Abstract
TMEM127 is an endosome-associated tumor suppressor gene in pheochromocytomas, neuroendocrine tumors that can co-occur with renal cell carcinomas (RCCs). TMEM127 loss leads to increased mTOR signaling. However, the spectrum of tumors with TMEM127 mutation and how TMEM127 and mTOR interact in tumorigenesis remains unknown. Here, we report that germline TMEM127 mutations occur in RCCs and that some mutant proteins, unlike wild-type (WT) TMEM127, fail to cooperate with activated early endosomal GTPase, Rab5, to inhibit mTOR signaling. Tmem127-null mouse embryonic fibroblasts (MEFs) are deficient in generating early-to-late hybrid endosomes upon constitutive Rab5 activation, a defect rescued by WT, but not mutant, TMEM127. This endosomal dysfunction results in diminished mTOR colocalization with Rab5-positive vesicles. Conversely, active, lysosomal-bound mTOR is increased in Tmem127-null MEFs, which also display enhanced lysosomal biogenesis. Our data map the tumor-suppressive properties of TMEM127 to modulation of mTOR function in the endolysosome, a feature that may contribute to both pheochromocytoma and RCC pathogenesis.
INTRODUCTION
Germline mutations in VHL, MET, FH, FLCN and SDHB genes account for a proportion of hereditary renal cell carcinomas (RCCs) (1–5). However, the cause of many familial forms of these tumors remains unknown. A link between RCCs and pheochromocytomas or paragangliomas, which are neural crest-derived, catecholamine-secreting tumors, has been well established in two hereditary tumor syndromes. In von Hippel-Lindau disease, a familial autosomal dominant disorder caused by mutations of the VHL gene, patients develop RCC, pheochromocytomas and hemangioblastomas of the central nervous system (6). In familial paraganglioma type (PGL4), caused by germline mutations in SDHB, patients can develop paraganglial tumors, some presenting with concurrent RCC (7), and, more recently SDHB mutations have also been reported in patients presenting with RCC alone (4,6). These genetic associations prompted us to investigate the role of the pheochromocytoma susceptibility gene TMEM127 in renal cancers.
Germline truncating or missense TMEM127 mutations occur in pheochromocytomas and paragangliomas with features of a prototypical tumor suppressor gene (8,9). TMEM127 is a ubiquitously expressed protein of unknown function which localizes to multiple endosomal domains. While recombinant wild-type (WT) TMEM127 displays a punctate appearance typical of endomembrane association, ectopically expressed mutant TMEM127 is diffusely distributed in the cytoplasm, suggesting that endomembrane localization is relevant for its tumor suppressor function (8,9). Furthermore, both TMEM127-mutant primary pheochromocytomas and cell lines with TMEM127 depletion by short-interfering (si)- or short-hairpin (sh)-RNA downregulation display increased phosphorylation of mTOR downstream targets, while overexpression of TMEM127 leads to low levels of target phosphorylation (8). However, the mechanism through which TMEM127 and mTOR interact is unknown.
One of the endomembrane domains to which TMEM127 associates is the early endosome, which controls the output signal of multiple growth factors and oncogenes through regulation of protein trafficking and turnover (8,10). The small guanosine triphosphate (GTP)ase Rab5 is required for assembly and function of the early endosome and, in its GTP-bound form, actively regulates membrane trafficking and modulates early endosome formation and maturation into late endosomes and lysosomes (11,12). Recently, signaling through the mTOR complex 1 (mTORC1) was shown to require intact endolysosome function. Activation of the early endosome by expression of a GTP-bound Rab5 mutant was found to inhibit mTOR signaling in response to amino acids (13–15). In addition, amino acid exposure promotes translocation of mTOR to the lysosome, its site of activation (16–18), while reduced access of mTOR to the lysosome limits its ability to bind to its activator Rheb, thus leading to reduced signaling (13,14,18).
Here, we report that loss of TMEM127 in vitro and in vivo lead to disrupted formation of hybrid early-to-late endosomal vesicles by Rab5 and to an increased association of mTOR with the lysosome. Together, these data provide a potential mechanism for the inhibitory effects of TMEM127 on mTOR. We also describe novel germline TMEM127 mutations in renal cell carcinoma and show that some of these mutations are defective for endosomal function. These results suggest that TMEM127 disruption contributes to both pheochromocytoma and RCC pathogenesis.
RESULTS
Mutations of conserved TMEM127 residues in RCC
We screened samples of 214 patients with RCC from two independent cohorts for TMEM127 gene mutations (Supplementary Material, Table S1). The first cohort comprised 104 germline samples from patients with features of RCC susceptibility, including early-onset disease, bilateral or multicentric tumors or a positive familial history of renal cancer, but without a detectable mutation in the known susceptibility genes (VHL, SDHB, FH and FLCN). The second cohort included 110 tumors from unselected patients with RCC, spanning a broad spectrum of histological subtypes and tumor grading (Supplementary Material, Table S1).
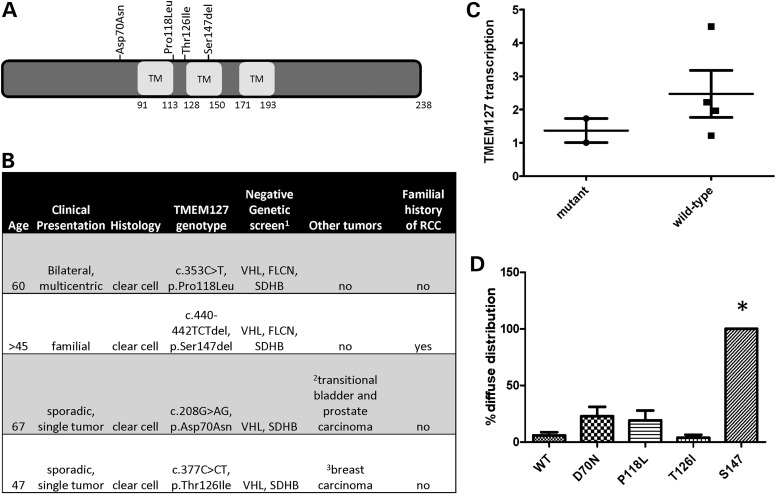
In total, four unique TMEM127 variants were identified (1.86%, 4/214, Fig. 1A), two from each cohort, and affected only conserved residues of the protein. One of these variants was an in-frame, single amino acid deletion targeting the second transmembrane domain of TMEM127 (c.440–442TCTdel, p.Ser147del, hereafter referred to as S147del), and the remainder were missense mutations. For two of the cases, only germline DNA was available (S147del and c.353C>T, p.Pro118Leu or P118L); in one case, we had both germline and tumor material (c.208G>AG, p.Asp70Asn or D70N) and the fourth sample had only tumor tissue (c.377C>CT, p.Thr126Ile or T126I). The mutations were present in the germline of all three samples with available constitutive material. One of the mutations (D70N) had previously been described in a patient with pheochromocytoma (9) and was recently reported at low frequency (minor allele frequency 0.002, SNP rs121908819, and NHLBI Exome Sequencing Project ss342069846) in databases which include samples from both healthy and non-healthy individuals. However, this variant was not detected in a group of 1000 normal, ethnically matched control alleles which we previously reported (9), thus its pathogenicity remains unclear. None of the other three RCC mutations were identified in the SNP database or our normal control group. One of the patients had a family history of RCC, and two of the mutation carriers had also been diagnosed with other neoplasias, either concurrently or prior to RCC, but no pheochromocytoma, paraganglioma or family history of either of these tumors was reported in these four patients (Fig. 1B). The reason for the lack of association with paraganglial tumors is not known, but could be due to incomplete penetrance of the mutant alleles, as suggested in SDHB-associated RCCs without co-occurring paragangliomas (19).
