and facilitative ecological interactions among parasites or infectious agents may influence infection and the establishment of different parasites within hosts [25], [26]. Among humans for example, commensal gut bacteria that colonize early may establish themselves as the dominant bacterial community, which may influence the establishment of late colonizers [25], [27]. Fourth, for environmentally transmitted agents, aspects of the environment may enhance or obscure socially structured transmission. For instance, habitats that are suitable for bacterial proliferation (e.g. moist, warm places) may increase the length of time that bacteria can survive in the environment and enhance the opportunities for transmission. Hence, socially structured transmission may be dominated by interactions within a few suitable transmission hotspots in the habitat [28], [29].
In this study, we used population genetic information on Escherichia coli to test the influence of social structure, environment, and host traits on patterns of bacterial transmission in wild African savannah elephants, Loxodonta africana (Blumenbach, 1870). African elephants live in fission-fusion societies where individuals belong to predictable social groups, but the strength of associations between individuals and groups can vary depending on ecological conditions [30], [31]. The basic social unit, called a “core” or “family” group, can include up to fifty adult females and their immature offspring [31], [32], [33]. At maturity, males disperse from their natal family and range widely across the population, never permanently joining another family group [34], [35]. Over the course of hours, days, or weeks, families may fission into subgroups, or they might fuse together with members of other families. Members of the same family typically spend 60% to 90% of their time together in the same group, while members of different families typically spend much less time together, ranging from zero to 40% [30], [32]. Elephants are not territorial, but families occupy predictable home ranges that overlap with a fraction of the other families in the population [36]. Thus, there are several behaviorally mediated traits that potentially influence transmission in this species.
E. coli is an enteric commensal and an occasional pathogen in many vertebrates. It is transmitted between hosts through both direct physical contact and the ingestion of fecal-contaminated food and water [37], [38]. E. coli is a common model organism to study patterns of bacterial transmission in wildlife because it is easy to sample individual E. coli isolates from fecal samples [10], [39], [40]. Furthermore, these isolates can be genotyped using multi-locus markers developed for strain typing and population genetics [41]. In addition, E. coli reproduce clonally, and recombination–i.e., the exchange of genes or gene segments with other bacteria–is usually too low to obscure its clonal structure and transmission patterns [42], [43]. With respect elephant management, disease dynamics in elephants is understudied and understanding the transmission of commensal E. coli may lend insight into the spread of other, more harmful fecal-oral transmitted microbes, such as Salmonella sp. [44], [45].
Prior studies have found that host social structure can be correlated with E. coli transmission patterns [9], [10], [46], [47]. For instance in giraffe, hosts that were more closely linked in a social network were more likely to be infected with similar sub-types of E. coli than more distantly connected hosts [9]. In humans, members of the same household are more likely to be infected with similar stains of E. coli than members of different households [47]. Similarly in wild gorillas, members of the same social group harbor more similar E. coli than members of different groups [10]. However, several variables may obscure the signal of socially structured transmission [48]. For instance, E. coli infects many vertebrate species; hence, its population structure may often not be heavily influenced by the social structure of a single host species [49], [50]. In addition, E. coli can persist for several days to several years in water, sediment, and soil [51], [52], and habitats that are suitable for bacterial proliferation may increase opportunities for recombination [53] and transmission. Furthermore, E. coli is a member of the gut microbiome, and host age and ecological interactions among bacteria may influence which isolates establish in a given host [54], [55].
Our main objective was to test whether host social structure plays a detectable role in shaping patterns of E. coli transmission among wild elephants. We examined the role of social structure in the context of several other factors that may also influence E. coli transmission, including aspects of the habitat, host sex, and host age. We inferred patterns of transmission using the population genetic structure of E. coli isolated from elephants living in two populations: the Amboseli ecosystem in southern Kenya, and the Samburu-Laikipia ecosystem in central northern Kenya. Although both populations occupy areas with a similar climate, they differ in that the Amboseli population has a permanent swamp in their core habitat, while the Samburu population does not. We addressed two main questions: 1) Do host social structure and patterns of habitat use predict the population genetic structure of E. coli? And 2) what other aspects of individual hosts or their environments are important in shaping the population genetic structure of E. coli? If social structure influences E. coli transmission, we predicted that: a) E. coli infecting elephants from the same family should be genetically more similar than E. coli infecting members of different families, and b) the degree of range overlap would predict the degree of E. coli genetic similarity between families. In terms of host traits, we tested whether male elephants, which have larger ranges than females and possible differences in immune function, harbored genetically more diverse or potentially more pathogenic E. coli than female elephants. Finally, we explored age-related effects on patterns of E. coli infection, and specifically tested the prediction that elephants more similar in age are infected with genetically more similar E. coli, compared to elephants more different in age.
Methods and Materials
Ethics Statement
All protocols were noninvasive and adhered to the guidelines approved by the Institutional Animal Care & Use Committee (IACUC) of the University of Notre Dame. In Kenya, permission to conduct research was granted by the Kenyan government through research permit number NCST RRI/12/1/MAS/118/4.
Study Area and Host Populations
Study subjects were wild elephants living in the Amboseli ecosystem (8,000 km2), in southern Kenya, and the Samburu-Laikipia ecosystem (37,360 km2), which lies about 390 km to the north of Amboseli, in central northern Kenya. Elephant migration between these parks is probably currently rare, given the level of urban development between these locations. Both populations have been subject of long-term research; elephants in Amboseli have been monitored continuously for over 40 years, while elephants in Samburu have been monitored continuously for over 15 years [56], [57]. In both populations, individuals can be reliably identified, and for most animals born since the onset of monitoring, age is known ±2 weeks. For animals older than the onset of monitoring (wild elephants can live 65 years or more), age is estimated using well-developed morphological metrics, including shoulder height, hind footprint lengths and body shape [58], [59]. Ages were considered accurate within a few months to a few years, depending on the age class of the animal [60], [61].
There are several environmental similarities between the two habitats. In both Amboseli and Samburu, the elephants use the protected areas as core areas while making regular forays outside these protected areas to forage [62], [63], [64]. The core range of the Amboseli elephant population (∼1400 elephants) is Amboseli National Park, which covers 390 km2 between 1° 37′–3°13′ S and 35° 49′–38° 00′ E [56]. The core range of the Samburu elephant population (∼900 elephants) is the Samburu and Buffalo Springs National Reserves, which covers 220 km2 between 00° 30′–00° 80′ N, and 37°–38°E [65]. Both habitats are classified as semi-arid savannah with mixed open and bushy grassland and sparse woodland. Both habitats also contain other species of vertebrate hosts that are likely to be infected with E. coli and that may influence transmission patterns. Both habitats have similar patterns of seasonality and rainfall; rainfall occurs between November and May, and average annual rainfall is 340 mm in Amboseli and 360 mm in Samburu [66], [67].
While there are several ecological similarities between Amboseli and Samburu, there is one difference that may have implications for E. coli transmission: Amboseli has perennial swamps that cover about 12% of the National Park, whereas the Samburu has a free-flowing river. The presence of extensive, permanent, water-fed swamps in Amboseli may provide a conducive environment for the proliferation and transmission of E.coli [53], [68].
Defining Family Groups and Measuring Range Overlap between Groups in Protected Areas
To understand how social structure influenced the transmission of E. coli, we measured the genetic structure of E. coli isolates sampled from elephants living in 10 families in Amboseli and 5 families in Samburu (Table S1). Families were defined as a collection of adult females and their juvenile offspring that exhibited consistent associations, coordinated activities, and high rates of affiliative behaviors exchanged exclusively among family members [32], [33], [69]. In Amboseli, the families in our study contained between 7 and 31 adult females and their juvenile offspring, with an average size of ∼21 individuals. In Samburu, families ranged in size from 10 to 21 animals, with an average size of ∼14 individuals.
