Abstract
Objective
This meta-analysis was performed to evaluate the role of toll-like receptor 4 (TLR-4) in colorectal carcinogenesis.
Methods
The PubMed, CISCOM, CINAHL, Web of Science, Google Scholar, EBSCO, Cochrane Library, and CBM databases were searched from inception through November 1st, 2013 without language restrictions. Odds ratios (ORs) or standardized mean differences (SMD) with their 95% confidence intervals (CI) were calculated.
Results
Fourteen case-control studies met the inclusion criteria for this meta-analysis. A total of 1,209 colorectal cancer (CRC) cases and 1,218 healthy controls were involved in this meta-analysis. Two common polymorphisms (299 A>G and 399 C>T) in the TLR-4 gene, TLR-4 mRNA and protein expression were assessed. Our meta-analysis results revealed that the TLR-4 399 C>T polymorphism might increase the risk of CRC (allele model: OR = 1.77, 95%CI = 1.32∼2.36, P<0.001; dominant model: OR = 1.83, 95%CI = 1.32∼2.52, P<0.001; respectively). However, we found no correlation between the TLR-4 299 A>G polymorphism and CRC risk (all P>0.05). A subgroup analysis by ethnicity suggested that TLR-4 genetic polymorphisms were associated with an increased risk of CRC among Asians (allele model: OR = 1.50, 95%CI = 1.19∼1.88, P = 0.001; dominant model: OR = 1.49, 95%CI = 1.16∼1.92, P = 0.002; respectively), but not among Caucasians and Africans (all P>0.05). Furthermore, our results showed that TLR-4 mRNA and protein levels in CRC patients were higher than those in healthy controls (TLR-4 mRNA: SMD = 2.51, 95%CI = 0.98∼4.05, P = 0.001; TLR-4 protein: OR = 4.75, 95%CI = 1.16∼19.36, P = 0.030; respectively).
Conclusion
Our findings provide empirical evidence that TLR-4 may play an important role in colorectal carcinogenesis. Thus, TLR-4 is a promising potential biomarker for the early diagnosis of CRC.
Introduction
Colorectal cancer (CRC), which includes colon and rectal cancers, is the third most commonly diagnosed cancer in males and the second in females [1], [2]. The symptoms of CRC typically include rectal bleeding and anemia, which are sometimes associated with weight loss and changes in bowel habits [3]. It has been recognized that colorectal cancer is a multifactorial disease caused by complex interactions between environmental and genetic factors [4]. Risk factors for CRC consist of high intakes of fat and alcohol, obesity, smoking and lack of physical exercise [5]. Nowadays, many candidate genes have been identified, such as toll-like receptor 4 (TLR-4), which may be implicated in the genesis of colorectal cancer [6], [7].
TLR-4 belongs to a family of Toll-like receptors (TLRs), which are receptors playing the main role in the recognition of a wide array of pathogens, including viruses, bacteria, protozoa, and fungi [8]. The human TLR-4 gene is located in chromosomal region 9 (9q32-q33) and consists of four exons and three introns with an overall length of approximately 19 kb [9]. As one of the most actively investigated TLR, TLR-4 has been implicated in signal transduction events induced by the lipopolysaccharide of gram-negative bacteria and its activation in the production of several pro-inflammatory, antiviral and anti-bacterial cytokines [10], [11]. Since TLR-4 is critical in immune and inflammation responses to various bacteria in the intestine [12], single nucleotide polymorphisms (SNPs) in TLR-4 may decrease the response to bacterial components, impact gut homeostasis, result in the impairment of TLRs activation, and thereby be conducive to the development of several inflammatory diseases including CRC [13], [14]. Furthermore, human CRC cell lines with high rates of microsatellite instability were found to stimulate the activation of TLR-4 through the release of cytokines, and elevate the level of TLR-4 mRNA, thereby inducing the pathogenesis of CRC [15]. Therefore, it was hypothesized that TLR-4 might play important roles in the development and progression of CRC [16]. Additionally, it is also worth noting that numerous studies have been conducted to investigate the potential associations between common polymorphisms in the TLR-4 gene and CRC risk, especially focusing on two SNPs, (299 A>G, rs4986790 and 399 C>T, rs4986791), which are located in the extracellular domain of TLR-4 [8], [10], [17]. These two important SNPs may alter the amino acid sequence of the TLR-4 protein, disrupting the normal structure of the extracellular domain of the TLR-4 and TLR-4 signaling [14]. The dysregulation of TLR-4 signaling may change the ligand binding and balance between pro- and anti-inflammatory cytokines, creating a pro-inflammatory environment that favors tumor growth, thereby modulating the risk of CRC [10]. However, previous studies have arrived at contradictory results [10], [13], [18]. Consequently, we performed the present meta-analysis to evaluate the exact role of TLR-4 in colorectal carcinogenesis.
Methods
Literature search
The PubMed, CISCOM, CINAHL, Web of Science, Google Scholar, EBSCO, Cochrane Library, and CBM databases were searched for relevant articles published before November 1st, 2013 without any language restrictions. The following keywords and MeSH terms were used: [“colorectal cancer” or “CRC” or “colorectal tumor” or “colorectal neoplasm” or “colorectal carcinogenesis” or “colon cancer” or “rectal cancer”] and [“toll-like receptor 4” or “TLR-4” or “toll-4 receptor” or “toll 4 receptor”]. We also performed a manual search to find other potential articles.
Selection criteria
The included studies had to meet all the following criteria: (1) the study must be clinical cohort or case-control study; (2) the study must relate to the role of TLR-4 in colorectal carcinogenesis; (3) all patients must conform to the diagnostic criteria of CRC; (4) the study must provide sufficient information about TLR-4 SNP frequencies, mRNA or protein expressions. If the study did not meet the inclusion criteria, it was excluded. When authors published several studies using the same subjects either the most recent or the largest sample size publication was included. The supporting PRISMA checklist is available as supplementary information; see Checklist S1.