Figure 1.
TMEM127 mutations in renal cell carcinoma. (A) Distribution of the TMEM127 mutations found in four RCCs along the TMEM127 protein structure; (B) main clinical and genetic features of these patients; (C) real-time PCR quantification of two TMEM127-mutant and four non-mutant RCC tumors. Each sample was run in triplicate in two separate experiments. Results were combined for each group and are presented as mean ± SEM; (D) pattern of intracellular distribution of four TMEM127 mutants and WT GFP-fusion constructs transfected in HeLa cells shown as the fraction of diffuse cytoplasmic localization by confocal microscopy from at least five independent experiments and an average of 220 cells counted per construct (*P < 0.05).
The two mutations detected in somatic tissue (D70N and T126I), along with a specimen from prostate cancer that was removed at the time of nephrectomy of one of these patients, retained heterozygosity at the mutation site, indicating that the WT TMEM127 allele was not deleted in these tissues. Motivated by our earlier observation that pheochromocytoma-associated TMEM127 mutations led to reduced TMEM127 transcription at the tumor level beyond that which would be expected by loss of a single allele in the somatic tissue (8), we measured TMEM127 mRNA expression by real-time PCR in the two available mutant RCC tumors. In these samples, TMEM127 transcription was about 50% of that of a group of RCCs with intact TMEM127 sequence (average ± SD, 1.37 ± 0.43 in mutants and 2.47 ± 1.33 in WT tumors; Fig. 1C), consistent with decreased transcription rate or instability of mutant TMEM127. Although only two mutant samples could be tested, these findings suggest that the TMEM127 mutations might function in haploinsufficiency in the RCC context.
RCC-derived TMEM127 mutation is deficient in endosomal localization
We had previously found that the intracellular distribution of pheochromocytoma-related mutant TMEM127 was distinct from that of WT constructs (9). To determine the status of the mutations identified in RCC patients with respect to their endomembrane association, we generated GFP-tagged versions of each mutant and examined the subcellular distribution after transfection of these constructs in HeLa cells. We found that the TMEM127-S147del construct, carrying the in-frame deletion, was diffusely distributed in the cytoplasm of all transfected cells, in contrast with the punctate localization of WT TMEM127 (Figs 1D and 2A). The other three RCC mutants had variable distribution: TMEM127-D70N was diffuse in approximately one-third of the cells, while both TMEM127-P118L and TMEM127-T126I had a predominant punctate appearance, similar to the WT construct (Figs 1D and 2A). These experiments indicate that the mutations had variable impact on TMEM127 intracellular distribution.
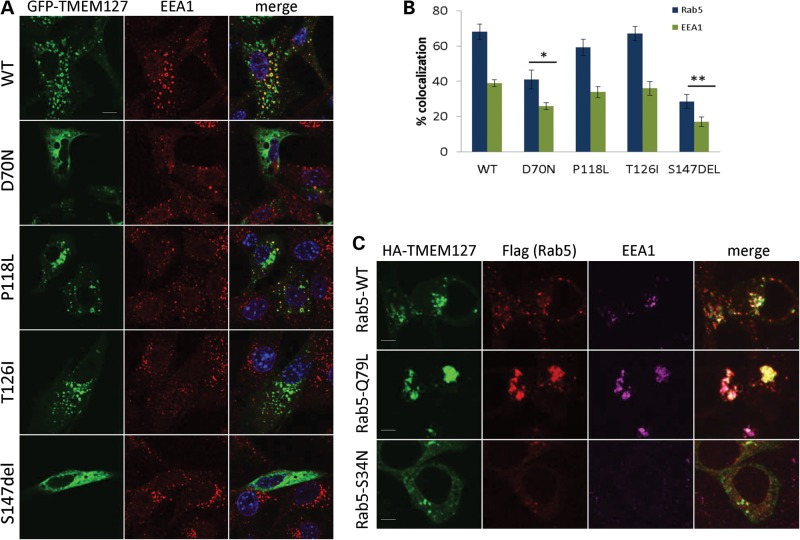
Figure 2.
TMEM127 mutants have reduced association with the early endosome. (A) Confocal microscopy of GFP-fusion constructs encoding each of the four mutants or WT TMEM127 (green) co-stained with the early endosomal marker EEA1 (red). Merged images are shown in yellow and nuclei are DAPI stained (blue). Scale bars are 10 µm. (B) Degree of colocalization of each construct shown in (a) with EEA1 or with Rab5 (images shown in Supplementary Material, Fig. S1). Data were quantified from at least three independent experiments with at least 14 cells analyzed for each construct (*P < 0.05; **P < 0.01). (C) HA-TMEM127 (green) co-transfected with Flag-tagged Rab5 constructs WT, Q79L or S34N (red) and stained with endogenous EEA1 (magenta). Merged signals are shown in white. TMEM127 colocalization with the early endosomal marker EEA1 is increased by Rab5-Q79L expression and decreased by Rab5S34N. Note the lack of EEA1 staining in Rab5-positive structures on the bottom panel, indicating dysfunctional early endosomal assembly in Rab5S34N-transfected cells. Scale bars are 5 µm.
Previously, recombinant TMEM127 was found to partially overlap with the early endosomal marker Rab5 by confocal microscopy (8). When the degree of overlap was quantified, 68 ± 4.2 and 38 ± 1.9% (mean ± SEM) of GFP-WT-TMEM127- positive structures colocalized with Rab5 and with the Rab5 effector EEA1, respectively (Fig. 2A, Supplementary Material, Fig. S1). The extent of colocalization was reduced in S147del and D70N mutants, but not in P118L and T126I (Fig. 2A and B). These results confirm our previous observations that TMEM127 localizes to early endosomal domains (8), and indicate that RCC-related mutations can decrease (D70N) or abolish (S147del) this association (Fig. 2A and B, Supplementary Material, Fig. S1). The two remaining mutations did not differ from the WT protein in their distribution under the conditions used in this assay, and it cannot be excluded that these mutants are defective for another aspect of endosomal function, or that they are simply rare non-pathogenic variants.
