Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is the leading cause of death from an infectious disease worldwide. Over the course of its life cycle in vivo, Mtb is exposed to a plethora of environmental stress conditions. Temporal regulation of genes involved in sensing and responding to such conditions is therefore crucial for Mtb to establish an infection. The Rv2745c (clgR) gene encodes a Clp protease gene regulator that is induced in response to a variety of stress conditions and potentially plays a role in Mtb pathogenesis. Our isogenic mutant, Mtb:ΔRv2745c, is significantly more sensitive to in vitro redox stress generated by diamide, relative to wild-type Mtb as well as to a complemented strain. Together with the fact that the expression of Rv2745c is strongly induced in response to redox stress, these results strongly implicate a role for ClgR in the management of intraphagosomal redox stress. Additionally, we observed that redox stress led to the dysregulation of the expression of the σH/σE regulon in the isogenic mutant, Mtb:ΔRv2745c. Furthermore, induction of clgR in Mtb and Mtb:ΔRv2745c (comp) did not lead to Clp protease induction, indicating that clgR has additional functions that need to be elucidated. Our data, when taken together with that obtained by other groups, indicates that ClgR plays diverse roles in multiple regulatory networks in response to different stress conditions. In addition to redox stress, the expression of Rv2745c correlates with the expression of genes involved in sulfate assimilation as well as in response to hypoxia and reaeration. Clearly, the Mtb Rv2745c-encoded ClgR performs different functions during stress response and is important for the pathogenicity of Mtb in-vivo, regardless of its induction of the Clp proteolytic pathway.
Introduction
One third of the population is infected with Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB) [1]. TB is the leading cause of death worldwide from an infectious disease and is responsible for approximately 1.4 million deaths annually [1]. Unfortunately, the current vaccine, Bacillus Calmette-Guérine (BCG) vaccine, has a low protective efficacy against adult pulmonary TB [2], [3]. Infected individuals require a long treatment regimen, ranging from 6 to 12 months that involve serious side effects leading to noncompliance, and ultimately the development of resistance. Consequently, understanding the interplay between host and pathogen becomes increasingly important in order to develop alternative treatments and more effective vaccines.
One critical step in Mtb pathogenesis is the ability of Mtb to reside within the macrophage (MΦ) phagolysosome, as infected alveolar macrophages (AMΦ) are responsible for antigen presentation to CD4+ T cells during Mtb infection [4], [5]. Residence within the phagolysosome leads to exposure of a plethora of environmental stressors, such as reactive nitrogen species (RNS) and reactive oxygen intermediates (ROI), low pH, and hydrolases, that Mtb must be able to sense and respond to in order to establish an infection [6]–[9]. Thus, the temporal regulation of genes involved in sensing and responding to environmental stressors becomes increasingly important for understanding the ability of Mtb to take residence within the host MΦ. Mtb encodes over 200 regulators of transcription including 13 different sigma factors and is well equipped to respond to rapid changes in its environment.
Upon residence within the host MΦ, Mtb intracellular proteins are targets of RNS and ROIs [8]. It is likely that these stress conditions lead to changes in protein conformation, such as misfolding and aggregation. Clearance of misfolded and/or aggregated proteins is necessary for maintenance of protein homeostasis, which is also crucial for infection establishment. One Mtb gene that may play a role in this process is Rv2745c (clgR), which encodes a Clp protease gene regulator that is induced under a variety of stress conditions [10]–[12]. The expression of Rv2745c appears to be under the control of either σE or σH, or both. However, the activation of Rv2745c and its role in signaling cascades may be context dependent, implicating additional moonlighting functions for clgR in response to different environmental stressors. To study the role of Rv2745c in Mtb pathogenesis, we generated an isogenic mutant, Mtb:ΔRv2745c, using allelic exchange. Furthermore, we complemented the isogenic mutant to generate a Mtb:ΔRv2745c (comp) strain. We analyzed the growth and survival phenotype of these strains, along with wild-type Mtb, in response to treatment with a thiol-oxidative agent, diamide, for up to 90 minutes. Further, by subjecting RNA from each strain and untreated controls to comparative transcriptomics, we analyzed the mechanisms by which the Rv2745c gene-product may help Mtb respond to redox stress. Our results, presented as part of this investigation, along with those from other groups, indicate that ClgR may fulfill diverse functions in response to the multitude of stress conditions that induce its expression. As such, the roles of clgR in signaling cascades in response to stressors is not yet fully defined, thus to attribute clp protease gene regulation as the sole function of Rv2745c is inaccurate.