To test whether host occupancy patterns influenced the transmission of E. coli between family groups, the range (km2) of each group within the protected areas and the percent of spatial overlap between ranges in the protected areas were estimated using GPS locations of sightings of family members collected over a five-year period prior to E. coli sampling (March 2005 to March 2010 in Amboseli and January 2006 to December 2011 in Samburu). We chose a five-year span to maximize the number of sightings for each family (Table S1). Elephant GPS locations were collected while driving on roads within either Amboseli National Park or the Samburu and Buffalo Springs National Reserves. When an elephant group was sighted, we left the road and approached the group to record a GPS point representing the group’s location. These range estimates under-represent the total area used by the elephants because they lack data on the elephants’ ranges outside the protected areas. However, in both Amboseli and Samburu, the protected areas contain reliable food and water and are relatively safe from human threats; hence they comprise a major proportion of each family’s range, especially in the dry season [62], [63], [64]. Moreover, in both populations, the elephants often follow a diurnal pattern whereby they range within the park during the day and radiate beyond the park’s borders at night, with lower range overlap between families outside than inside the park [62], [63], [64]. As a result, the elephant ranging patterns we measured represent the areas of highest elephant density, greatest spatial overlap between elephant families, and perhaps the highest potential for bacterial transmission [62], [64]; however we cannot exclude the possibility that ranging patterns outside the protected areas also influenced the transmission patterns we observed.
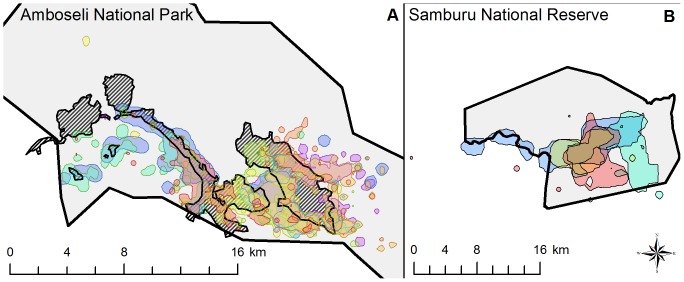
From these GPS data, we used a kernel density estimator in BIOTAS software (Version 2.0a.3.8) to calculate a “protected area range” for each family from 95% of GPS locations by eliminating 5% of the locations that were spatial outliers (Figure 1). To minimize the influence of large differences in range size between families, we calculated percent overlap in protected areas as the ratio of area shared by two families divided by the geometric mean of the two ranges. Compared to the arithmetic mean, the geometric mean minimizes bias in percent overlap caused by large differences in a pair of ranges [70], [71]. In Amboseli, we also estimated the area (km2) of each family’s range in swamp habitat, and the percent overlap among pairs of families in swamp habitat (Samburu lacked swamp habitat).
Figure 1. Ranges of elephant families within (A) Amboseli NP and (B) Samburu NR.
The outline of each protected area is depicted by a thick black line, and the protected area is shaded in light grey. The ranging patterns for each family are shown by different colors, and areas of overlap are shown as a blend of colors of the different overlapping family groups. Swamps in Amboseli are represented by a black striped pattern.
Escherichia coli Collection, DNA Extraction, and Genotyping
E. coli was cultured from fecal samples collected from known elephants during intensive 1-month sampling efforts during July 2010 in Amboseli and June 2011 in Samburu (Table S1). When members of a study group were located, we approached them in a vehicle and waited to collect fecal samples. As soon as a sample was produced by a known individual, we drove close to the sample, donned sterile gloves, and used sterile tongue depressors to collect pieces from the outside of the fecal bolus into sterile tubes (we sampled from the outside of the bolus to maximize the region in contact with the intestinal wall, where we suspected E. coli might be most likely to be found). Samples were typically collected within 10 minutes of defecation, and we avoided all sections of the fecal sample that may have made contact with the ground. Samples were kept at ambient temperature for 1 to 4 hours until we cultured and collected individual E. coli isolates from each sample. Specifically, we lit a candle in the workspace to create an up-draft. We then lightly touched the surface of the fecal sample with a flame-sterilized metal loop to collect a small amount of feces (an amount barely visible to the naked eye), and streaked the loop onto individual MacConkey agar plates (MacConkey agar is selective for gram negative bacteria and indicates the presence of lactose fermentation). Samples were incubated at 37°C for 12–24 h to allow colonies to grow. We collected one fecal sample per individual, and for each fecal sample, we used sterile toothpicks to collect 4 to 10 putative E. coli colonies (i.e. colonies matching the expected colony phenotype for E. coli on MacConkey agar: round, uniform, bright pink colonies). After collection, each putative E. coli colony was transferred to its own tube, containing 95% ethanol. We refer to these preserved colonies as “isolates”. Each putative E. coli isolate was stored at room temperature for up to four weeks prior to genetic analysis.
We genotyped each putative E. coli isolate using seven multi-locus sequence typing (MLST) loci [41] consisting of the following genes: adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase) icd (isocitrate dehydrogenase), mdh (malate dehydrogenase), uidA (beta-glucuronidase) and ClpX (caseinolytic protease × homolog) [72], [73]. The combined length of these sequences was ∼3700 base pairs. We choose these genes because they have sufficient variation for discriminating bacterial isolates and have been used to track and monitor population genetics and epidemiology of a number of bacterial pathogens, including pathogenic E. coli. We extracted DNA from 2 to 4 individual E. coli isolates per Amboseli elephant and 4 E. coli isolates per Samburu elephant. DNA was extracted using DNAeasy (Qiagen tissue kit) following the manufacturer’s instructions. We amplified each MLST marker using the polymerase chain reaction (PCR) conditions described in [72], [73]. Positive PCR reactions were sequenced in the reverse and forward directions using Dye Terminator Cycle Sequencing (Applied Biosystems). Sequences were inspected and cleaned using Sequencher software (version 4.9). A small fraction of sequences with multiple ambiguous nucleotides were discarded from further analyses. Clean sequences were subjected to NCBI BLAST to confirm that the isolates were E. coli (>95% of isolates were assigned to E. coli; others were excluded from further analyses).
Population Genetic and Statistical Analyses
We concatenated the sequences from each MLST gene for each E. coli isolate. An E. coli isolate with a distinct concatenated sequence is henceforth referred to as a “haplotype”. Prior research has shown that E. coli haplotypes can belong to one of four major phylogenetic groups, referred to as A, B1, B2, and D [74]. Phylogroups A, B2, and D, are common in humans, while phylogroup B1 is common in animals and abiotic environments [43], [68], [75]. Members of phylogroups B2 and D are most often commensal, but are more likely to be carriers of extra-intestinal virulence factors than are phylogroups A or B1 [76], [77]. We assigned our haplotypes to phylogroups by comparing our haplotypes with an E. coli Reference Collection (ECOR) consisting of 72 sequences with known phylogroup membership [78]. We did this by constructing a neighbor-joining tree including our isolates and the 72 ECOR haplotypes in MEGA software (MEGA5.2.1).
We examined basic patterns of genetic diversity, demographic history, and recombination rates since these forces might influence the population structure of E. coli and hence our ability to detect transmission patterns. We calculated nucleotide diversities and Tajima’s D in Arlequin (Version 3.5.1.3) [79]. We estimated mutation rates, recombination rates, and population history using Bayesian inference implemented in ClonalFrame (Version 1.1) [80]. We calculated two measures of the recombination rate: ρ/θ, which measures the relative frequency of occurrence of recombination and mutation in the population, and R/M, which measures the relative contribution of recombination and mutation to nucleotide substitution. Four independent runs of ClonalFrame were performed each consisting of 400,000 MCMC iterations, with the first half discarded as burn-in. Convergence and mixing of the MCMC for each simulation run was found to be satisfactory after visual inspection and after conducting the Gelman-Rubins statistics for all model parameters for convergence between any pair of runs.
To test the influence of host social structure on E. coli population genetic structure, we used analysis of molecular variance (AMOVA) in Arlequin to partition genetic variance in E. coli sampled from adult female and juvenile elephants within and between host populations, family groups and individual hosts. Measuring the population genetic structure of bacteria can be challenging because the clonal structure may vary depending on the rate of recombination. Hence, we performed AMOVA and calculated FST using two different measures of genetic distance, each with their own assumptions. First, to account for evolutionary distance between haplotypes caused by mutations and recombination events, we calculated genetic distance based on the ClonalFrame analysis. Specifically, we measured “patristic distance” or the total branch lengths between each pair of isolates calculated from a 50% consensus evolutionary tree, assuming uniform nucleotide substitution and recombination using a Bayesian analysis in ClonalFrame. This approach to inferring transmission assumes that genetically more similar haplotypes share more recent common ancestors and more recent transmission events. Second, we measured “haplotype distance” by performing AMOVA using classic haplotype frequency-based measures of genetic variance, which ignore phylogenetic distance between haplotypes. This measure of genetic distance, which is based on the proportion of shared haplotypes, may be better at capturing patterns of transmission when evolutionary rates are high.