TMEM127 associates with Rab5 in a GTP-status-dependent manner
Next, we examined whether the association of TMEM127 with Rab5-positive vesicles was dependent on the Rab5 GTP status. A WT version of Rab5 (Rab5WT), a constitutively active version of this construct, locked in its GTP-bound conformation, Rab5Q79L, and a Rab5S34N inactive mutant, which maintains Rab5 bound to GDP (11), all tagged with Flag, were co-transfected with WT-TMEM127 in HeLa cells. Expression of Rab5Q79L leads to enlargement of early endosomes due to increased homo- and heterotypic fusion, while Rab5S34N has a predominant non-membrane localization pattern (11). Accordingly, co-staining of these cells with endogenous EEA1 indicates the assembly of a functional early endosome in the presence of Rab5WT and Rab5Q79L, but not in the presence of the GDP-bound mutant, S34N (Fig. 2C). Co-expression of HA-TMEM127 with Flag-Rab5S34N led to dispersion of both the Rab5 and TMEM127 signals (Fig. 2C), while expression of the constitutively active Flag-Rab5Q79L mutant increased the fraction of TMEM127 that associates with Rab5-positive enlarged vesicles (Fig. 2C). These data suggest that TMEM127 early endosome localization is enhanced by an active Rab5 endosomal domain. In further support of an association between TMEM127 and Rab5, HA-TMEM127 was found to co-immunoprecipitate with all three Rab5 plasmids in HEK293T cells, although this association was increased in cells expressing the GTP-bound mutant (Fig. 3A and B). These results were replicated by immunoprecipitation with endogenous TMEM127, indicating that the association was not due to tag effects (Fig. 3B). These findings suggest that TMEM127 is a component of Rab5-containing protein complexes, and that this association is GTP dependent.
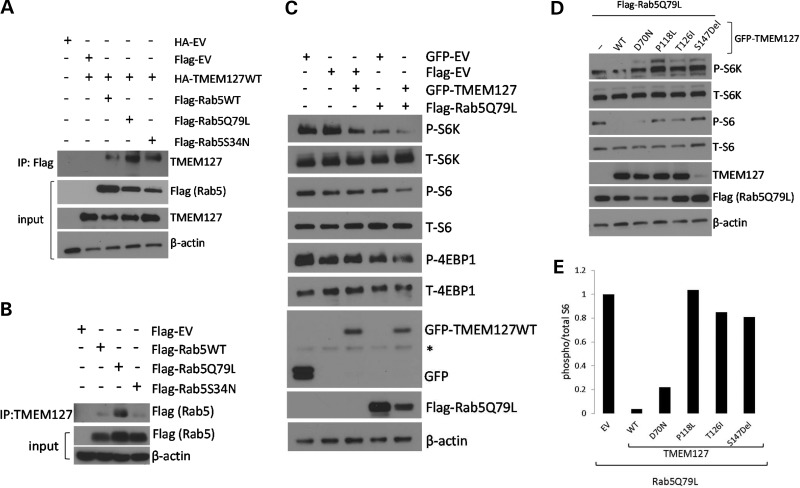
Figure 3.
TMEM127 potentiates Rab5 inhibition of mTOR signaling. (A) Lysates from 293T cells expressing the Flag-Rab5 WT, Q79L or S34N and HA-TMEM127 or an HA-EV constructs were immunoprecipitated (IP) with Flag antibody and immunoblotted with TMEM127 antibody. Input lysates are shown after probing with Flag (for the Rab5 constructs), TMEM127 or β-actin as a loading control. (B) Lysates from 293T cells expressing the indicated Flag-Rab5 constructs or empty vector were immunoprecipitated (IP) with an endogenous TMEM127 polyclonal antibody and probed with Flag antibody and β-actin. (C) 293T cells were transfected with GFP-TMEM127, Flag-Rab5Q79L, both constructs or their respective empty vectors. Whole cell lysates were harvested at 48 h, run on western blots and probed with antibodies for both phosphorylated (P) and total (T) mTOR targets, S6K, S6 or 4EBP1, and for GFP, Flag antibody and β-actin (asterisk indicates non-specific band). (D) 293T cells were transfected with empty GFP-vector (EV), GFP-TMEM127WT (WT) or each of the four RCC TMEM127 mutations, along with Rab5Q79L, and harvested after 48 h. Blots were probed with antibodies targeting phosphorylated (P) and total (T) S6K, S6, Flag, TMEM127 and β-actin. (E) Relative levels of P/T S6 from lysates shown in (d) were quantified by Image J and normalized to cells expressing empty vector (−) .
TMEM127 mutations disrupt Rab5-mediated mTOR signaling inhibition
Based on the inhibitory effects of Rab5 on mTOR (13,14), we postulated that the effect of TMEM127 on reducing mTOR target phosphorylation that we reported previously (8) might be mediated by its association with Rab5. We thus examined whether TMEM127 overexpression could potentiate the effect of the constitutively active Rab5 mutant, Rab5Q79L, on mTOR downstream signaling. In agreement with previous data, expression of either WT GFP-TMEM127 or the constitutively active Flag-Rab5Q79L in cells exposed to amino acids led to reduced phosphorylation of the mTOR targets S6K, S6 and 4EBP1 (Fig. 3C). These effects were enhanced in cells that co-expressed both constructs, suggesting that TMEM127 cooperated with activated Rab5 to reduce mTOR signaling (Fig. 3C). In contrast, when co-expressed with Rab5Q79L, three of the constructs encoding RCC-associated TMEM127 mutations did not reduce mTOR target phosphorylation under the same culture conditions (Fig. 3D and E). Notably, mutant S147del was expressed at markedly lower levels than the WT construct, suggesting instability of this variant, similar to pheochromocytoma-related TMEM127 mutations (8). Levels of the co-transfected Flag-Rab5Q79L product tended to be lower in cells expressing three of the RCC mutants compared with WT-TMEM127, suggesting a possible effect of these TMEM127 mutations in reducing abundance or stability of the Rab5 construct (Fig. 3D). These data are consistent with an effect of TMEM127 on Rab5-mediated mTOR inhibition.
Loss of TMEM127 disrupts Rab5-mediated endosomal fusion in vitro and in vivo
To characterize further the role of TMEM127 in the context of endosomal function, we expressed Rab5Q79L in HeLa cells that had been depleted of TMEM127 by short-interfering (si)-RNA (si-TMEM127#2, Supplementary Material, Fig. S2). Confocal microscopy of cells stained with Flag (for Rab5Q79L) showed the expected enlarged vesicles indicative of fused early-late endosomes in control knockdown cells (Fig. 4A). However, these vesicles were markedly reduced, or even absent, in cells depleted of TMEM127 (P < 0.05, Fig. 4A). Similar findings were seen with an independent siRNA sequence targeting TMEM127, siTMEM127#4 (Fig. 4A, Supplementary Material, Fig. S2) and were rescued by re-expression of a TMEM127 construct which had been made resistant to siRNA by mutation of the target nucleotide sequence without changing the resulting amino acid (Fig. 4B). These results suggest that TMEM127 is required for Rab5-mediated endosomal fusion.