Data extraction
Relevant data were systematically extracted from all included studies by two researchers using a standardized form. The researchers collected the following data: language of publication, publication year, the first author's surname, geographical location, design of study, sample size, the source of the subjects, SNP frequencies, mRNA/protein levels, source of samples, genotyping method, mRNA/protein detection method, etc.
Quality assessment
Methodological quality was independently assessed by two researchers according to the Newcastle-Ottawa Scale (NOS) criteria [19]. The NOS criteria assigns scores based on three aspects: (1) subject selection: 0∼4; (2) comparability of subject: 0∼2; (3) clinical outcome: 0∼3. Total NOS scores range from 0 to 9 with a score ≥7 indicating high quality. The supporting NOS score criterion is available in Supplement S1.
Statistical analysis
The STATA version 12.0 (Stata Corp, College Station, TX, USA) software was used for this meta-analysis. We calculated crude odds ratio (OR) with their 95% confidence interval (95%CI) to evaluate the specified relationships. The Z test was used to estimate the statistical significance of pooled statistics. The Cochran's Q-statistic and I2 test were used to evaluate potential heterogeneity between studies [20]. If Q-test showed a P<0.05 or I2 test exhibited >50%, indicating significant heterogeneity, the random-effect model was conducted; otherwise the fixed-effects model was used. We also performed subgroup and meta-regression analyses to investigate potential sources of heterogeneity. In order to evaluate the influence of single studies on overall estimates, a sensitivity analysis was performed. We also conducted Begger's funnel plots and Egger's linear regression test to investigate publication bias [21].
Results
Characteristics of included studies
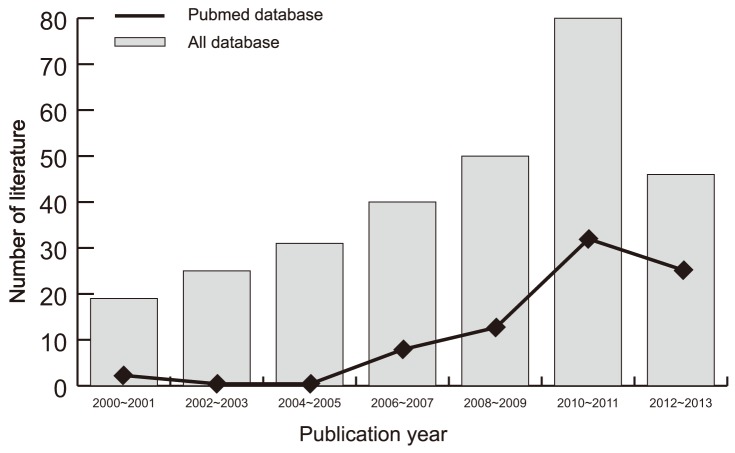
Initially, the searched keywords identified 291 articles. We reviewed the titles and abstracts of all articles and excluded 156 articles; full texts and data integrity were then reviewed and a further 121 articles were excluded. Finally, 14 case-control studies were included in this meta-analysis [10], [13], [14], [18], [22]–[31]. Publication years of the eligible studies range from 2006 to 2013. Figure 1 shows the selection process of eligible articles. The distribution of the number of topic-related literature in electronic databases over the last decade is shown in Figure 2. A total of 2,427 subjects were involved in this meta-analysis, including 1,209 CRC patients and 1,218 healthy controls. Eleven studies were performed in Asian populations, two studies in Caucasian populations and only one study in an African population. The NOS scores of all included studies were ≥5 (moderate-high quality). We summarized the study characteristics and methodological quality in Table 1.
Figure 1. Flow chart of literature search and study selection.
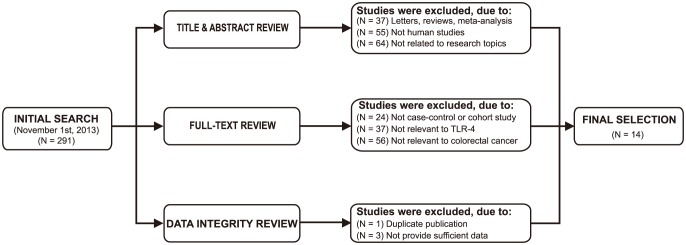
Fourteen case-control studies were included in this meta-analysis.
Figure 2. Distribution of the number of topic-related literatures in electronic databases over the last decade.
Table 1. Main characteristics and methodological quality of all eligible studies.