We next tested whether range overlap in protected areas influenced E. coli genetic similarity between family groups. As in the AMOVA’s above, E. coli isolates sampled from adult males were excluded from these analyses. Mantel tests were used to assess the correlation between matrices of the percent of range overlap and matrices of genetic differentiation (FST) based on patristic distance and haplotype distance between E. coli sampled from different family groups. In Amboseli we also conducted a second analysis using percent range overlap in swamps alone.
We next performed three analyses to understand the associations between host traits (sex and age) and E. coli genetic and phylogroup structure. First, using samples from Samburu only, we tested whether independent adult males (N = 7; males who were no longer regularly associated with a single family) were infected with more diverse E. coli than adult females. Specifically, we performed a regression analysis with host sex and age as predictor variables and average nucleotide diversity among E. coli isolates from within a host as the dependent variable. When a covariance analysis showed that age was not an important predictor of E. coli nucleotide diversity within individuals, but sex was, we carried out an independent sample t-test between genetic distances of E. coli isolates from adult male and adult female hosts.
Second, we used generalized linear mixed modeling (GLMM) to test whether host age or sex influenced the tendency for individuals to be infected with each phylogroup. We expected males to be more often infected with phylogroups associated with greater pathogenic potential (phylogroups B2 and D). We constructed a Poisson GLMM for each E. coli phylogroup. To test whether the proportion of isolates belonging to a given phylogroup changed as a function of host age and sex, we used the number of E. coli isolates belonging to a phylogroup from each individual elephant as the response variable and the total number of E. coli isolates obtained from that individual host as an offset variable. The use of the offset variable allowed us to model counts of each phylogroup as proportions. We used host sex and age as fixed factors, and host population as a random factor. The effect of age and sex on the proportion of isolates for some phylogroups did not vary by host population (e.g. A and B1). For these phylogroups, we instead ran a generalized linear model (GLM) that retained all the fixed factors but without including population as a random factor.
Third, we tested the prediction that same-aged elephants are infected with genetically more similar strains of E. coli, as compared to animals more disparate in age. Specifically, we used Mantel tests to correlate pairwise matrices of difference in age with matrices of genetic distance between E. coli isolates sampled from that pair of elephants.
Results
Basic Patterns of Genetic Diversity, Phylogroup Membership, and Sequence Evolution
We assessed the population genetic structure of E. coli using 210 E. coli isolates from 85 adult female and juvenile Amboseli elephants, and 143 E. coli isolates from 36 Samburu elephants (7 males and 29 adult females and juveniles; Table S1; NCBI GenBank Accession numbers KJ078651- KJ081101). We observed 140 total haplotypes; 41 of these haplotypes (29%) occurred in multiple hosts. In Amboseli, we found 93 distinct haplotypes in 85 elephants; in Samburu we found 60 haplotypes in 36 elephants. Two-thirds of E. coli isolates were assigned to phylogroup B1, while the remaining third was divided among phylogroups A, B2, D, and a small unassigned class (Table S2). Nucleotide diversities were similar in Amboseli and Samburu, with an average percent of nucleotide differentiation around 1.2% (Table 1). ClonalFrame analyses indicated that recombination was the dominant evolutionary force in E. coli, and the relative contribution of recombination was about 3 times the contribution of mutation (Table S3). In support for the idea that aquatic environments promote recombination, the relative contribution of recombination versus mutation was about 1.5 times higher in Amboseli as compared to Samburu (Table S3).
Table 1. Basic genetic diversity statistics for E. coli sampled from elephants in Amboseli and Samburu.
| Parameters | Amboseli | Samburu | Combined |
| Number of isolates | 210 | 143 | 353 |
| Number of segregating sites | 277 | 330 | 388 |
| Number of haplotypes | 93 | 60 | 140 |
| Mean population Nucleotide diversity ± SD | 0.012±0.006 | 0.013±0.006 | 0.012±0.006 |
| Mean Nucleotide diversity within individuals ± SD | 0.009±0.007 | 0.007±0.005 | 0.008±0.007 |
| Tajima’s D (p-value) | −0.574 (0.331) | −0.686 (0.287) | −0.795 (0.228) |
Neither Host Population nor Social Group were Significant Barriers to E. coli Transmission
Before exploring the effects of elephant social structure on E. coli genetic structure, we first investigated how genetic variation in E. coli was partitioned within and between host populations and individual hosts (Table 2). We performed AMOVA on two measures of genetic distance: “patristic distance” from ClonalFrame, which estimates phylogenetic distance between haplotypes controlling for recombination, and “haplotype distance”, which ignores phylogenetic distances between haplotypes (see methods). As expected for E. coli [49], [50], we found little genetic differentiation in E. coli sampled from these two populations, despite being separated by nearly 400 km (Table 2). For haplotype distance, around 1% of genetic variance was explained by host population, indicating that there are small but significant differences in the frequencies of some E. coli haplotypes in Amboseli versus Samburu (Table 2). For instance, families of elephants from the same population (i.e. either Amboseli or Samburu) tended to share a greater proportion of haplotypes compared to families from different populations (average proportion of haplotypes shared between families in the same population ± SE = 7.6% ±0.6%; average proportion of haplotypes shared between families in different populations ± SE = 3.8% ±0.6%). However, the great majority of genetic variance in E. coli (99%) was found within and between individual elephants living in the same population. Indeed, individual hosts tended to contain diverse haplotypes; for instance, within-host nucleotide diversities were around 1%, and the average number of pairwise nucleotide differences between isolates sampled from the same host was 30.8±25.1 in Amboseli and 24.4±18.8 in Samburu (Table 1).
Table 2. Results of an AMOVA depicting the contribution of host populations and host individuals to the partitioning of genetic variation in E. coli isolates.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage variation | FST | P |
| Patristic distance | ||||||
| Among host populations | 1 | 0.753 | 0.002 | 0.68 | 0.007 | 0.094 |
| Among individual hosts within host populations | 119 | 49.492 | 0.095 | 40.46 | 0.407 | 0.000 |
| Within individual hosts | 232 | 32.181 | 0.139 | 58.86 | 0.411 | 0.000 |
| Haplotype distance | ||||||
| Among host populations | 1 | 1.608 | 0.004 | 0.89 | 0.009 | 0.000 |
| Among individual hosts within host populations | 119 | 90.265 | 0.138 | 27.57 | 0.278 | 0.000 |
| Within individual hosts | 232 | 82.983 | 0.358 | 71.54 | 0.285 | 0.000 |
We next tested whether families represented significant barriers to E. coli gene flow. If elephants are more likely to be infected with E. coli from members of their own family, as compared to members of other families, then E. coli sampled from members of the same family should be genetically more similar than E. coli sampled from members of different families. However, we found no support for this prediction. Instead, family groups did not explain a significant fraction of the genetic variance in E. coli in either Amboseli or Samburu, regardless of the measure of genetic distance (Tables 3 and 4; P = 0.688 in Amboseli; P = 0.142 in Samburu). In support, individuals were just as likely to share haplotypes with members of different families as they were with members of their own family (average proportion of haplotypes shared between members of the same family ± SE = 8.3% ±1.4%; average proportion of haplotypes shared between members of different families, but from the same population ± SE = 7.6% ±0.7%).