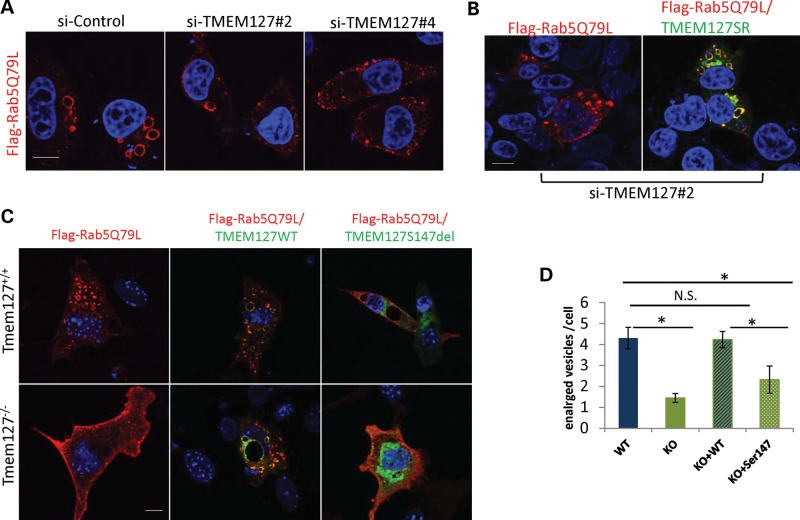
Figure 4.
The enlarged vesicle phenotype of Rab5 activation is reduced by TMEM127 depletion or absence. (A) HeLa cells expressing two independent TMEM127 siRNAs (#2 and #4) or a control siRNA (si-ctl) were transfected with Flag-tagged Rab5Q79L and fixed after 24 h for staining and confocal microscopy. Flag-Rab5Q79L mutant is shown in red and DAPI-stained nuclei are in blue. (B) Rab5Q79L staining of HeLa cells expressing siTMEM127#2 without (left) or after transfection of a TMEM127 construct engineered to be resistant to siRNA#2 (WT-TMEM127SR). (C) MEFs from Tmem127−/− or Tmem127+/+ embryos were transfected with Rab5Q79L and a GFP empty vector (left), GFP-WT TMEM127 (middle) or GFP-Ser147del TMEM127 (right). (D) Quantification of the number of enlarged vesicles per cell of each cell type shown in (C) is indicated. Data are representative from at least three independent experiments (P < 0.05; NS = non-significant). Scale bars are 10 mm. Data are representative from at least three independent experiments (P < 0.05; NS = non-significant). Scale bars are 10 µm.
To confirm these findings in a more physiological setting, we generated fibroblast lines from mouse embryos lacking the Tmem127 gene (Tmem127−/−) or from control WT (Tmem127+/+) or heterozygous (Tmem127+/−) littermates (Supplementary Material, Fig. S3; detailed description of the Tmem127−/− mice will be reported elsewhere). These mouse embryonic fibroblasts (MEFs) were prepared from E13.5 embryos and cultured for no more than seven passages. Tmem127 null or control MEFs were transfected with the same Flag-Rab5Q79L construct and stained as described above. Null MEFs from multiple independent batches showed a striking decrease in the number of enlarged Rab5Q79L-positive vesicles, in contrast with WT cells, thus recapitulating the findings observed following TMEM127 knockdown in HeLa cells (Fig. 4C). Ectopic expression of WT-TMEM127 rescued this phenotype, while the TMEM127S147del mutant did not, confirming that this mutant is defective for Rab5-mediated early endosomal regulation (Fig. 4C). In contrast with the morphological differences noted on Rab5-positive vesicles, no changes were seen in the equivalent GTP- and GDP-bound mutants of the recycling GTPase Rab4 (Supplementary Material, Fig. S4), further indicating a specific effect of TMEM127 toward Rab5.
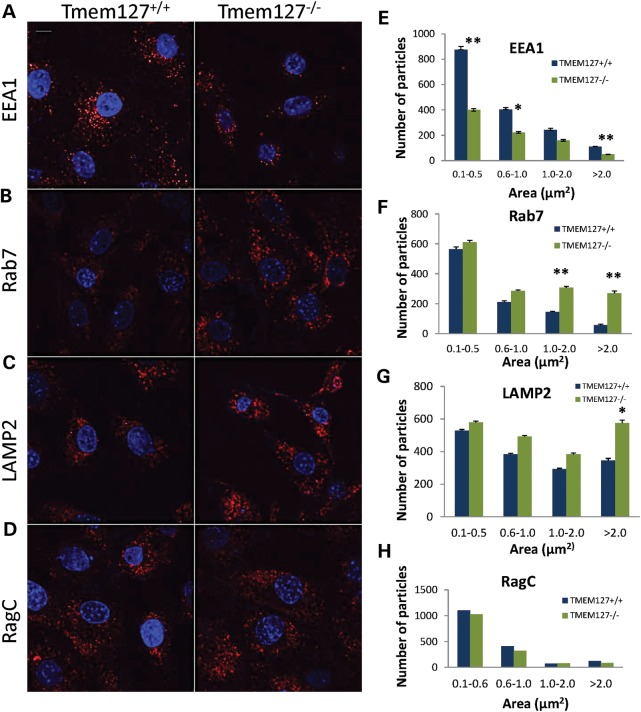
To determine whether loss of Tmem127 affects endogenous endosomal markers, we stained null or WT MEFs. Tmem127−/− MEFs had lower overall EEA1-positive puncta than did control MEFs (Fig. 5A). In contrast, the number of late endosomal and lysosomal vesicles, identified by Rab7-positive (Fig. 5B) and LAMP2-positive (Fig. 5C) puncta, respectively, was higher in null compared with WT MEFs. Other endomembrane-localized proteins such as RagC (Fig. 5D) showed no difference between WT and null cells, indicating that the differential number of puncta was selective, rather than an indiscriminant phenomenon affecting all endomembrane domains. Often, the puncta size difference was more accentuated in the larger vesicle size range (Fig. 5E–H). These findings are consistent with a potential effect of Tmem127 in fusion events of multiple endosomal domains (20), and suggest that in its absence, some domains downstream of the early endosome, i.e. late endosome and lysosome, may be enhanced in a compensatory manner. Although marked depletion of Rab5 in vivo has been shown to severely impair endolysosomal biogenesis downstream of the early endosome, even partial Rab5 function is capable of mounting a robust recovery mechanism (21), which may explain the profile observed in Tmem127-deficient cells. Taken together, these results suggest that TMEM127 is required for regulation of Rab5 vesicle fusion and early-to-late endosomal transition, leading to a redistribution of particle organization.
Figure 5.