Materials and Methods
Bacterial Strains and Culture Conditions
Liquid cultures of Mtb CDC1551 (referred to as Mtb), Mtb:ΔRv2745c, and Mtb:ΔRv2745c (comp) were grown in Middlebrook 7H9 broth (BD Diagnostic Systems) supplemented with 0.1% glycerol, 0.05% Tween-80, 10% Albumin Dextrose Catalase (ADC). Selective antibiotics were used for culturing the isogenic mutant [75 μg/mL hygromycin B (hygro) (Roche Applied Sciences)] as well as the complemented strain [hygro+50 μg/mL kanamycin (kan) (Sigma)].
Allelic Exchange
Bacteriophage supernatant containing ΔRv2745c::HygR was a kind gift from of Drs William R Jacobs, Jr. and Michelle Larsen, Albert Einstein College of Medicine, Bronx, NY. The supernatant was used to transduce Mtb CDC1551 to generate the mutant, Mtb:ΔRV2745c following previously published protocols [13].
Complemented Strain Generation
Rv2745c coding sequence and an additional 481 base pairs upstream were amplified such that they contained NdeI and PacI restriction sites and cloned into NdeI and PacI (New England Biolabs) digested pSCW38 integration vector [14], [15]. Following transformation into E. coli DH5α, the plasmid was screened for the correct insertion via NdeI and PacI double digest, PCR screening, and DNA sequencing. After insertion verification, the plasmid was electroporated into Mtb:ΔRv2745c to generate Mtb:ΔRv2745c (comp) as previously published [13].
PCR
PCR was performed as per manufacturers’ instructions using the GC-RICH PCR System (Roche Applied Science). PCR was performed using an initial denaturation for 3 min at 95°C followed by 10 cycles of: 30 s at 95°C, 30 s at 65°C, 45 s at 72°C; then 25 cycles of: 95°C, 30 s at 65°C, 45 s (or 1 min depending upon the amplified region) at 72°C; a final elongation for 7 min at 72°C, and then stopped at 4°C. Primer sequences are listed in Table S1 (ST1).
DNA Extraction
Mtb strains were cultured to late exponential phase (OD260 0.8–1.0) in ADC supplemented Middlebrook 7H9 media containing 0.1% Tween-80. Briefly, cells were lysed at 37°C overnight in Lysis Buffer (50 mM Tris, pH 8.0; 150 mM NaCl;10 mM EDTA, pH 8.0; 0.5% SDS) containing Proteinase K (20 μg/mL). Bead beating and subsequent phenol extraction was employed as previously described [14]. DNA concentration was determined using Nanodrop 2000 (ThermoScientific).
Southern Blot
Southern Blot was performed as previously described by Manganelli, et al. and Wang, et al. using NcoI digested genomic DNA and biotin labeled DNA probes [16], [17].
Diamide Disc Diffusion Assay
The susceptibility of all three strains were compared via diamide disc diffusion assay as previously published [16].
In vitro Diamide Treatment
Cultures were grown to mid-log phase (OD260 0.39–0.45) without antibiotics. At time zero, 25 mL of culture was removed for RNA extraction. Upon time zero, cultures were treated with a final concentration of 10 mM diamide as previously described [14]. After treatment, 25 mL of culture was removed for each time point, t = 30, 60, and 90 min. Absorbance readings were taken at each time point.
Western Blot
Whole cell lysates were extracted from diamide treated cultures at t = 60 minutes post-diamide treatment. A total amount of 10 μg protein was run on an 18% Tris-glycine gel. Protein was transferred and blotted as previously described, with the following changes: anti-Rv2745c antibody was used at a dilution of 1∶500 and goat anti-rabbit was used at a dilution of 1∶200 [14].
Bacterial RNA Extraction
25 mL of culture was used to extract RNA upon cell lysis via the Trizol bead beater method and phenol extraction [14]. RNA concentrations were quantified using a Nanodrop 2000 (NanoDrop Technologies).