Table 3. Results of an AMOVA depicting the contribution of elephant social groups (adult females and juveniles only) to the partitioning of genetic variation of the E. coli isolates from Amboseli elephants.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | FST | P |
| Patristic distance | ||||||
| Among family groups | 9 | 4.374 | −0.001 | −0.40 | −0.004 | 0.688 |
| Among individual hosts within family groups | 75 | 35.457 | 0.111 | 35.51 | 0.354 | <0.001 |
| Within individual hosts | 125 | 25.383 | 0.203 | 64.89 | 0.351 | <0.001 |
| Haplotype distance | ||||||
| Among family groups | 9 | 5.975 | 0.000 | −0.09 | −0.001 | 0.605 |
| Among individual hosts within family groups | 75 | 47.844 | 0.099 | 19.99 | 0.200 | <0.001 |
| Within individual hosts | 125 | 49.667 | 0.397 | 80.1 | 0.199 | <0.001 |
Table 4. Results of an AMOVA depicting the contribution of elephant social groups (adult females and juveniles only) to the partitioning of genetic variation of E. coli isolates from Samburu elephants.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | FST | P |
| Patristic distance | ||||||
| Among family groups | 4 | 4.251 | 0.009 | 2.57 | 0.026 | 0.142 |
| Among individual hosts within family groups | 24 | 20.565 | 0.175 | 51.11 | 0.525 | 0.000 |
| Within individual hosts | 87 | 13.781 | 0.158 | 46.33 | 0.537 | 0.000 |
| Haplotype distance | ||||||
| Among family groups | 4 | 4.782 | 0.006 | 1.12 | 0.011 | 0.116 |
| Among individual hosts within family groups | 24 | 25.539 | 0.192 | 38.84 | 0.393 | 0.000 |
| Within individual hosts | 87 | 25.817 | 0.297 | 60.04 | 0.400 | 0.000 |
Range Overlap between Family Groups in Protected Areas is a Mixed Predictor of E. coli Gene Flow
To test whether range overlap in protected areas was correlated with E. coli genetic similarity, we first examined the degree of range overlap between elephant families within the two reserves. Within protected areas, family ranges were larger and had higher percent overlap with each other in Samburu compared to Amboseli (Figure 1; Samburu: average ± SD size = 53.1±16.3 km2; average percent ± SD overlap = 50.2% ±10.1%; Amboseli: average ± SD size = 20.7±7.0 km2; average percent ± SD overlap = 22.6% ±18.7%). In Amboseli, elephant families had considerable overlap with swamp, which constituted, on average 70% of a family’s range within this protected area (Figure 1; Average area in swamp ± SD = 2.425±2.253 km2; range = 0.051 to 8.240 km2). In terms of range overlap between families, the mean ± SD percent overlap in swamp was 27.86% ±22.87% (range: 0.95% to 80.84%).
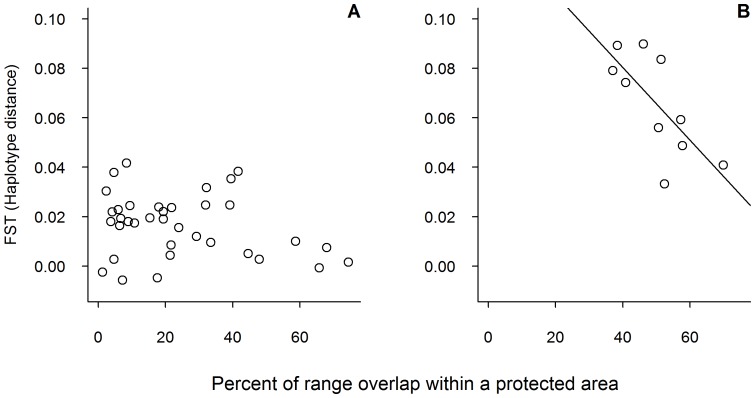
We expected that higher range overlap in protected areas would be associated with greater E. coli genetic similarity between families. We found mixed support for this prediction. In Amboseli, the percent of range overlap was not correlated with E. coli genetic similarity, whether we tested this relationship for complete ranges (Figure 2a; patristic FST: rs = 0.085, P = 0.612; haplotype FST: rs = −0.148, P = 0.387), or only for range overlap in swamp (patristic FST: rs = −0.048, P = 0.777; haplotype FST, rs = −0.264, P = 0.116). However, we did observe a significant relationship between range overlap between families and genetic similarity of E. coli in Samburu (Figure 2b; patristic FST: rs = −0.770, P = 0.013; and haplotype FST: rs = −0.697, P = 0.031).
Figure 2. E. coli genetic distance as a function of range overlap between elephant families.
(A) Depicts data from Amboseli and (B) depicts data from Samburu. The relationship between percent overlap and FST was statistically significant in Samburu, but not Amboseli. Plots are for visualization purposes only; statistical analyses were performed using Mantel tests (see text for details). Analyses include data from adult female and juvenile elephants only.
Adult Male Elephants are Infected with Genetically More Diverse and Potentially More Pathogenic E. coli than Adult Females
We predicted that adult male elephants might harbor greater genetic diversity of E. coli than adult females. In support, we found that adult males were infected with genetically more diverse strains than females as measured by nucleotide diversity (mean nucleotide diversity for females = 0.006±0.005, for males = 0.011±0.005; t = −2.743, df = 25 P = 0.011, Figure S1). We also predicted that males might be more likely to be infected with phylogroups B2 and D, which are more likely to carry virulence factors than other phylogroups. In support, we found that adult males harbored a significantly higher proportion of phylogroup D than adult females (Table 5). We also observed a non-significant trend for males to be infected with a higher proportion of phylogroup B2 (Table 5).
Table 5. Results of generalized linear models (model 1 & 2) and generalized linear mixed effects models (models 3, 4 & 5) showing the influence of host age and sex on the proportion of each phylogenetic group of E. coli in elephants.
| Model | Phylogroup | Covariate | Estimate | Standard error | z value | P |
| Model 1 | A | |||||
| Age | −0.025 | 0.016 | −1.530 | 0.126 | ||
| Sex | −0.005 | 0.405 | −0.012 | 0.990 | ||
| Model 2 | B1 | |||||
| Age | −0.003 | 0.005 | −0.607 | 0.544 | ||
| Sex | −0.225 | 0.164 | −1.374 | 0.169 | ||
| Model 3 | B2 | |||||
| Age | 0.027 | 0.019 | 1.428 | 0.153 | ||
| Sex | 0.926 | 0.554 | 1.671 | 0.095 | ||
| Model 4 | D | |||||
| Age | −0.036 | 0.023 | −1.598 | 0.110 | ||
| Sex | 1.069 | 0.451 | 2.368 | 0.018 | ||
| Model 5 | unclassified | |||||
| Age | 0.035 | 0.012 | 2.813 | 0.005 | ||
| Sex | −0.251 | 0.514 | −0.487 | 0.626 |
Elephants from Similar Age Cohorts were Infected with Genetically More Similar E. coli
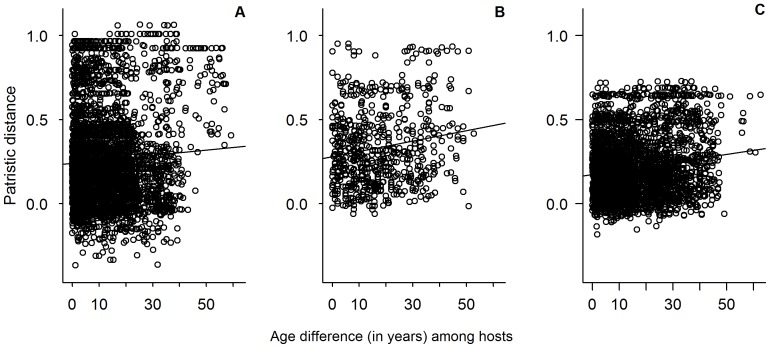
For many pathogens, host age plays an important role in transmission, and there is some evidence that age can influence patterns of E. coli infection in humans [55], [81], [82], [83]. We tested for age effects on patterns of E. coli infection by correlating a matrix of pairwise difference in age between pairs of elephants with a matrix of pairwise genetic distance between E. coli isolates infecting those elephants. We found some support for genetic structuring in E. coli populations based on host age. In both Amboseli and Samburu, difference in age explained a small but significant fraction of the genetic variance in E. coli (Figure 3; patristic distance in Amboseli: r = 0.062, P = 0.0003; patristic distance in Samburu: r = 0.170, P<0.0001; haplotype distance in Amboseli: r = 0.040, P = 0.019; haplotype distance in Samburu: r = 0.186, P<0.0001). Specifically, elephants closer in age were more likely to be infected with genetically more similar E. coli than elephants more different in age (Figure 3).
Figure 3. E. coli genetic distance as a function of age-difference among hosts.