Differential staining of endogenous endosomal markers in Tmem127 null MEFs. Confocal microscopy of Tmem127+/+ or Tmem127−/− MEFs stained with EEA1 (A), Rab7 (B), LAMP2 (C) and RagC (D) is shown in red. Nuclei are stained with DAPI (blue). Scale bars are 10 µm. (E–H) Quantification of puncta of the indicated size ranges for each endosomal marker from 20 to 40 cells analyzed from two to four independent experiments per marker is shown as indicated. Data are plotted as mean ± SEM (*P < 0.05, **P < 0.01).
mTOR associates with the lysosome domain in a TMEM127-dependent manner
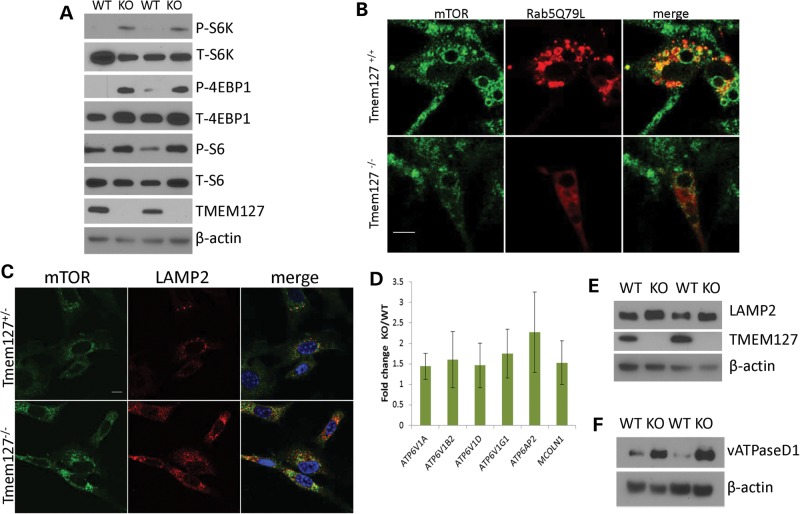
Similar to our previous findings in mutant pheochromocytomas and cells with TMEM127 knockdown (8), lysates from Tmem127-null MEFs showed increased mTOR target phosphorylation compared with WT cells (Fig. 6A), indicating that the murine model recapitulated, though to a lesser extent, the earlier findings in human cells. The relatively modest effect on phosphorylation downstream of mTOR may, at least in part, reflect the emergence of partial compensatory mechanisms in vivo, in contrast with the ‘acute’ inhibition due to siRNA-induced depletion. Overexpression of the Rab5Q79L constitutively active mutant has been suggested to trap amino acid-stimulated mTOR into Rab5-positive enlarged endosomal vesicles, thus leading to reduced mTOR access to subsequent domains of the endosomal cycle (i.e. late endosome/lysosome) and ultimately resulting in decreased mTOR downstream activation (13,15). To determine, at the cellular level, whether TMEM127 influenced mTOR association with Rab5-positive enlarged vesicles, we stained mTOR in Tmem127 null or control MEFs transfected with Flag-Rab5Q79L and examined these cells by confocal microscopy after they had been exposed to amino acids for 15 min following 50 min of amino acid deprivation, as reported (13,15). The reduced number of Rab5-positive enlarged vesicles in TMEM127 null cells posed a technical challenge for this quantification. However, in the cells in which both Rab5Q79L and mTOR staining could be assessed, the degree of colocalization was lower in Tmem127KO than in control MEFs (Fig. 6B, Supplementary Material, Fig. S5). Similar findings were noted in MEFs under regular culture conditions (Supplementary Material, Fig. S6). These results suggest that in the context of constitutive Rab5 activation, loss of Tmem127 disrupts mTOR distribution to an activated early endosome.
Figure 6.
mTOR association with the lysosomal domain is increased in Tmem127 KO cells. (A) Western blot lysates from two independent Tmem127 null (KO) or WT MEFs probed for phosphorylated (P) or total (T) S6K, S6 and 4EBP1, and also TMEM127 and β-actin. (B) Confocal microscopy images of endogenous mTOR staining (green) of WT (Tmem127+/+) or KO (Tmem127−/−) MEFs after transfection with Flag-Rab5Q79L (red) and amino acid exposure for 15 min following a 50 min amino acid deprivation. Merged images are shown in yellow. Scale bar is 10 µm. Quantification of colocalization of mTOR/Rab5Q79L is shown in Supplementary Material, Figure S6. (C) Confocal microscopy of endogenous mTOR (green), LAMP2 (red) and merged images in KO or WT MEFs maintained under steady-state culture conditions (Supplementary Material, Fig. S7a shows quantification of the degree of colocalization). Scale bar is 10 µm; (D) Real-time PCR quantification of lysosomal genes vacuolar ATPase (vATPase) subunits V1a, V1b2, V1d, V1g1, AP2 and the gene encoding for Mucolipin 1 (Mcoln1) in MEFs. Results represent the fold change of KO in relation to WT MEFs, generated from triplicate data (±SEM) from experiments performed in independent pairs of MEFs. (E) Western blot of lysates from two independent Tmem127 KO or WT MEF pairs, probed with LAMP2, TMEM127 and β-actin. (F) Western blot of MEF lysates from two independent Tmem127 KO or WT MEF pairs, probed with vATPase D1, TMEM127 and β-actin.
If less mTOR is associated with early endosomal, Rab5Q79L-positive vesicles, we reasoned that in cells devoid of Tmem127 a larger fraction of mTOR might become available to associate with the lysosome. Indeed, we found that the amount of mTOR- and LAMP2-double positive vesicles was increased in Tmem127 null cells both under steady-state culture conditions (Fig. 6C, Supplementary Material, Fig. S7A) and after amino acid exposure (Supplementary Material, Fig. S7B and C). To further characterize the status of lysosomal activity in Tmem127KO we also examined the expression of lysosomal genes and proteins in these cells. We found that transcription of several lysosomal genes, including those encoding for vacuolar ATPase (vATPase) subunits, which are components of the lysosome-tethered complex that regulates amino acid-dependent mTOR activation, and Mcoln1, a non-selective cation channel that is involved in lysosomal fusion with other membranes, was upregulated in Tmem127-null MEFs compared with WT cells (Fig. 6D). In addition, protein levels of vATPase D1 subunit (vATPaseD1, Fig. 6E) and LAMP2 (Fig. 6F) were more abundant in null Tmem127 MEFs compared with WT controls. Together with the increased lysosomal mass described above (Fig. 5C), these data support an activation of lysosomal function in the null MEFs. Taken together, these data suggest that in the absence of Tmem127, lysosomal biogenesis is enhanced and there is an increased proportion of lysosomal-bound, active mTOR.
DISCUSSION
We had previously found that TMEM127 mutations predispose to pheochromocytomas and lead to increased mTOR signaling (8). Here, we expand these findings by showing evidence that TMEM127 is required for normal Rab5 function in early-to-late endosomal transition and that the resulting disruption is associated with enhanced lysosomal-bound mTOR. We also report novel germline TMEM127 mutations in patients with RCC, and show that some of these mutants are deficient in cooperating with Rab5 to inhibit mTOR. The degree of disruption was variable across the mutants and it is not possible to exclude that some of these variants had a subtler defect or are not pathogenic. However, one of the mutants, carrying an in-frame deletion of the second transmembrane domain, was incapable of rescuing the disrupted endosomal fusion defect of TMEM127 null cells. These results expand the spectrum of tumors with TMEM127 dysfunction and provide insights into the role of TMEM127 in the endolysosomal system. These findings also support a novel model through which mTOR signals can be modulated in cancer.