| First author [Ref] | Year | Country | Ethnicity | Sample size | Gender (male/female) | Age (years) | Marker | Detection method | NOS score | |||
| Case | Control | Case | Control | Case | Control | |||||||
| Pimentel-Nunes P [13] | 2013 | Portugal | Caucasian | 193 | 278 | 123/70 | 176/102 | 62.0±7.0 | 56.0±4.0 | 299 A>G | PCR-RFLP | 7 |
| Omrane I [10] | 2013 | Tunisia | African | 100 | 140 | 53/47 | 45/95 | 58.2±14.4 | 56.2±15.7 | 299 A>G | Direct sequencing | 8 |
| 399 C>T | Direct sequencing | 8 | ||||||||||
| Dai Q [21] | 2012 | China | Asian | 268 | 268 | - | - | - | - | 299 A>G | PCR-RFLP | 6 |
| Yang GG [22] | 2011 | China | Asian | 102 | 87 | 64/38 | 52/35 | 52 | 50 | 299 A>G | Direct sequencing | 6 |
| 399 C>T | Direct sequencing | 6 | ||||||||||
| Boraska Jelavic T [17] | 2006 | Croatia | Caucasian | 89 | 88 | 61/28 | 61/67 | 61.5±9.5 | 61.5±9.5 | 299 A>G | PCR-RFLP | 6 |
| 399 C>T | PCR-RFLP | 6 | ||||||||||
| Davoodi H [14] | 2011 | Malaysia | Asian | 60 | 50 | 30/30 | 25/25 | 30∼50 | 30∼50 | 299 A>G | PCR-RFLP | 6 |
| 399 C>T | PCR-RFLP | 6 | ||||||||||
| Nihon-Yanagi Y [28] | 2012 | Japan | Asian | 50 | 50 | 22/28 | 68 (52∼90) | mRNA | RT-PCR | 7 | ||
| Yu YH [30] | 2011 | China | Asian | 62 | 25 | - | - | mRNA | RT-PCR | 6 | ||
| Huang HY [26] | 2010 | China | Asian | 63 | 63 | 35/28 | - | mRNA | RT-PCR | 7 | ||
| Jin HM [27] | 2009 | China | Asian | 24 | 24 | 11/13 | 67 (35∼86) | mRNA | RT-PCR | 8 | ||
| Tian F [29] | 2008 | China | Asian | 30 | 30 | 18/12 | 64 (51∼82) | mRNA | RT-PCR | 8 | ||
| Hu KF [25] | 2013 | China | Asian | 40 | 20 | 27/13 | 32∼80 | Protein | SP | 8 | ||
| Cheng ZL [23] | 2012 | China | Asian | 58 | 25 | 38/20 | 22∼83 | Protein | SP | 8 | ||
| Guo YW [24] | 2007 | China | Asian | 70 | 70 | 42/28 | 33∼71 | Protein | Flow cytometry | 8 | ||
Legend: PCR-RFLP - polymerase chain reaction-restriction fragment length polymorphism; NOS - the Newcastle-Ottawa Scale.
TLR-4 genetic polymorphisms with CRC risk
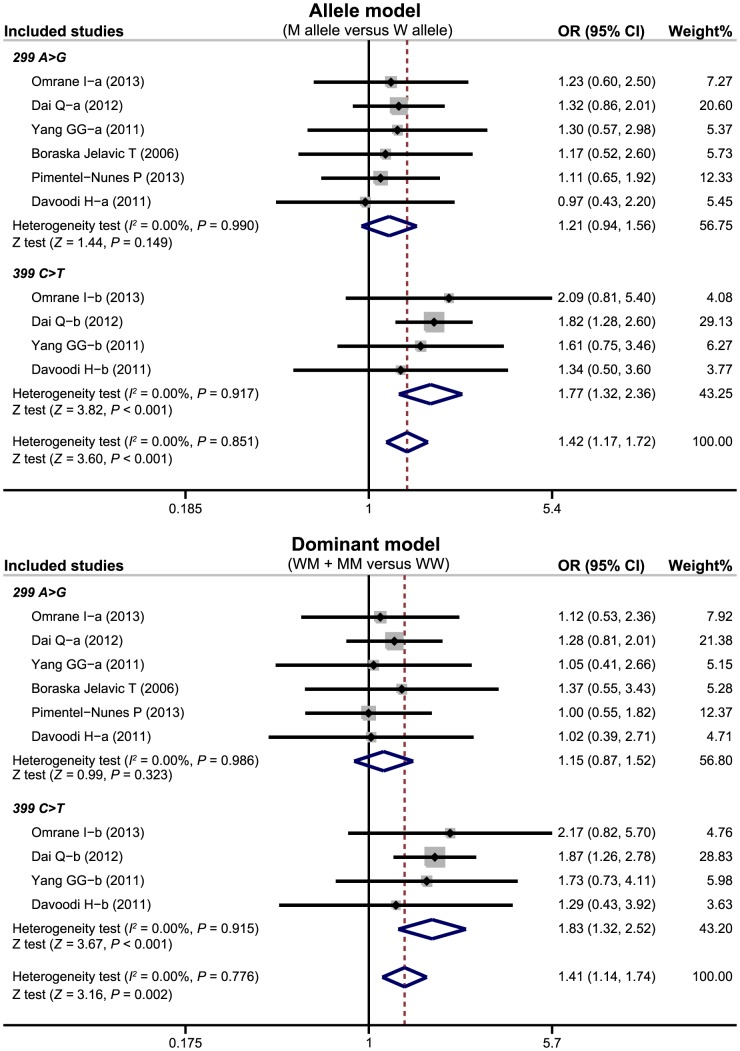
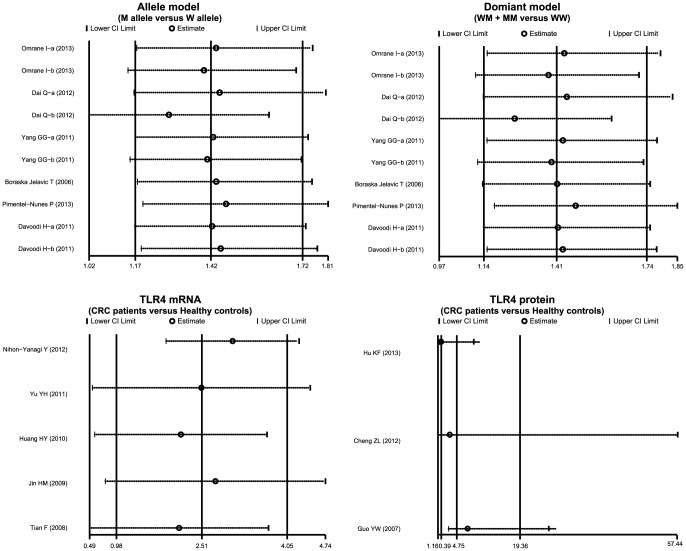
There were six studies focused on the correlation of TLR-4 genetic polymorphisms with susceptibility to CRC. Our meta-analysis findings on the relationships between TLR-4 genetic polymorphisms and the risk of CRC are shown in Table 2. The random effects model was conducted due to the existence of significant heterogeneity between studies. Two common polymorphisms in the TLR-4 gene (299 A>G, rs4986790; 399 C>T, rs4986791) were assessed. Our meta-analysis results revealed that the TLR-4 399 C>T polymorphism might increase the risk of CRC (allele model: OR = 1.77, 95%CI: 1.32∼2.36, P<0.001; dominant model: OR = 1.83, 95%CI: 1.32∼2.52, P<0.001; respectively) (Figure 3). Nevertheless, there was no evidence to support any association between the TLR-4 299 A>G polymorphism and CRC risk (all P>0.05).