(A) Depicts the relationship between age similarity and E. coli genetic similarity as measured by patristic distance for pairs of elephants from Amboseli; (B) depicts these relationships for pairs of elephants from Samburu; (C) depicts these relationships for pairs of elephants where one member was drawn from each population. Plots are for visualization purposes only; statistical analyses were performed using Mantel tests (see text for details).
We also examined the correlation between E. coli genetic distance and host age for E. coli isolated from pairs of elephants from the two different populations. Interestingly, the effect of age similarity cut across elephant populations (Figure 3c). Specifically, using only pairs of elephants from the two different populations, age similarity still predicted genetic similarity of E. coli (patristic distance: r = 0.160, P<0.0001; haplotype distance: r = 0.124, P<0.0001). One explanation for these results is that age might predict a host’s probability of infection with a given phlyogroup, perhaps through age-specific patterns of susceptibility [55], [81], [82], [83]. However, we found only limited support for this idea; age did not predicted the probability that elephants were infected with a given phylogroup, and phylogroups were evenly distributed across hosts of different ages, but we observed that the proportion of strains that did not cluster with a specific phylogroup increased with age across both elephant populations (Table 5; Figure S2).
Discussion
Social organization and behavior can influence the transmission of both directly [5]–[7] and environmentally transmitted infectious agents [1], [2], [4], [8], [9], [10], [11], [84]. However in natural populations, several other factors may also influence transmission, including aspects of the abiotic environment that promote or impede transmission, and host traits such as sex or age that influence individual exposure and the probability of infection [19], [22]. In this study, we used population genetic tools to explore the effects of social structure, environment, and host traits on the transmission of E. coli infecting wild elephants. We found little evidence for socially structured transmission; instead, the population genetic structure of E. coli was heavily influenced by processes occurring in the environment and at the level of individual hosts. Below we discuss our results, starting with the effects of habitat on patterns of E. coli transmission.
The Influence of Social-structure and Habitat on Transmission
Many researchers have hypothesized that exposure to infectious disease is an important evolutionary cost of group living [1], [85], [86]. This hypothesis assumes that directly and environmentally transmitted agents are more likely to spread among members of the same social group than between members of different groups. While there is considerable evidence for this assumption, even for environmentally transmitted infectious agents [8], [9], [10], [11], [84], in our study we found no evidence that elephants were more likely to be infected with E. coli from members of their own family group than members of other families. Instead, E. coli isolates sampled from members of the same family were not genetically more similar compared to E. coli from different families, and families did not represent major barriers to E. coli gene flow.
These results differ somewhat from prior studies on other social species, including giraffes, baboons, gorillas, and humans, which found that individuals with more frequent social contact were more likely to be infected with more similar strains of E. coli [9], [10], [46], [47], [87], [88], [89]. At least four factors may explain why our results differ from some prior studies. First, variation in social organization and behavior may explain why socially structured patterns of E. coli transmission are more common in some species than others. During rainy periods, elephants aggregate in large groups and have physical contact between members of different families, especially during play or mating, which may promote between-group transmission. In addition, elephants have high range overlap around water sources, which may serve as environmental reservoirs for E. coli and promote between-group transmission. By contrast, aspects of human behavior may make E. coli transmission within households much more common than between households, such as high rates of physical contact between family members, hand washing, cooking and eating at home, and the use of toilets and latrines.
Second, differences in study design may have led to higher rates of within- versus between-group contact rates in prior studies compared to our study. For instance, in the studies on baboons and gorillas, researchers chose social groups with non-overlapping home ranges, which should reduce the probability that individuals would encounter fecal contamination from non-group members [10], [89]. Third, different E. coli phylogroups vary in their prevalence across host species and abiotic environments, and the likelihood of socially structured transmission may depend on phylogroup identity. For instance, nearly 70% of isolates from the elephants in our study were attributed to phylogroup B1, a commensal form that is known to be environmentally ubiquitous, to infect a wide variety of animals, and to have high turnover within hosts [75], [90], [91]. These traits may make it less likely to detect the effects of host social structure on transmission. In contrast, most human studies focus on pathogenic or virulent strains. Such strains are more likely to be epidemic and clonal in nature [92], [93], [94], making it easier to detect patterns of socially structured transmission.
Fourth, differences in genotyping methods may also explain why our results differed from some prior studies. In particular, two prior studies that found effects of social structure on E. coli transmission used the BOX-PCR method of Cesaris et al. [95] to define genetically similar isolates of E. coli in giraffes. Studies on E. coli infecting gorillas and humans used a similar approach called repetitive-element PCR [10], [96]. These approaches rely on gel- or capillary-based banding patterns, and there are fewer statistical tools to analyze such data, compared to DNA sequences of MLST markers. However, BOX-PCR and repetitive-element PCR may provide genetic information across a greater proportion of the E. coli genome, which may lend more power to distinguish relationships among strains. Future studies may find it valuable to compare results from these two methods of E. coli genotyping.
While our results indicate that elephants were not more likely to be infected by E. coli from members of their own family than from members of other families, we observed some support for socially structured transmission via habitat overlap [10], [40], [97]. However, this evidence was equivocal because we only observed significant patterns in one of our study populations (i.e. Samburu). One possible explanation for this mixed result is that elephant families may have substantially different patterns of habitat overlap inside versus outside protected areas; hence our measure of habitat overlap may not have accurately captured patterns of environmental exposure. However, at the population level elephant families exhibit the highest level of habitat overlap within protected areas, so we think this explanation is unlikely [63].
Another possible explanation for these mixed results is that differences in habitat moisture and the degree of habitat overlap between elephant family groups favored socially structured transmission via range overlap in Samburu, but not in Amboseli. Specifically, in Samburu, the habitat was drier, and the elephant groups exhibited a greater degree of range overlap than in Amboseli. These two forces may act in concert to increase the strength of socially structured transmission; harsh habitat may reduce E. coli persistence times [52], [98], while higher range overlap will increase contact between groups, ultimately leading to relatively high rates of E. coli transmission between elephant groups.
One final observation on socially structured transmission via range overlap: while we observed support for this hypothesis in Samburu, it is as yet unclear whether the patterns of transmission we observed between elephant groups were driven by socially mediated factors (i.e., contact with fecal material from members of different elephant families) or were instead driven by the fact that these elephant groups used the same areas and so were exposed to the same environmental sources of E. coli (i.e., shared habitat with alternative host species that also transmit E. coli). Hence, despite possible support for socially structured transmission, these results are far from conclusive about the role of social behavior in the transmission of E. coli between elephant groups.
The Influence of Host Sex and Age on the Structure of E. coli Populations within Hosts
Most of the genetic variation in E. coli populations was structured within and between individual hosts, not family groups or host populations. This population structure–i.e., high gene flow between geographically distinct populations, but strong genetic differentiation between hosts–is typical of E. coli. For instance, researchers often find high levels of E. coli gene flow between host populations separated by large geographic distances [50]. Moreover, many studies have found that the majority of genetic variation in E. coli is structured within and between individual hosts [40], [89], [99].
For the elephants in Amboseli and Samburu, most individuals were infected with multiple, genetically diverse haplotypes, which often differed in identity and frequency from the haplotypes found infecting other members of their family or population. These patterns suggest that processes occurring at the level of individual hosts, as opposed to families or host populations, are most important in influencing the structure of E. coli populations. In particular, E. coli is a normal member of the gut microbiome, and the process of microbiome assembly–including bacterial colonization, interactions among bacterial species, and interactions with the host’s genome and immune system–may influence which E. coli haplotypes occur in a given host [25], [27], [100]. We identified two specific host traits that were associated with the structure of E. coli populations in individual hosts: host sex and age. With respect to sex, we found that adult males were infected with more genetically diverse E. coli and were more likely to be infected with strains from phylogroup D than were adult females. This result is based on a relatively small sample size, and it should be interpreted with caution. Moreover, phylogroup proportions are not independent of each other; however, we observed no other significant relationships between sex and phylogroup proportions, suggesting that sex is the primary predictor of phylogroup D. If true, one explanation for this result is that adult male elephants range more widely than adult females, which may expose males to more diverse E. coli strains. Indeed, similar effects have been described for other infectious agents [23]. In addition, differences in immune responses between males and females due to the hormonal challenges of male reproductive states may also underlie sex differences in E. coli infection [20], [21], [22]. That said, two prior studies of marsupials found no sex differences in E. coli infection patterns [55], [101] and it remains to be seen whether sex differences in E. coli infection are common across vertebrate species.