The endosome is known to regulate turnover of various proteins, including growth factor receptors, and its disruption can lead to their accumulation and increased signaling through downstream pathways (22–24). Loss of the tumor suppressor gene VHL, which predisposes to pheochromocytoma and RCC (1), leads to deficient formation of enlarged fused endosomal vesicles after expression of Rab5Q79L, similar to TMEM127 deletion (25). These effects of VHL-null cells occur in a manner dependent on the VHL target, hypoxia inducible factor (HIF), and result in reduced degradation of the epidermal growth factor receptor (EGFR), leading to increased activation of EGFR downstream signaling (25). In the present study, we show that cells deficient in TMEM127 disrupt Rab5-induced endosomal fusion and lead to increased lysosomal-centered mTOR signaling. These findings are compatible with a model whereby TMEM127 might enhance mTOR association with the Rab5-positive domain of the early endosome. In the absence of functional TMEM127, a larger fraction of mTOR may become available to associate with the lysosomal surface, which facilitates mTOR activation through a multiprotein complex known as the ragulator (17). The exact mechanisms through which TMEM127 regulates early-to-late endosomal transition and, possibly, lysosomal function, need to be further explored; nevertheless, our results corroborate earlier evidence that mTOR signaling is modulated by the endolysosome (13–15).
An active role of the lysosome on protein signaling, besides its well-known function in protein degradation, has recently been revealed by the identification of feedback loops between the lysosome and transcription regulation of multiple targets (26,27). In particular, mTOR was found to reciprocally modulate lysosomal function (28–31). Our data suggest that in cells lacking TMEM127 lysosomal activity is increased, in agreement with findings in Tsc2-null cells, which have constitutive activation of the mTOR pathway (28). A key component of mTOR-mediated lysosomal modulation is the transcription factor TFEB, a master regulator of lysosomal biogenesis (27,31). It remains to be determined whether TMEM127 acts through TFEB to modulate lysosomal function. Interestingly, TFEB has been reported to be a target of translocation in RCC (32,33), and more recently, another RCC susceptibility gene, FLCN, was found to interact with TFEB at the lysosome(34). It is, however, unclear whether genetic lesions that lead to disrupted endolysosomal function favor renal cell proliferation.
Although rare, identification of germline TMEM127 mutations in RCC provides additional evidence for the molecular heterogeneity of hereditary forms of these tumors. TMEM127 mutations were not reported in recent large-scale surveys of RCC genomic alterations (35–37). However, this is not surprising, as only somatic changes were examined in these large cohorts and germline variants were excluded from the analyses. In pheochromocytomas and paragangliomas, TMEM127 mutations have also been restricted to the germline, suggesting that disruption during development may be a distinctive feature of this gene (9,38). Finally, the present findings add to an increasing body of literature that recognizes the contribution of endolysosomal dysregulation to cancer (34,39–42) and lend support to the investigation of endosomal modulators as potential therapeutic targets in tumors, similar to those proposed to address mutations in another RCC susceptibility gene, MET (43).
MATERIALS AND METHODS
Patients and samples
Blood and/or tumor samples were obtained from patients with histologically confirmed RCCs. Informed consent was obtained from all subjects or waived for discarded tumor material according to Institutional Review Board-approved protocols of the University of Texas Health Science Center at San Antonio, Texas, USA and University of Birmingham, UK. Germline samples from RCC patients were screened (and found to be negative) for mutations in the VHL, MET, SDHB, FH and FLCN genes, as previously described (44). Hereditary RCC was defined by the diagnosis of an RCC in at least one first-degree relative. In all, 214 samples, including 104 germline samples and 110 tumors from the UTHSCSA tumor bank were included in this study. Main clinical and histological features of the samples are summarized in Supplementary Material, Table S1.
Methods
Reagents
Reagents were obtained from the following sources: antibodies to TMEM127 and HA (goat) for immunoprecipitation from Bethyl Laboratories (Montgomery, TX, USA), phospho-T37-46-4EBP1, phospho-T389-S6K, phospho-S235/236-S6, Rab7, EEA1 and RagC from Cell Signaling (Beverly, MA, USA); LAMP2 antibody from Abcam (Cambridge, MA, USA); HA and GFP antibody from Covance (Princeton, NJ, USA); DAPI from Life Technologies (Foster City, CA, USA); Cy3 conjugated secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA); HRP-labeled anti-mouse and anti-rabbit secondary antibodies and SybrGreen PCR Master Mix from Life Technologies; FLAG-M2 and β-actin antibody, Dulbecco-PBS, DMEM, 50× MEM amino acid mixture, sodium orthovanadate, phenylmethylsulfonyl fluoride, rapamycin, DNA and siRNA oligonucleotides from Sigma-Aldrich (St. Louis, MO, USA); fetal calf serum, penicillin, streptomycin, trypsin, HEPES from Cell-Gro (Manassas, VA, USA), Complete-miniEDTA Protease Cocktail from Roche (Oceanside, CA, USA); RNAsin, random hexamers and reverse transcriptase from Promega (Madison, WI, USA); deoxynucleotides, HotMaster Taq polymerase, Trizol, Lipofectamine 2000, Immobilon PVDF membrane and WestPico Chemiluminescence kit from ThermoFisher (Hudson, NH, USA).
DNA and RNA isolation
Blood, tumor and MEF DNA were isolated using the Qiagen Genomic tip or the Qiagen Gentra Puregene Cell for paraffin-embedded samples (Qiagen, Valencia, CA, USA) following the manufacturer's protocols. Total RNA was prepared from frozen tumor specimens or MEFs with Trizol (Life Technologies) according to the manufacturer's instructions and reverse transcribed to cDNA using Superscript II reverse transcriptase kit (Life Technologies).
TMEM127 gene sequencing
Primers spanning the three coding exons of TMEM127 gene were used to amplify germline or tumor DNA from 214 RCC cases, as previously reported (8). PCR products were Sanger sequenced by Beckman Genomics (Danvers, MA, USA). Sequence analysis and estimation of heterozygosity was performed using the Mutation Surveyor Software (Softgenetics, College Station, PA, USA).
Quantitative real-time PCR
Quantitative real-time PCR was performed in cDNA isolated from two TMEM127-mutant RCCs and four RCCs without TMEM127 mutations for comparison, and from MEFs. Quantitative real-time-quantitative PCR was performed in triplicate using the StepOnePlus System (Applied Biosystems/Life Technologies). The TATA-box binding protein (TBP) gene was used as a reference for calculating 2−ΔΔ Ct as previously reported (45). Expression levels from non-TMEM127-mutant samples were normalized to one for plotting. t-student's test was used for comparison. Primer sequences are available upon request.
Clones and constructs
The four mutations detected in RCC were cloned in fusion with GFP (pEGFP-C2 plasmid) using a PCR-based site-directed mutagenesis protocol using Phusion High-Fidelity DNA Polymerase (ThermoScientific) (8). A WT GFP-TMEM127 construct has been previously described (9). A modified WT GFP-TMEM127 construct was generated by site-directed mutagenesis as above. The primer sequence included mutagenized nucleotides (shown in lower case, GCCGAAGCATCCTGCTCTGAAaATaACaCGaCGCTATGCCTTCGCCCATATC) that span the target site for TMEM127 siRNA oligo#2, while maintaining the same amino acid sequence as the WT coding sequence (GFP-TMEM127SR). Flag-tagged Rab5-WT, Rab5-Q79L (GTP-bound mutant), Rab5-S34N (GDP-bound mutant) and YFP-tagged Rab4-WT constructs were a kind gift from Marino Zerial (Max Planck Institute, Dresden, Germany). Rab4-Q67L (GTP-bound) and Rab4-S22N (GDP-bound mutant) constructs were generated by applying the same mutagenesis protocol described above, using YFP-Rab4WT as template. HA-TMEM127, TMEM127 siRNA and GFP siRNA (Sigma-Aldrich) were as previously described (8).