Table 2. Meta-analysis of the associations between TLR-4 genetic polymorphisms and colorectal cancer risk.
| M allele vs. W allele | WM + MM vs. WW | MM vs. WW + WM | MM vs. WW | MM vs. WM | |||||||||||
| (Allele model) | (Dominant model) | (Recessive model) | (Homozygous model) | (Heterozygous model) | |||||||||||
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| Overall | 1.42 | 1.17∼1.72 | <0.001 | 1.41 | 1.14∼1.74 | 0.002 | 1.85 | 1.00∼3.44 | 0.051 | 1.93 | 1.04∼3.59 | 0.038 | 1.61 | 0.82∼3.13 | 0.163 |
| SNP type | |||||||||||||||
| 299 A>G | 1.21 | 0.94∼1.56 | 0.149 | 1.15 | 0.87∼1.52 | 0.323 | 1.65 | 0.77∼3.54 | 0.198 | 1.67 | 0.78∼3.59 | 0.187 | 1.61 | 0.70∼3.69 | 0.257 |
| 399 C>T | 1.77 | 1.32∼2.36 | <0.001 | 1.83 | 1.32∼2.52 | <0.001 | 2.32 | 0.80∼6.72 | 0.120 | 2.54 | 0.88∼7.38 | 0.086 | 1.60 | 0.52∼4.93 | 0.417 |
| Ethnicity | |||||||||||||||
| Africa | 1.49 | 0.84∼2.62 | 0.173 | 1.44 | 0.77∼2.72 | 0.256 | 3.63 | 0.17∼76.40 | 0.407 | 3.63 | 0.17∼76.57 | 0.407 | 3.65 | 0.16∼82.33 | 0.416 |
| Asians | 1.50 | 1.19∼1.88 | 0.001 | 1.49 | 1.19∼1.92 | 0.002 | 2.01 | 0.96∼4.21 | 0.065 | 2.11 | 1.01∼4.44 | 0.048 | 1.68 | 0.76∼3.72 | 0.202 |
| Caucasians | 1.13 | 0.72∼1.78 | 0.539 | 1.10 | 0.66∼1.81 | 0.719 | 1.33 | 0.37∼4.82 | 0.664 | 1.36 | 0.40∼4.61 | 0.625 | 1.14 | 0.17∼7.46 | 0.895 |
| Genotyping method | |||||||||||||||
| Non-PCR-RFLP | 1.37 | 0.88∼2.12 | 0.166 | 1.26 | 0.78∼2.04 | 0.351 | 2.15 | 0.60∼7.69 | 0.238 | 2.21 | 0.62∼7.90 | 0.223 | 1.85 | 0.46∼7.38 | 0.382 |
| PCR-RFLP | 1.44 | 1.16∼1.78 | 0.001 | 1.44 | 1.14∼1.82 | 0.002 | 1.77 | 0.87∼3.60 | 0.115 | 1.85 | 0.91∼3.76 | 0.090 | 1.54 | 0.72∼3.30 | 0.267 |
| Sample size | |||||||||||||||
| Large (n > 200) | 1.48 | 1.19∼1.86 | 0.001 | 1.43 | 1.10∼1.87 | 0.008 | 3.02 | 1.24∼7.38 | 0.015 | 3.17 | 1.30∼7.76 | 0.011 | 2.60 | 1.03∼6.54 | 0.043 |
| Small (n ≤ 200) | 1.26 | 0.87∼1.83 | 0.217 | 1.29 | 0.84∼1.97 | 0.246 | 1.18 | 0.50∼2.78 | 0.709 | 1.21 | 0.51∼2.87 | 0.660 | 0.95 | 0.36∼2.50 | 0.916 |
Legend: W - wild allele; M - mutant allele; WW - wild homozygote; WM - heterozygote; MM - mutant homozygote; OR - odds ratio; 95%CI - 95% confidence interval; SNP - single nucleotide polymorphism; PCR-RFLP - polymerase chain reaction-restriction fragment length polymorphism.
Figure 3. Forest plots for the relationships of TLR-4 genetic polymorphisms with the risk of colorectal cancer under the allele and dominant models.
(a) TLR-4 299 A>G; (b) TLR-4 399 C>T.