We also found effects of host age on E. coli populations. Specifically, elephants born around the same time were infected with genetically more similar E. coli than pairs of elephants further apart in age. This pattern occurred for pairs of elephants drawn from the same host population, as well as for pairs drawn from the two different populations (i.e., Amboseli and Samburu). One explanation for these effects is that they are caused by age-related changes in gut morphology or immune function leading to differences in the composition of phylogroups in younger versus older elephants. While this explanation is possible, it is not well supported by our data, as we found few changes in phylogroup composition as a function of age.
Instead, we think our results are more consistent with the idea that age-related patterns of E. coli infection reflect temporal structure in environmental E. coli. Specifically, individual hosts may be infected with E. coli when in the first few months of life, and while individuals are continually exposed to E. coli throughout life, most of these later strains may fail to establish [50]. In support, several studies have shown that temporal effects on genetic variation appear to be a dominant force in the population structure of E. coli within hosts and in the environment [55], [99], [102], [103]. For instance, when E. coli was sampled from several locations in a lake repeatedly for several days or years, there was strong genetic similarity among isolates collected on the same day or the same year across different locations separated by distances of 50 kilometers, as compared to E. coli collected on different days or years in the same location [102], [104]. Similar effects have been observed in E. coli populations infecting animal hosts [99], [101], [102], [103]. Our results appear to be novel with respect to the time scale as temporal effects on E. coli population structure have typically been explored over the scale of weeks or months, not decades. Given that elephant births are often seasonally clustered, and habitat and rainfall conditions can vary markedly between birth cohorts over their first years of life, these variations may affect transmission during the early acquisition of E. coli. That said, the time scale over which we observed temporal patterns on E. coli communities is surprising, given that B1 strains, which were the most common in our study, are among the most transient members of the gut. It is possible that B1 phylogroups behave differently in elephants as compared to other hosts. However, our results require further confirmation to see if they will be upheld. We encourage future researchers to test for similar temporal patterns within host populations.
Conclusions
Social structure is often proposed to be an important conduit for the transmission of directly and environmentally transmitted infectious agents. However, our results indicate that social structure plays, at most, a weak role in E. coli transmission in wild elephants. Instead, transmission patterns were dominated by host habitat and aspects of individual hosts, such as individual sex and age. Strong patterns of socially mediated transmission may be limited to infectious agents with high host specificity, transmitted only via physical contact between hosts, and to hosts with social structures that minimize habitat overlap and contact between different group members. Generalist infectious agents that can be transmitted through the environment, such as E. coli, are common in wildlife, and they may be some of the most important from an evolutionary and a management perspective. In the future, we encourage researchers to incorporate multiple aspects of hosts and their environment, as well as social contacts, to gain the greatest insight into transmission patterns.
Supporting Information
Nucleotide diversity in E. coli as a function of host sex. Boxplots depict nucleotide diversity in E. coli infecting individual elephants as a function of host sex. This analysis was performed on adult animals from Samburu NR only.
(TIF)
The distribution of E. coli phylogroups groups as a function of host age. Plot depicts a cross-sectional analysis of the proportion of each phylogroup type found infecting elephant hosts of different ages. Relationships demonstrate that unclassified E. coli isolates increased in older age groups.
(TIF)
Sample information, including the number of elephant hosts per family group, the number of E. coli isolates genotyped per family group, and the number of GPS sightings per family group.
(DOCX)
Percent of E. coli isolates assigned to different phylogroups in Amboseli and Samburu. Unassigned isolates did not cluster with any known ECOR sequences.
(DOCX)
Basic evolutionary parameter estimates for concatenated E. coli sequences.
(DOCX)
Acknowledgments
We thank the Office of the President of the Government of Kenya for permission to conduct research. We also thank Kenya Wildlife Services and the National Park staff for their assistance during the study. In Amboseli, we thank, N. Njiraini, K. Sayialel and S. Sayialel, who provided invaluable assistance in data collection. In Samburu, we thank the Samburu and Buffalo-Springs national reserves’ county councils, wardens, and rangers. We also thank C. Leadisimo, D. Daballen, and G. Sabinga and the rest of the Save the Elephants team. We further acknowledge support provided by the Clare Boothe Luce Foundation to EAA, as well as support provided by the Amboseli Trust for Elephants.
Funding Statement
This research was supported by funds from the Clare Boothe Luce Foundation to EAA, as well as the Amboseli Trust for Elephants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Freeland WJ (1979) Primate social groups as biological islands. Ecology 60: 719–728. [Google Scholar]
- 2.Anderson RM, May RM (1991) Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press. 757 p. [Google Scholar]
- 3. Nunn CL, Thrall PH, Leendertz FH, Boesch C (2011) The spread of fecally transmitted parasites in socially-structured populations. PLoS ONE 6: e21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, et al. (2003) Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu Rev Ecol Syst 34: 517–547. [Google Scholar]
- 5. Caillaud D, Levero F, Cristescu R, Gatti S, Dewas M, et al. (2006) Gorilla susceptibility to the Ebola virus: the cost of sociality. Curr Biol 16: R489–R491. [DOI] [PubMed] [Google Scholar]
- 6. Hamede RK, Bashford J, McCallum H, Jones M (2009) Contact networks in a wild Tasmanian devil (Sarcophilus harrisii) population: using social network analysis to reveal seasonal variability in social behaviour and its implications for transmission of devil facial tumour disease. Ecol Lett 12: 1147–1157. [DOI] [PubMed] [Google Scholar]
- 7. Corner LAL, Pfeiffer DU, Morris RS (2003) Social-network analysis of Mycobacterium bovis transmission among captive brushtail possums (Trichosurus vulpecula). Prev Vet Med 59: 147–167. [DOI] [PubMed] [Google Scholar]
- 8. Godfrey SS, Bull CM, James R, Murray K (2009) Network structure and parasite transmission in a group living lizard, the gidgee skink, Egernia stokesii . Behav Ecol Sociobiol 63: 1045–1056. [Google Scholar]
- 9. VanderWaal KL, Atwill ER, Isbell LA, McCowan B (2013) Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa camelopardalis). J Anim Ecol83: 406–414. [DOI] [PubMed] [Google Scholar]
- 10. Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL (2008) Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv Biol 22: 1600–1607. [DOI] [PubMed] [Google Scholar]
- 11. Bull CM, Godfrey SS, Gordon DM (2012) Social networks and the spread of Salmonella in a sleepy lizard population. Mol Ecol 21: 4386–4392. [DOI] [PubMed] [Google Scholar]
- 12. Eames KTD (2008) Modelling disease spread through random and regular contacts in clustered populations. Theor Popul Biol 73: 104–111. [DOI] [PubMed] [Google Scholar]
- 13. Keeling MJ (1999) The effects of local spatial structure on epidemiological invasions. P Roy Soc B 266: 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bansal S, Grenfell BT, Meyers LA (2007) When individual behaviour matters: homogeneous and network models in epidemiology. J Roy Soc Interface 4: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drewe JA, Eames KTD, Madden JR, Pearce GP (2011) Integrating contact network structure into tuberculosis epidemiology in meerkats in South Africa: Implications for control. Prev Vet Med 101: 113–120. [DOI] [PubMed] [Google Scholar]
- 16. Freeland WJ (1976) Pathogens and the evolution of primate sociality. Biotropica 8: 12–24. [Google Scholar]
- 17. Cote IM, Poulin R (1995) Parasitism and group-size in social animals: a meta analysis. Behav Ecol 6: 159–165. [Google Scholar]
- 18. Dean EA, Whipp SC, Moon HW (1989) Age-specific colonization of porcine intestinal epithelium by 987P-piliated enterotoxigenic Escherichia coli . Infect Immun 57: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steens A, Waaijenborg S, Teunis PFM, Reimerink JHJ, Meijer A, et al. (2011) Age-dependent patterns of infection and severity explaining the low impact of 2009 influenza A (H1N1): Evidence from serial serologic surveys in the Netherlands. Am J Epidemiol 174: 1307–1315. [DOI] [PubMed] [Google Scholar]
- 20. Folstad I, Karter AJ (1992) Parasites, bright males, and the immunocompetence handicap. Am Nat 139: 603–622. [Google Scholar]
- 21. Skorping A, Jensen KH (2004) Disease dynamics: all caused by males? Trends Ecol Evol 19: 219–220. [DOI] [PubMed] [Google Scholar]
- 22. Ferrari N, Cattadori IM, Nespereira J, Rizzoli A, Hudson PJ (2004) The role of host sex in parasite dynamics: field experiments on the yellow-necked mouse Apodemus flavicollis . Ecol Lett 7: 88–94. [Google Scholar]
- 23. Caillaud D, Prugnolle F, Durand P, Theron A, de Meeus T (2006) Host sex and parasite genetic diversity. Microbes Infect 8: 2477–2483. [DOI] [PubMed] [Google Scholar]
- 24. Nunn CL, Lindenfors P, Pursall ER, Rolff J (2009) On sexual dimorphism in immune function. Phil T Roy Soc B 364: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graham AL (2008) Ecological rules governing helminth-microparasite coinfection. P Natl Acad Sci USA 105: 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walter J, Ley R (2011) The human gut microbiome: ecology and recent evolutionary changes. Annul Rev Microbiol 65: 411–429. [DOI] [PubMed] [Google Scholar]
- 28. Judge J, Kyriazakis I, Greig A, Allcroft DJ, Hutchings MR (2005) Clustering of Mycobacterium avium subsp. paratuberculosis in rabbits and the environment: How hot is a hot spot? Appl Environ Microb 71: 6033–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bousema T, Drakeley C, Gesase S, Hashim R, Magesa S, et al. (2010) Identification of hot spots of malaria transmission for targeted malaria control. J Infect Dis 201: 1764–1774. [DOI] [PubMed] [Google Scholar]
- 30. Wittemyer G, Douglas-Hamilton I, Getz WM (2005) The socioecology of elephants: analysis of the processes creating multitiered social structures. Anim Behav 69: 1357–1371. [Google Scholar]
- 31.Moss CJ, Poole JH (1983) Relationships and social structure of African elephants. In: Hinde RA, editor. Primate Social Relationships. Sunderland, MA: Sinauer. 315–325.