DNA Microarrays and RT-PCR
Mtb specific DNA microarrays (MYcroarrays, Biodiscovery Llc.) were used to compare transcriptome-wide responses in Mtb, the mutant and the complemented strains to redox stress by diamide. Detailed protocols for array procedures have been described earlier [10], [14], [16]. Genes were considered to have a perturbed expression level if they exhibited a 2- or a 4-fold higher or lower expression in the mycobacterial strain (wild-type, mutant or the complemented strain) at a given time point, relative to control samples in each of the three biological replicate arrays and in every technical replicate spot on each array. Raw and processed microarray data has been submitted to the Gene Expression Omnibus and can be retrieved using the GEO platform number GPL18320. For real-time (RT) PCR, RNA was treated with DNase as previously described [14]. RNA was reverse transcribed following the manufacturers’ instructions using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) [14]. RT-PCR was performed as per manufacturers’ instructions using Power SYBR Green PCR Master Mix (Applied Biosystems) and as previously described [14]. Expression levels were normalized to sigA levels. For primer sequences, see ST1.
Statistical Analysis
Statistical analyses were performed using an ANOVA using GraphPad Prism. Microarray statistical analyses were performed using a t test script in the Spotfire DecisionSite/S+ Array Analyzer.
Regulatory Compliance
The investigators received approval from the Tulane Institutional Biosafety Committee for all procedures involving Mtb.
Results
Isogenic Mutant and Complemented Strain Generation
In order to better understand the role played by the product of the Rv2745c gene in the management of host stress during Mtb infection, we generated an isogenic mutant in this gene, using Mtb CDC1551 as the parental strain. Allelic exchange was employed to generate the isogenic mutant, Mtb:ΔRv2745c [13]. Upon selection of isolated colonies from hyg containing plates, genomic DNA isolation and subsequent PCR screening confirms that Rv2745c was successfully replaced with a hygromycin resistant cassette (hygr) upon transduction of Mtb with ΔRv2745c::HygR bacteriophage lysate (Figure S1a & b). Replacement of Rv2745c with hygr was further confirmed via sequencing (data not shown) and Southern Blot (data not shown). Upon generation of the isogenic mutant, an integration vector, pSCW35, containing Rv2745c, was used to complement the deletion mutant, generating Mtb:ΔRv2745c (comp). Since the exact location of the promoter element(s) for Rv2745c is unknown, 481 base pairs upstream of Rv2745c on the coding strand were included in the integration vector as this contains both the intergenic region as well as base pairs within the adjacent upstream gene. Upon selection of isolated colonies for genomic DNA isolation, PCR screening identified several candidates that had successful integration of Rv2745c into the att site, which was further confirmed via sequencing (Figure S1c and data not shown). To confirm that ClgR protein levels were restored, Rv2745c was induced using diamide treatment and clgR levels were compared between the complemented and wild-type strains via Western Blot (Figure S2). After screening, we selected one out of several of the isolated colonies that had ClgR levels comparable to that of the wild-type Mtb upon induction via diamide treatment (Figure S2).
Diamide Susceptibility
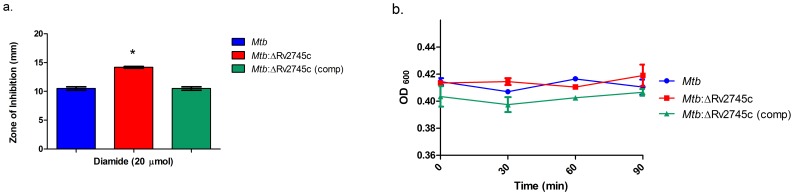
The current understanding of how Rv2745c contributes to Mtb stress responses is largely unknown; specifically, the phenotypic changes associated with loss of Rv2745c function are unknown, as this has not been studied. The expression of Rv2745c is significantly induced in Mtb by redox stress in-vitro. Hence, upon successful generation of the isogenic mutant and complemented strain, phenotypic changes associated with deletion of Rv275c were assessed via diamide disc diffusion assay. The Mtb:ΔRv2745c mutant was more sensitive to redox stress via diamide treatment relative to Mtb and Mtb:ΔRv2745c (comp), as the zone of inhibition was significantly larger for the isogenic mutant when compared to wild-type and the complemented strains (Figure 1a). Prolonged exposure to redox stress leads to cell death, however when comparing the early response there is not a significant difference between strains (Figure 1b). Thus, it is the initial disruption of signaling cascades that ultimately lead to higher levels of cell death in the isogenic mutant at later time points.