Cell culture and transfections
Cell lines: 293T, HeLa and MEFs were cultured in DMEM and 10% fetal bovine serum supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin. Amino acid response was measured as previously described (8). Plasmid transfections in 293T, HeLa or MEFs were performed with Lipofectamine 2000 (Life Technologies), following the manufacturer's guidelines. TMEM127 si-RNA oligonucleotides were transfected in 293T or HeLa with RNAiMAX Transfection Reagent (Life Technologies) according to manufacturer's instructions. Knockdown >70% (calculated by real-time PCR, as before (8) or by western blot inspection) in relation to control siRNA or shRNA was obtained 48 or 72 h post-transfection.
Immunoblots and immunoprecipitation
Cells were rinsed once with ice-cold PBS and lysed in ice-cold lysis buffer [40 mm HEPES (pH 7.4)], 2 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate and 0.3% CHAPS or 1% Triton X-100, and one tablet of EDTA-free protease inhibitors per 25 ml). The soluble fractions of cell lysates were isolated by centrifugation. For immunoprecipitations, primary antibodies were added to the lysates and incubated with rotation for 1.5 h at 4°C. Then 60 µl of a 50% slurry of protein G-sepharose was added and the incubation continued for an additional 1 h. This was followed by three washes of the immunoprecipitates that were washed with lysis buffer containing 150 mm NaCl. Immunoprecipitated proteins were denatured by the addition of sample buffer and resolved by SDS-PAGE, and analyzed by immunoblotting as described (8).
Immunofluorescence microscopy
293T, HeLa cells or MEFs were plated on poly-d-lysine-coated glass coverslips, fixed, stained and imaged as previously described (8). Image acquisition of the fluorescence intensity was performed with the Zeiss LSM510 Software 3.2 SP2. Microscope images are representative of at least 50 cells examined in at least three independent experiments. For counting Rab5Q79L-derived enlarged vesicles, images were acquired at 2× zoom. The raw images were converted to TIF files and processed using Metamorph program (Molecular Devices, Downingtown, PA, USA). Enlarged vesicles (defined as ring-like structures >2 µm) were outlined manually. For analysis of endogenous markers EEA1, Rab5, Rab7, RagC and LAMP2, images were smoothened using a 3 × 3 low pass filter and the background was corrected by subtracting 32 × 32 median filtered images from the smoothened images. A threshold was applied to the images and the areas of each segmented objects in the specified range displayed in the graphs were determined using a size filter in the ImageJ program (46). For these analyses 25–80 cells were evaluated from at least three independent experiments. In experiments performed with transfected GFP-TMEM127 mutants, we only chose cells expressing moderately intense levels of the respective construct, and 10–19 cells per condition in three independent experiments.
Generation of Tmem127 knockout mouse
Tmem127 was targeted by introduction of a neomycin (neo) cassette flanked by frt sites and a loxP recognition sequence in intron 2, and an IRES-GFP cassette and loxP site with flanking diagnostic BglII site in the 3′UTR, (Supplementary Material, Fig. S3a), resulting in the removal of exons 3 and 4, a region that encodes Tmem127′s three transmembrane domains (generated by Vector Biolabs, Philadelphia, PA, USA). A 14.9-kbp fragment was electroporated into the embryonic stem (ES) cell line Bruce4 (C57BL/6-derived) ES cells using standard procedures at the University of Michigan Transgenic Core facilities. ES cell colonies were selected and identified by Southern blot and confirmed by PCR amplification (Supplementary Material, Fig. S3b). A recombinant clone was identified and used for microinjection into blastocysts and transferred into pseudo-pregnant females to obtain chimeric mice. Germline transmission was achieved from breeding of two founder male chimeras. By crossbreeding the ubiquitously expressed CMV-Cre transgenic mice of C57BL/6J background (47) with Tmem127loxP/loxP mice and subsequent breeding of CMV-Cre;Tmem127−loxP/+ F1 offspring with Tmem127loxP/loxP mice, biallelic and monoallelic all-tissue Tmem127 knockout mice were generated and verified by PCR (Supplementary Material, Fig. S3c). Cre-negative Tmem127+/+, or in some cases, Tmem127+/− littermates were used as controls. Mice were housed according to UTHSCSA guidelines, and procedures were approved by the Institutional Animal Care and Use Committee (IACUC), in compliance with the standards for the use of laboratory animals.
MEF generation
MEFs from E13.5 embryos of Tmem127+/+ or Tmem127−/− genotype, originated from breeding of Cre- negative Tmem127−/+ heterozygote pairs, were prepared by digestion with trypsin and collagenase, followed by mechanical disaggregation, as previously described (48). Genotypes were assigned by PCR using the primers shown in Supplementary Material, Figure S3. All experiments were performed between passages 3 and 7 of culture. Genotypes were confirmed in cell lysates from WT and null cells (Supplementary Material, Fig. S3d).
Statistical analysis
Error bars are depicted as SEM. Statistical significance was determined via Student's t-test calculated using Prism 5 (Graph Pad). For multiple comparison analyses an ANOVA was first performed followed by a Tukey difference test. Statistical significance, defined by P values of < 0.05 or < 0.01, is denoted in figures by one or two asterisks, respectively. Results are representative of at least three independent experiments or as indicated in the figures.
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by grants from the Cancer Prevention and Research Institute of Texas (CPRIT #RP110202), Department of Defense CDMRP (#W81XWH-12-1-0508), Voelcker Fund, Alex's Lemonade Foundation, Concern Foundation and Greehey Children's Cancer Research Institute (GCCRI) to P.L.M.D. Images were generated in the Core Optical Imaging Facility which is supported by UTHSCSA, NIH-NCI P30 CA54174 (CTRC at UTHSCSA) and NIH-NIA P01AG19316.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ricardo Aguiar for many insights and helpful suggestions, and Jim Brugarolas and Kris Vogel for their valuable comments. We are grateful to Shoulei Jiang and Li Yao for technical assistance, Vickie Frohlich and Jim Wewer for optical imaging advice, Bruce Nicholson and Eddy Kalmykov for assistance with the Metamorph software and Christian Bowman-Colin for advice with the mouse colony. Rab4 and Rab5 constructs were kind gifts from Marino Zerial, Max Planck Institute, Dresden, Germany.
Conflict of Interest statement. The authors report no conflict of interests.