- 32. Archie EA, Moss CJ, Alberts SC (2006) The ties that bind: genetic relatedness predicts the fission and fusion of social groups in wild African elephants. P Roy Soc B 273: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wittemyer G, Okello JBA, Rasmussen HB, Arctander P, Nyakaana S, et al. (2009) Where sociality and relatedness diverge: the genetic basis for hierarchical social organization in African elephants. P Roy Soc B 276: 3513–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee PC, Poole JC, Njiraini N, Sayialel CK, Moss CJ (2011) Male social dynamics: independence and beyond. In: Moss CJ, Croze H, Lee PC, editors. The Amboseli elephants: a long-term perspective on a long-lived mammal Chicago: University of Chicago Press.
- 35.Poole JH, Moss CJ (1989) Elephant mate searching: Group dynamics and vocal and olfactory communication. In: Jewell PA, Maloiy GMO, editors. Biology of large African mammals in their environment. 111–125.
- 36. Charif R, Ramey RII, Langbauer W Jr, Payne K, Martin R, et al. (2005) Spatial relationships and matrilineal kinship in African savanna elephant (Loxodonta africana) clans. Behav Ecol Sociobiol 57: 327–338. [Google Scholar]
- 37. Boudailliez B, Berquin P, Mariani-Kurkdjian P, Ilef DD, Cuvelier B, et al. (1997) Possible person-to-person transmission of Escherichia coli O111– associated hemolytic uremic syndrome. Pediatr Nephrol 11: 36–39. [DOI] [PubMed] [Google Scholar]
- 38.Wilson M (2008) Bacteriology of humans. Malden, MA: Wiley-Blackwell.
- 40. Goldberg TL, Gillespie TR, Rwego IB, Wheeler E, Estoff EL, et al. (2007) Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol Conserv 135: 511–517. [Google Scholar]
- 41. Maiden MCJ (2006) Multilocus sequence typing of bacteria. Annul Rev Microbiol 60: 561–588. [DOI] [PubMed] [Google Scholar]
- 42. Feil EJ, Spratt BG (2001) Recombination and the population structures of bacterial pathogens. Annul Rev Microbiol 55: 561–590. [DOI] [PubMed] [Google Scholar]
- 43. Tenaillon O, Skurnik D, Picard B, Denamur E (2010) The population genetics of commensal Escherichia coli . Nat Rev Microbiol 8: 207–217. [DOI] [PubMed] [Google Scholar]
- 44. Mbise AN, Mlengeya TDK, Mollel JO (1998) Septicaemic salmonellosis of elephants in Tanzania. Bulletin of Animal Health and Production in Africa 46: 95–100. [Google Scholar]
- 45.Fowler ME, Mikota SK (2006) Biology, medicine and surgery of elephants. Ames, Iowa: Blackwell Publishing. 565 p. [Google Scholar]
- 46. Caugant DA, Levin B, Selander R (1984) Distribution of multilocus genotypes of Escherichia coli within and between host families. Epidemiol Infect 92: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson JR, Owens K, Gajewski A, Clabots C (2008) Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J Infect Dis 197: 218–224. [DOI] [PubMed] [Google Scholar]
- 48. Bergholz PW, Noar JD, Buckley DH (2011) Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl Environ Microb 77: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gordon DM, Cowling A (2003) The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149: 3575–3586. [DOI] [PubMed] [Google Scholar]
- 50. Gordon DM (2001) Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147: 1079–1085. [DOI] [PubMed] [Google Scholar]
- 51. Brennan FP, O’Flaherty V, Kramers G, Grant J, Richards KG (2010) Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl Environ Microb 76: 1449–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiménez L, Muñiz I, Toranzos GA, Hazen TC (1989) Survival and activity of Salmonella typhimurium and Escherichia coli in tropical freshwater. J Appl Microbiol 67: 61–69. [DOI] [PubMed] [Google Scholar]
- 53. Whittam TS (1992) Sex in the soil. Current Biology 2: 676–678. [DOI] [PubMed] [Google Scholar]
- 54. Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, et al. (2012) Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. P Natl Acad Sci USA 109: 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blyton MDJ, Banks SC, Peakall R, Gordon DM (2013) High temporal variability in commensal Escherichia coli strain communities of a herbivorous marsupial. Env Microbiol 15: 2162–2172. [DOI] [PubMed] [Google Scholar]
- 56.Moss CJ, Croze H, Lee PC (2011) The Amboseli elephants: a long-term perspective on a long-lived mammal. Chicago: University of Chicago Press. 400 p. [Google Scholar]
- 57. Wittemyer G, Daballen D, Douglas-Hamilton I (2013) Comparative demography of an at-risk African elephant population. PloS ONE 8: e53726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jachmann H (1988) Estimating age in African elephants: a revision of Laws molar evaluation technique. Afr J Ecol 26: 51–56. [Google Scholar]
- 59. Lee PC, Moss CJ (1995) Statural growth in known-age African elephants (Loxodonta africana). J Zool 236: 29–41. [Google Scholar]
- 60. Rasmussen HB, Wittemyer G, Douglas-Hamilton I (2005) Estimating age of immobilized elephants from teeth impressions using dental silicon. Afr J Ecol 43: 215–219. [Google Scholar]
- 61. Lee PC, Sayialel S, Lindsay WK, Moss CJ (2012) African elephant age determination from teeth: validation from known individuals. Afr J Ecol 50: 9–20. [Google Scholar]
- 62.Croze H, Moss CJ (2011) Patterns of occupancy in time and space. In: Moss CJ, Croze H, editors. Amboseli Elephants: A long-term perspective on a long-lived mammal Chicago University of Chicago Press.