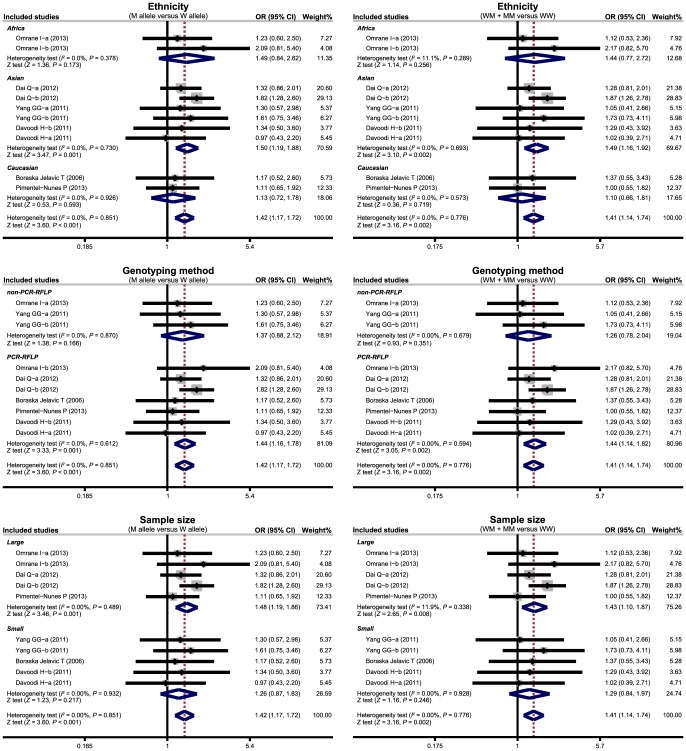
Subgroup analyses were conducted based on ethnicity, genotyping method and sample size to investigate potential sources of heterogeneity. Our findings suggested that genetic polymorphisms in TLR-4 genes were associated with an increased risk of CRC among Asian populations (allele model: OR = 1.50, 95%CI = 1.19∼1.88, P = 0.001; dominant model: OR = 1.49, 95%CI = 1.16∼1.92, P = 0.002; respectively), but not among Caucasian or African populations (all P>0.05) (Figure 4). We also found that TLR-4 genetic polymorphisms were closely linked to CRC risk in the PCR-RFLP subgroup (allele model: OR = 1.44, 95%CI = 1.16∼1.78, P = 0.001; dominant model: OR = 1.44, 95%CI = 1.14∼1.82, P = 0.002; respectively) and the large sample-size subgroup (allele model: OR = 1.48, 95%CI = 1.19∼1.86, P = 0.001; dominant model: OR = 1.43, 95%CI = 1.10∼1.87, P = 0.008; respectively), but no similar correlations were observed in non-PCR-RFLP or small sample-size subgroups (all P>0.05). Our meta-regression analyses suggested that ethnicity was a major source of heterogeneity (as shown in Table 3).
Figure 4. Subgroup analyses by ethnicity and genotyping method of the relationships of TLR-4 genetic polymorphisms with the risk of colorectal cancer under the allele and dominant models.
(a) TLR-4 299 A>G; (b) TLR-4 399 C>T.
Table 3. Univariate and multivariate meta-regression analyses of potential source of heterogeneity.
| Heterogeneity factors | Coefficient | SE | Z | P | 95%CI | |
| LL | UL | |||||
| Publication year | ||||||
| Univariate | 0.026 | 0.063 | 0.42 | 0.678 | −0.097 | 0.149 |
| Multivariate | 0.040 | 0.097 | −0.42 | 0.678 | −0.230 | 0.150 |
| SNP type | ||||||
| Univariate | 0.381 | 0.197 | 1.93 | 0.053 | −0.006 | 0.781 |
| Multivariate | 0.376 | 0.208 | 1.81 | 0.071 | −0.032 | 0.783 |
| Ethnicity | ||||||
| Univariate | −0.067 | 0.143 | −0.47 | 0.638 | −0.347 | 0.213 |
| Multivariate | 0.031 | 0.163 | −0.19 | 0.851 | −0.350 | 0.289 |
| Genotyping method | ||||||
| Univariate | −0.050 | 0.250 | −0.20 | 0.840 | −0.540 | 0.439 |
| Multivariate | 0.137 | 0.323 | 0.42 | 0.672 | −0.496 | 0.770 |
| Sample size | ||||||
| Univariate | −0.161 | 0.221 | −0.73 | 0.467 | −0.595 | 0.273 |
| Multivariate | −0.284 | 0.382 | −0.74 | 0.457 | −1.032 | 0.467 |
Legend: SE - standard error; 95%CI - 95% confidence interval; UL - upper limit; LL - lower limit; SNP - single nucleotide polymorphism.
Expression of TLR-4 mRNA and protein in colorectal carcinogenesis
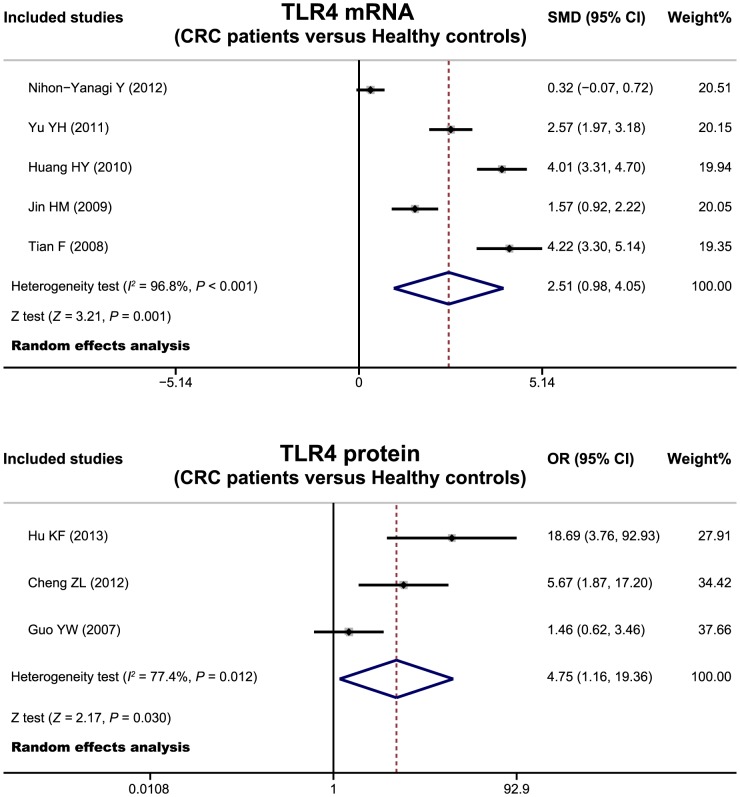
Five studies evaluated the potential role of TLR-4 mRNA expression in colorectal carcinogenesis. The pooled results of these studies indicated that CRC patients had a higher levels of TLR-4 mRNA than those of healthy controls (SMD = 2.51, 95%CI = 0.98∼4.05, P = 0.001) (Figure 5). In addition, three studies reported correlations between TLR-4 protein expression and colorectal carcinogenesis. Meta-analysis results also showed a significant difference in TLR-4 protein levels between CRC patients and healthy controls (OR = 4.75, 95%CI = 1.16∼19.36, P = 0.030).