Figure 1. Diamide Susceptibility.
a.) Disc diffusion assay was performed using discs containing 20 μmol diamide. The zone of inhibition of Mtb:ΔRv2745c (red bar) is significantly larger when compared to both Mtb (blue bar) and Mtb:ΔRv2745c (comp) (green bar) indicating that the isogenic mutant is more susceptible to redox stress. n = 3. *p<0.001 b.) OD graph of diamide treated cultures. In the initial stages of treatment, there is no significant difference in growth between the different groups.
Transcriptomic Changes Post-Diamide Treatment
Rv2745c is predicted to encode a transcriptional regulator. Although the expression of the Rv2745c gene is induced by a variety of in-vitro stress conditions, the role played by its gene-product in regulating downstream signaling cascades is poorly understood [10], [18]. Thus, Rv2745c expression is induced in Mtb upon redox stress by diamide [14]; by membrane damage due to SDS [19] and thioridazine [20], as well as by hypoxia and reaeration [12]. Reaeration and hypoxia but not diamide and thioridazine appear to result in the induction of Clp protease genes clpP1, clpP2 and clpC1. This poses a surprising conundrum and indicates that the product of Rv2745c gene performs different functions in response to host stress, some of which do not require the deployment of the ATP-dependent Clp protease system [10], [11], [18], [21]. We have therefore taken the approach of studying Rv2745c expression and phenotype in a variety of in-vitro and in-vivo