- 63. Wittemyer G, Getz WM, Vollrath F, Douglas-Hamilton I (2007) Social dominance, seasonal movements, and spatial segregation in African elephants: a contribution to conservation behavior. Behav Ecol Sociobiol 61: 1919–1931. [Google Scholar]
- 64. Douglas-Hamilton I, Krink T, Vollrath F (2005) Movements and corridors of African elephants in relation to protected areas. Naturwissenschaften 92: 158–163. [DOI] [PubMed] [Google Scholar]
- 65. Wittemyer G, Daballen D, Rasmussen H, Kahindi O, Douglas-Hamilton I (2005) Demographic status of elephants in the Samburu and Buffalo Springs National Reserves, Kenya. Afr J Ecol 43: 44–47. [Google Scholar]
- 66.Alberts SC, Hollister-Smith JA, Mututua RS, Sayialel SN, Muruthi PM, et al. (2005) Seasonality and long-term change in a savanna environment. In: Brockman DK, van Schaik CP, editors. Seasonality in primates: studies of liviing and extinct extinct human and non-human primates Cambridge: Cambridge University Press. 157–196.
- 67. Wittemyer G (2001) The elephant population of Samburu and Buffalo Springs National Reserves, Kenya. Afr J Ecol 39: 357–365. [Google Scholar]
- 68. Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS (2007) Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Env Microbiol 9: 2274–2288. [DOI] [PubMed] [Google Scholar]
- 69.Moss CJ, Lee PC (2011) Female social dynamics: fidelity and flexibility. In: Moss CJ, Croze H, Lee PC, editors. The Amboseli elephants: a long-term perspective on a long-lived mammal Chicago: University of Chicago Press. 205–245.
- 70. Janmaat KRL, Olupot W, Chancellor RL, Arlet ME, Waser PM (2009) Long-term site fidelity and individual home range shifts in Lophocebus albigena . Int J Primatol 30: 443–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Young JK, Glasscock SN, Shivik JA (2008) Does spatial structure persist despite resource and population changes? Effects of experimental manipulations on coyotes. J Mammal 89: 1094–1104. [Google Scholar]
- 72. Wirth T, Falush D, Lan R, Colles F, Mensa P, et al. (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60: 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Contreras CA, Ochoa TJ, Ruiz J, Lacher DW, Rivera FP, et al. (2011) Phylogenetic relationships of Shiga toxin-producing Escherichia coli isolated from Peruvian children. J Med Microbiol 60: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gordon DM, Clermont O, Tolley H, Denamur E (2008) Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Env Microbiol 10: 2484–2496. [DOI] [PubMed] [Google Scholar]
- 75. Méric G, Kemsley EK, Falush D, Saggers EJ, Lucchini S (2013) Phylogenetic distribution of traits associated with plant colonization in Escherichia coli . Env Microbiol 15: 487–501. [DOI] [PubMed] [Google Scholar]
- 76. Johnson JR, Delavari P, Kuskowski M, Stell AL (2001) Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli . J Infect Dis 183: 78–88. [DOI] [PubMed] [Google Scholar]
- 77. Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, et al. (1999) The link between phylogeny and virulence in Escherichia coli extra-intestinal infection. Infect Immun 67: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ochman H, Selander RK (1984) Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157: 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 80. Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Su C, Brandt LJ (1995) Escherichia coli O157:H7 infection in humans. Ann Intern Med 123: 698–714. [DOI] [PubMed] [Google Scholar]
- 82. Slutsker L, Ries AA, Greene KD, Wells JG, Hutwagner L, et al. (1997) Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann Intern Med 126: 505–513. [DOI] [PubMed] [Google Scholar]
- 83. Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, et al. (1993) Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol 138: 849–869. [DOI] [PubMed] [Google Scholar]
- 84. Ezenwa VO (2004) Host social behavior and parasitic infection: a multifactorial approach. Behav Ecol 15: 446–454. [Google Scholar]
- 85. Loehle C (1995) Social barriers to pathogen transmission in wild animal populations. Ecology 76: 326–335. [Google Scholar]
- 86. Alexander RD (1974) The evolution of social behavior. Ann Rev Ecol Syst 5: 325–383. [Google Scholar]
- 87. Tandé D, Boisramé-Gastrin S, Münck MR, Héry-Arnaud G, Gouriou S, et al. (2010) Intrafamilial transmission of extended-spectrum-β-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemoth 65: 859–865. [DOI] [PubMed] [Google Scholar]
- 88. Valverde A, Grill F, Coque TM, Pintado V, Baquero F, et al. (2008) High rate of intestinal colonization with Extended-Spectrum-β-Lactamase-Producing organisms in household contacts of infected community patients. J Clin Microbiol 46: 2796–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Routman E, Miller RD, Phillips-Conroy J, Hartl DL (1985) Antibiotic resistance and population structure in Escherichia coli from free-ranging African yellow baboons. Appl Environ Microb 50: 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Carlos C, Pires M, Stoppe N, Hachich E, Sato M, et al. (2010) Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol 10: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F (2013) Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microb 79: 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Smith JM, Feil EJ, Smith NH (2000) Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays 22: 1115–1122. [DOI] [PubMed] [Google Scholar]
- 93. Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, et al. (2009) Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Ch 53: 2733–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fraser C, Hanage WP, Spratt BG (2005) Neutral microepidemic evolution of bacterial pathogens. P Natl Acad Sci USA 102: 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cesaris L, Gillespie BE, Srinivasan V, Almeida RA, Zecconi A, et al. (2007) Discriminating between strains of Escherichia coli using pulsed-field gel electrophoresis and BOX-PCR. Foodborne Pathog Dis 4: 473–480. [DOI] [PubMed] [Google Scholar]
- 96. Goldberg TL, Gillespie TR, Singer RS (2006) Optimization of analytical parameters for inferring relationships among Escherichia coli isolates from repetitive-element PCR by maximizing correspondence with multilocus sequence typing data. Appl Environ Microb 72: 6049–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ezenwa VO (2003) Habitat overlap and gastrointestinal parasitism in sympatric African bovids. Parasitology 126: 379–388. [DOI] [PubMed] [Google Scholar]
- 98.Alm EW, Walk ST, Gordon DM (2011) The niche of Escherichia coli. In: Walk ST, Feng PCH, editors. Population genetics of bacteria: A tribute to Thomas S Whittam. Washington DC: ASM Press. 69–89.
- 99. Whittam TS (1989) Clonal dynamics of Escherichia coli in its natural habitat. Antonie Van Leeuwenhoek 55: 23–32. [DOI] [PubMed] [Google Scholar]
- 100. Ley RE, Peterson DA, Gordon JI (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848. [DOI] [PubMed] [Google Scholar]
- 101. Gordon DM (1997) The genetic structure of Escherichia coli populations in feral house mice. Microbiology 143: 2039–2046. [DOI] [PubMed] [Google Scholar]
- 102. Badgley BD, Ferguson J, Heuvel AV, Kleinheinz GT, McDermott CM, et al. (2011) Multi-scale temporal and spatial variation in genotypic composition of Cladophora-borne Escherichia coli populations in Lake Michigan. Water Res 45: 721–731. [DOI] [PubMed] [Google Scholar]
- 103. Hansen DL, Ishii S, Sadowsky MJ, Hicks RE (2009) Escherichia coli Populations in Great Lakes Waterfowl Exhibit Spatial Stability and Temporal Shifting. Appl Environ Microb 75: 1546–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Byappanahalli MN, Sawdey R, Ishii S, Shively DA, Ferguson JA, et al. (2009) Seasonal stability of Cladophora-associated Salmonella in Lake Michigan watersheds. Water Res 43: 806–814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide diversity in E. coli as a function of host sex. Boxplots depict nucleotide diversity in E. coli infecting individual elephants as a function of host sex. This analysis was performed on adult animals from Samburu NR only.
(TIF)
The distribution of E. coli phylogroups groups as a function of host age. Plot depicts a cross-sectional analysis of the proportion of each phylogroup type found infecting elephant hosts of different ages. Relationships demonstrate that unclassified E. coli isolates increased in older age groups.
(TIF)
Sample information, including the number of elephant hosts per family group, the number of E. coli isolates genotyped per family group, and the number of GPS sightings per family group.
(DOCX)
Percent of E. coli isolates assigned to different phylogroups in Amboseli and Samburu. Unassigned isolates did not cluster with any known ECOR sequences.
(DOCX)
Basic evolutionary parameter estimates for concatenated E. coli sequences.
(DOCX)