REFERENCES
- 1.Latif F., Tory K., Gnarra J., Yao M., Duh F.-M., Orcutt M.-L., Stackhouse T., Kuzmin I., Modi W., Geil L., et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 2.Nickerson M.L., Warren M.B., Toro J.R., Matrosova V., Glenn G., Turner M.L., Duray P., Merino M., Choyke P., Pavlovich C.P., et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt L., Duh F.M., Chen F., Kishida T., Glenn G., Choyke P., Scherer S.W., Zhuang Z., Lubensky I., Dean M., et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 4.Ricketts C., Woodward E.R., Killick P., Morris M.R., Astuti D., Latif F., Maher E.R. Germline SDHB mutations and familial renal cell carcinoma. J. Natl.Cancer. Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson I.P., Alam N.A., Rowan A.J., Barclay E., Jaeger E.E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 6.Maher E.R. Genetics of familial renal cancers. Nephron Exp. Nephrol. 2011;118:e21–e26. doi: 10.1159/000320892. [DOI] [PubMed] [Google Scholar]
- 7.Vanharanta S., Buchta M., McWhinney S.R., Virta S.K., Peczkowska M., Morrison C.D., Lehtonen R., Januszewicz A., Jarvinen H., Juhola M., et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am. J. Hum. Genet. 2004;74:153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y., Yao L., King E.E., Buddavarapu K., Lenci R.E., Chocron E.S., Lechleiter J.D., Sass M., Aronin N., Schiavi F., et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat. Genet. 2010;42:229–233. doi: 10.1038/ng.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao L., Schiavi F., Cascon A., Qin Y., Inglada-Perez L., King E.E., Toledo R.A., Ercolino T., Rapizzi E., Ricketts C.J., et al. Spectrum and prevalence of FP/TMEM127 gene mutations in pheochromocytomas and paragangliomas. JAMA. 2010;304:2611–2619. doi: 10.1001/jama.2010.1830. [DOI] [PubMed] [Google Scholar]
- 10.Casaletto J.B., McClatchey A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer. 2012;12:387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenmark H., Parton R.G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 13.Flinn R.J., Yan Y., Goswami S., Parker P.J., Backer J.M. The late endosome is essential for mTORC1 signaling. Mol. Biol. Cell. 2010;21:833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., Kim E., Yuan H., Inoki K., Goraksha-Hicks P., Schiesher R.L., Neufeld T.P., Guan K.L. Regulation of mTORC1 by the Rab and Arf GTPases. J. Biol. Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges D., Fisher K., Zolov S.N., Xiong T., Inoki K., Weisman L.S., Saltiel A.R. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. J. Biol. Chem. 2012;287:20913–20921. doi: 10.1074/jbc.M111.334060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng C. SDHB—a gene for all tumors? J. Natl. Cancer Inst. 2008;100:1193–1195. doi: 10.1093/jnci/djn263. [DOI] [PubMed] [Google Scholar]
- 20.McBride H.M., Rybin V., Murphy C., Giner A., Teasdale R., Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 21.Zeigerer A., Gilleron J., Bogorad R.L., Marsico G., Nonaka H., Seifert S., Epstein-Barash H., Kuchimanchi S., Peng C.G., Ruda V.M., et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 22.Miaczynska M., Pelkmans L., Zerial M. Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell. Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Wiley H.S., Burke P.M. Regulation of receptor tyrosine kinase signaling by endocytic trafficking. Traffic. 2001;2:12–18. doi: 10.1034/j.1600-0854.2001.020103.x. [DOI] [PubMed] [Google Scholar]
- 24.Lanzetti L., Rybin V., Malabarba M.G., Christoforidis S., Scita G., Zerial M., Di Fiore P.P. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Roche O., Yan M.S., Finak G., Evans A.J., Metcalf J.L., Hast B.E., Hanna S.C., Wondergem B., Furge K.A., et al. Regulation of endocytosis via the oxygen-sensing pathway. Nat. Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- 26.Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell. Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 28.Pena-Llopis S., Vega-Rubin-de-Celis S., Schwartz J.C., Wolff N.C., Tran T.A., Zou L., Xie X.J., Corey D.R., Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., Facchinetti V., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cang C., Zhou Y., Navarro B., Seo Y.J., Aranda K., Shi L., Battaglia-Hsu S., Nissim I., Clapham D.E., Ren D. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell. 2013;152:778–790. doi: 10.1016/j.cell.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis I.J., Hsi B.L., Arroyo J.D., Vargas S.O., Yeh Y.A., Motyckova G., Valencia P., Perez-Atayde A.R., Argani P., Ladanyi M., et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc. Natl Acad. Sci. USA. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuiper R.P., Schepens M., Thijssen J., van Asseldonk M., van den Berg E., Bridge J., Schuuring E., Schoenmakers E.F., van Kessel A.G. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 2003;12:1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 34.Petit C.S., Roczniak-Ferguson A., Ferguson S.M. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell. Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creighton C.J., Morgan M., Gunaratne P.H., Wheeler D.A., Gibbs R.A., Gordon Robertson A., Chu A., Beroukhim R., Cibulskis K., Signoretti S., et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalgliesh G.L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C., et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 38.Burnichon N., Vescovo L., Amar L., Libe R., de Reynies A., Venisse A., Jouanno E., Laurendeau I., Parfait B., Bertherat J., et al. Integrative genomic analysis reveals somatic mutations in pheochromocytoma and paraganglioma. Hum. Mol. Genet. 2011;20:3974–3985. doi: 10.1093/hmg/ddr324. [DOI] [PubMed] [Google Scholar]
- 39.Scott K.L., Kabbarah O., Liang M.C., Ivanova E., Anagnostou V., Wu J., Dhakal S., Wu M., Chen S., Feinberg T., et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collinet C., Stoter M., Bradshaw C.R., Samusik N., Rink J.C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M.S., et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y., Hedman A.C., Tan X., Schill N.J., Anderson R.A. Endosomal type Igamma PIP 5-kinase controls EGF receptor lysosomal sorting. Dev Cell. 2013;25:144–155. doi: 10.1016/j.devcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Er E.E., Mendoza M.C., Mackey A.M., Rameh L.E., Blenis J. AKT facilitates EGFR trafficking and degradation by phosphorylating and activating PIKfyve. Sci. Signal. 2013;6:ra45. doi: 10.1126/scisignal.2004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joffre C., Barrow R., Menard L., Calleja V., Hart I.R., Kermorgant S. A direct role for Met endocytosis in tumorigenesis. Nat. Cell. Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- 44.Astuti D., Ricketts C.J., Chowdhury R., McDonough M.A., Gentle D., Kirby G., Schlisio S., Kenchappa R.S., Carter B.D., Kaelin W.G., Jr., et al. Mutation analysis of HIF prolyl hydroxylases (PHD/EGLN) in individuals with features of phaeochromocytoma and renal cell carcinoma susceptibility. Endocr. Relat. Cancer. 2011;18:73–83. doi: 10.1677/ERC-10-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Rondanino C., Rojas R., Ruiz W.G., Wang E., Hughey R.P., Dunn K.W., Apodaca G. RhoB-dependent modulation of postendocytic traffic in polarized Madin-Darby canine kidney cells. Traffic. 2007;8:932–949. doi: 10.1111/j.1600-0854.2007.00575.x. [DOI] [PubMed] [Google Scholar]
- 47.Schwenk F., Baron U., Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. In: Frederick M.A., editor. Current Protocols in Molecular Biology. 2005. Wiley Copyright by John Wiley & Sons, Inc., or related companies, Chapter 28, Unit 28 21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.