Figure 5. Forest plots for the relationships of TLR-4 mRNA and protein expression with colorectal carcinogenesis.
Sensitivity analysis and publication bias evaluation
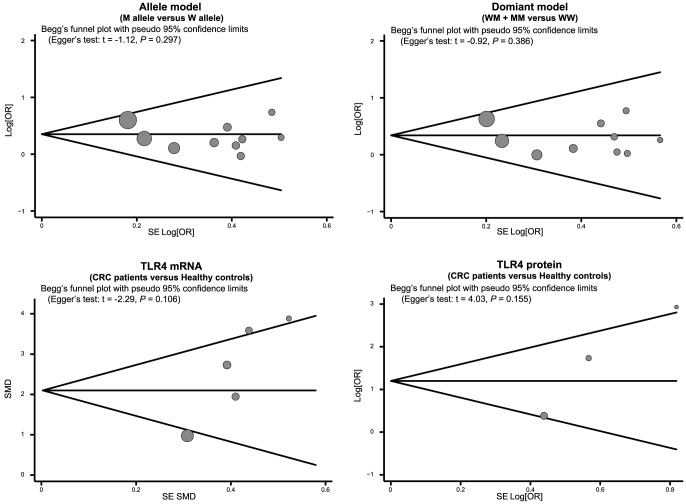
The results of a sensitivity analysis suggested that no single study significantly influenced overall pooled estimates (Figure 6). We also found no evidence of obvious asymmetry in Begger's funnel plots (Figure 7). Egger's test also did not display strong statistical evidence of publication bias (allele mode: t = −1.12, P = 0.297; dominant model: t = −0.92, P = 0.386; TLR-4 mRNA: t = −2.29, P = 0.106; TLR-4 protein: t = 4.03, P = 0.155; respectively).
Figure 6. Sensitivity analysis of the summary odds ratio coefficients on the relationships of TLR-4 genetic polymorphisms, TLR-4 mRNA and protein expression with colorectal carcinogenesis.
(a) TLR-4 299 A>G; (b) TLR-4 399 C>T.
Figure 7. Begger's funnel plot of publication biases on the relationships of TLR-4 genetic polymorphisms, TLR-4 mRNA and protein expression with colorectal carcinogenesis.
Discussion
TLR-4, a member of the IL-1R (IL-1 receptor) superfamily, shares the cytoplasmic region known as the TIR (Toll/IL-1R) domain [32]. TLRs have been suggested to play a key role in the recognition of pathogens, including protozoa, viruses, bacteria, and fungi, and thus are involved in innate and adaptive immune response processes [33], [34]. In general, TLR-4 is highly expressed in monocytes, macrophages, lymphocytes, and dendritic cells, and can also be observed in many types of epithelial and endothelial cells [35], [36]. It is widely accepted that epithelial cells of the intestine are responsible for regulating the immune response to bacterial antigen, while the sustained activation of the TLR-4 signaling pathway accompanied with an elevated levels of TLR-4 mRNA may stimulate the expression of TLR-4 and in turn act as a critical mechanism for the acquisition of malignant phenotypes by epithelial cancer cells [37]. Consequently, genetic polymorphisms in TLR-4 may promote the synthesis of proinflammatory cytokines, which are causative factors in the pathogenesis of CRC 38–40. Furthermore, the expression of TLR-4 mRNA and protein has also been postulated to be strongly associated with colorectal carcinogenesis [41].
In order to evaluate the exact role of TLR-4 in colorectal carcinogenesis, we performed a meta-analysis of 14 case-control studies with a total of 1,209 CRC patients and 1,218 healthy controls. Our meta-analysis results indicated that the TLR-4 399 C>T polymorphism was significantly associated with the risk of CRC, which suggests that this polymorphism may be involved in the development of CRC. However, we found no evidence of any associations between the TLR-4 299 A>G polymorphism and CRC risk, revealing that this polymorphism may not be an important determinant of colorectal carcinogenesis. Although the exact function of TLR-4 genetic polymorphisms in colorectal carcinogenesis is still not well understood, on possible explanation is that mutations in the TLR-4 gene could decrease cyclooxygenase-2 (COX-2) expression, which is responsible for increasing commensal bacteria, thereby causing inflammation and leading to CRC [42]. In addition, it is well established that SNPs may contribute to amino acid substitutions altering protein function and may also change transcription factor binding motifs, promoting an alternative translation initiation codon, and thereby resulting in a wild-type transcript down-regulation in the promoter region [43], [44]. In fact, 1196C/T and 896A/G are the most studied genetic variations of TLR-4. The nonsynonymous polymorphisms TLR-4 1196C/T (major allele C, minor allele T, rs4986791) and TLR-4 896A/G (major allele A, minor allele G, rs4986790), which are located in the fourth exon, may affect the extracellular domain of TLR4 and lead to the transitions of cytosine-thymine (C-T) and adenine-guanine (A–G), respectively [8]. In turn, such progressions may cause substitutions in amino acids: isoleucine instead of threonine at 399 position (Thr399Ile) and glycine instead of aspartic acid at 299 position (Asp299Gly) [45]. In this regard, TLR-4 SNPs of the transmembrane domain may lead to defects in intracellular receptor transfer that prevent the receptor to be converted to the cell surface where it works, and thereby may correlate with disorders in the immune system related to cancer of the intestinal mucosa [41]. A subgroup analysis indicates that TLR-4 genetic polymorphisms were associated with an increased risk of CRC among Asians, but not among Caucasians or Africans, which suggests that ethnicity might have been a source of heterogeneity. While the molecular basis of ethnic differences in susceptibility to CRC is currently not fully understood, possible sources of such differences could be natural selection and random genetic drift. A meta-regression analysis also confirms that ethnicity might have been a major source of heterogeneity. In addition, we found that TLR-4 mRNA and protein levels in CRC patients were higher than those in healthy controls, revealing that TLR-4 mRNA and protein may be implicated in the development and progression of CRC. These results may be explained by the fact that TLR-4 is a crucial regulator that helps maintain the balance between commensal bacteria in the gut and the mucosal immune system, while the abnormal expression of TLR-4 leads to the breakdown of homeostasis, which may be a key feature in the pathogenesis of CRC [41]. Furthermore, TLR-4 activation may promote the development of CRC by including enhanced COX-2 expression and increased EGFR signaling [46]. All in all, our findings were consistent with previous studies that TLR-4 may play an important role in colorectal carcinogenesis and may be a potential biomarker for the early diagnosis of CRC.
As the first meta-analysis focusing on the role of TLR-4 in colorectal carcinogenesis, our study has some limitations. First, our results lacked sufficient statistical power to assess the exact roles of TLR-4 SNPs, mRNA and protein expression in colorectal carcinogenesis due to relatively small sample sizes. Second, meta-analysis is a retrospective study that may lead to subject selection bias, which thus may have affected the reliability of our results. Third, our meta-analysis failed to obtain original data from included studies, which may have limited a further evaluation of the potential role of TLR-4 in the development and progression of CRC. A fourth limitation of this article is the fact that the significant P-values reported were driven by one single study with a small sample size, which may seriously affect the accuracy and comprehensiveness of this result. In addition, the inclusion criteria of cases and controls were not well defined in all included studies, which might also influence our results. Most importantly, this study was based on a small number of studies with very small sample sizes, especially for epidemiological research, which constrains the general applicability of our findings.
In conclusion, our findings provide empirical evidence that TLR-4 may play an important role in colorectal carcinogenesis. Thus, TLR-4 is a promising potential biomarker for the early diagnosis of CRC. However, due to the limitations mentioned above, more research studies with larger sample size are needed to achieve a more precise statistical analysis.
Supporting Information
PRISMA Checklist.
(DOC)
The Newcastle-Ottawa Scale for assessing methodological quality.
(DOC)
Flow chart of literature search and study selection. Fourteen case-control studies were included in this meta-analysis.
(DOC)
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Funding Statement
This work was supported by the Foundation of Education Department of Liaoning Province (No. 2008781) and the Natural Science Foundation of Liaoning Province (Nos. 201102281 and 20062053). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stein KB, Snyder CF, Barone BB, Yeh HC, Peairs KS, et al. (2010) Colorectal cancer outcomes, recurrence, and complications in persons with and without diabetes mellitus: a systematic review and meta-analysis. Dig Dis Sci 55(7): 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90. [DOI] [PubMed] [Google Scholar]
- 3. Spiller RC, Thompson WG (2010) Bowel disorders. Am J Gastroenterol 105(4): 775–785. [DOI] [PubMed] [Google Scholar]
- 4. Mlecnik B, Tosolini M, Charoentong P, Kirilovsky A, Bindea G, et al. (2010) Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology 138(4): 1429–1440. [DOI] [PubMed] [Google Scholar]
- 5. Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, et al. (2009) The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer 125(1): 171–180. [DOI] [PubMed] [Google Scholar]
- 6. Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, et al. (2009) PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res 69(5): 1851–1857. [DOI] [PubMed] [Google Scholar]
- 7. Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, et al. (2009) Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 9(7): 489–499. [DOI] [PubMed] [Google Scholar]
- 8. Kutikhin AG (2011) Impact of Toll-like receptor 4 polymorphisms on risk of cancer. Hum Immunol 72(2): 193–206. [DOI] [PubMed] [Google Scholar]
- 9. Horie Y, Meguro A, Ota M, Kitaichi N, Katsuyama Y, et al. (2009) Association of TLR4 polymorphisms with Behcet's disease in a Korean population. Rheumatology (Oxford) 48(6): 638–642. [DOI] [PubMed] [Google Scholar]
- 10. Omrane I, Baroudi O, Kourda N, Bignon YJ, Uhrhammer N, et al. (2014) Positive link between variant Toll-like receptor 4 (Asp299Gly and Thr399Ile) and colorectal cancer patients with advanced stage and lymph node metastasis. Tumour Biol 35: 545–551. [DOI] [PubMed] [Google Scholar]
- 11. Soares JB, Pimentel-Nunes P, Roncon-Albuquerque R, Leite-Moreira A (2010) The role of lipopolysaccharide/toll-like receptor 4 signaling in chronic liver diseases. Hepatol Int 4(4): 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hormannsperger G, Haller D (2010) Molecular crosstalk of probiotic bacteria with the intestinal immune system: clinical relevance in the context of inflammatory bowel disease. Int J Med Microbiol 300(1): 63–73. [DOI] [PubMed] [Google Scholar]
- 13. Pimentel-Nunes P, Teixeira AL, Pereira C, Gomes M, Brandao C, et al. (2013) Functional polymorphisms of Toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Dig Liver Dis 45(1): 63–69. [DOI] [PubMed] [Google Scholar]
- 14. Davoodi H, Seow HF (2011) Variant Toll-like receptor4 (Asp299Gly and Thr399Ile alleles) and Toll-like receptor2 (Arg753Gln and Arg677Trp alleles) in colorectal cancer. Iran J Allergy Asthma Immunol 10(2): 91–99. [PubMed] [Google Scholar]
- 15. Xu Y, Xu Q, Yang L, Liu F, Ye X, et al. (2013) Gene expression analysis of peripheral blood cells reveals Toll-like receptor pathway deregulation in colorectal cancer. PLoS One 8(5): e62870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, et al. (2010) High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer 102(5): 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moossavi S, Rezaei N (2013) Toll-like receptor signalling and their therapeutic targeting in colorectal cancer. Int Immunopharmacol 16(2): 199–209. [DOI] [PubMed] [Google Scholar]
- 18. Boraska Jelavic T, Barisic M, Drmic Hofman I, Boraska V, Vrdoljak E, et al. (2006) Microsatelite GT polymorphism in the toll-like receptor 2 is associated with colorectal cancer. Clin Genet 70(2): 156–160. [DOI] [PubMed] [Google Scholar]
- 19. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9): 603–605. [DOI] [PubMed] [Google Scholar]
- 20. Zintzaras E, Ioannidis JP (2005) HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 21(18): 3672–3673. [DOI] [PubMed] [Google Scholar]
- 21. Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L (2006) Comparison of two methods to detect publication bias in meta-analysis. JAMA 295(6): 676–680. [DOI] [PubMed] [Google Scholar]
- 22. Dai Q, Zhong L, Zhang X, Yang ZH (2012) Expression of IL-1α and TNF-α modified by Toll-like receptor 4 genetic polymorphism is associated with colorectal carcinoma. Chin J Pathpphysiol 28(11): 1976–1978. [Google Scholar]
- 23. Yang GG, Qiu JM, Wang X, Shen Z, Shao SX, et al. (2011) Variant Toll-like receptor 4 in colorectal cancer. Chin J Gastrointestin Surg 14(10): 814–815. [Google Scholar]
- 24.Cheng ZL, Li XM, Yang TT, Zhu Q (2012) Expression of TLR 4 in colorectal cancer and its relationships with FasL and EGFR. J Shandong Univ (Health Sci), 50(5): : 92–94,98. [Google Scholar]
- 25. Guo YW, Wen ZF, Zheng FP (2007) Expression of TLR2 and TLR4in colorectal carcinoma. Guangdong Med J 28(9): 1435–1437. [Google Scholar]
- 26. Hu KF, Chang JC, Liu Y, Zhu JS, Zhou ZY, et al. (2013) Expressions of TLR4 and TLR9 in Colorectal Carcinoma Tissues. J Basic Clin Oncol 26(3): 198–201. [Google Scholar]
- 27. Huang HY, Y CW, Liu FF, Zhang QY, Li XZ, et al. (2010) The expression and correlation of TLR4 and NF-κB p65 in colorectal carcinoma. Chin J Labor Med 33(10): 953–957. [Google Scholar]
- 28. Jin HM, Meng QY (2009) A study on the expression of TLR4 in humen colorental cancer by RT-PCR. Modern Practic Med 21(7): 682–684. [Google Scholar]
- 29. Nihon-Yanagi Y, Terai K, Murano T, Matsumoto T, Okazumi S (2012) Tissue expression of Toll-like receptors 2 and 4 in sporadic human colorectal cancer. Cancer Immunol Immunother 61(1): 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian F, Zhang L, Guo QX, Cui B, Zhang J, et al. (2008) Expression and significance of heme oxygenase-1 and Toll-like receptor 2/4 in rectal carcinoma. Chin J Current Adv Gener Surg, 11(3): 248–250,254.
- 31. Yu YH, Qian L, Chen W (2011) Toll-like receptor4 mRNA and myeloid differentiation factor 88 in colorectal cancer. Hainan Med J 22(12): 9–11. [Google Scholar]
- 32. Kang JY, Lee JO (2011) Structural biology of the Toll-like receptor family. Annu Rev Biochem 80: 917–941. [DOI] [PubMed] [Google Scholar]
- 33. Kawai T, Akira S (2009) The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 21(4): 317–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11(5): 373–384. [DOI] [PubMed] [Google Scholar]
- 35. Yu L, Wang L, Li M, Zhong J, Wang Z, et al. (2010) Expression of toll-like receptor 4 is down-regulated during progression of cervical neoplasia. Cancer Immunol Immunother 59(7): 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagos D, Vart RJ, Gratrix F, Westrop SJ, Emuss V, et al. (2008) Toll-like receptor 4 mediates innate immunity to Kaposi sarcoma herpesvirus. Cell Host Microbe 4(5): 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eissmann P, Evans JH, Mehrabi M, Rose EL, Nedvetzki S, et al. (2010) Multiple mechanisms downstream of TLR-4 stimulation allow expression of NKG2D ligands to facilitate macrophage/NK cell crosstalk. J Immunol 184(12): 6901–6909. [DOI] [PubMed] [Google Scholar]
- 38. Iwasaki A, Medzhitov R (2010) Regulation of adaptive immunity by the innate immune system. Science 327(5963): 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gorina R, Font-Nieves M, Marquez-Kisinousky L, Santalucia T, Planas AM (2011) Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFkappaB signaling, MAPK, and Jak1/Stat1 pathways. Glia 59(2): 242–255. [DOI] [PubMed] [Google Scholar]
- 40. Cario E (2010) Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis 16(9): 1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Slattery ML, Herrick JS, Bondurant KL, Wolff RK (2012) Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer 130(12): 2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee JY, Plakidas A, Lee WH, Heikkinen A, Chanmugam P, et al. (2003) Differential modulation of Toll-like receptors by fatty acids: preferential inhibition by n-3 polyunsaturated fatty acids. J Lipid Res 44(3): 479–486. [DOI] [PubMed] [Google Scholar]
- 43. Thomas KH, Meyn P, Suttorp N (2006) Single nucleotide polymorphism in 5'-flanking region reduces transcription of surfactant protein B gene in H441 cells. Am J Physiol Lung Cell Mol Physiol 291(3): L386–390. [DOI] [PubMed] [Google Scholar]
- 44. Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C (2009) Silent (synonymous) SNPs: should we care about them? Methods Mol Biol 578: 23–39. [DOI] [PubMed] [Google Scholar]
- 45. Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ, et al. (2008) Functional consequences of toll-like receptor 4 polymorphisms. Mol Med 14(5-6): 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, et al. (2007) Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology 133(6): 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)
The Newcastle-Ottawa Scale for assessing methodological quality.
(DOC)
Flow chart of literature search and study selection. Fourteen case-control studies were included in this meta-analysis.
(DOC)