conditions, one at a time, to clearly delineate the role played by Rv2745c-encoded protein. Here we studied expression changes in Mtb, the Mtb:ΔRv2745c mutant and the complemented strain, in response to diamide stress, as the Mtb:ΔRv2745c mutant is clearly susceptible to this condition. We analyzed genome-wide transcriptome responses in Mtb, the mutant and the complemented strain to diamide over the course of a 90-minute time period with 30 min intervals. All comparisons were performed relative to untreated controls of the representative strain (Figure 2a).
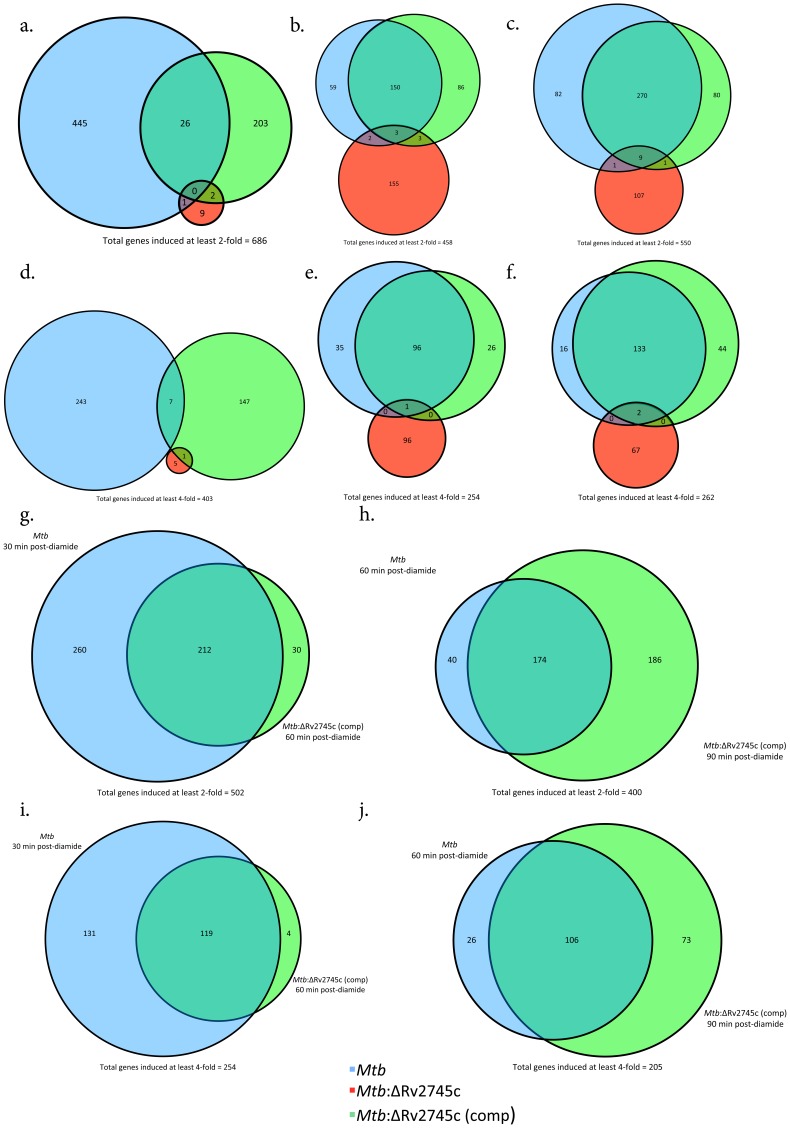
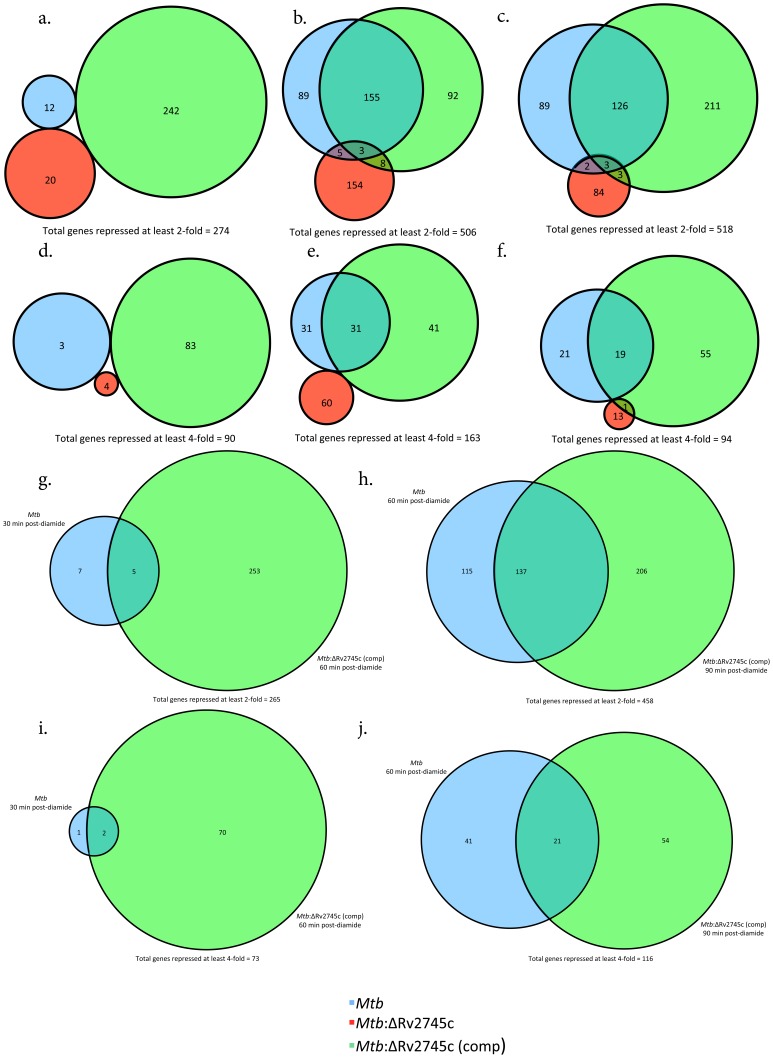
Figure 2. Venn Diagrams of Diamide Induced Genes.
Venn diagrams describe the extent of overlap between gene-expression upon diamide treatment in Mtb (blue circles), Mtb:ΔRv2745c (red circles) and Mtb:ΔRv2745c (comp) (green circles). Genes induced at least two-fold at: a.) 30, b.) 60, and c.) 90 min post-diamide treatment are shown. Genes induced at least four-fold at: d.) 30, e.) 60, and f.) 90 min post-diamide treatment. g–j.): Delayed response of Mtb:ΔRv2745c (comp). g.) Genes induced at least two-fold comparing Mtb at 30 min to Mtb:ΔRv2745c (comp) at 60 min and h.) Genes induced at least two-fold comparing Mtb at 60 min to Mtb:ΔRv2745c (comp) at 90 min. i.) Genes induced at least four-fold comparing Mtb at 30 min to Mtb:ΔRv2745c (comp) at 60 min. j.) Genes induced at least four-fold comparing Mtb at 60 min to Mtb:ΔRv2745c (comp) at 90 min. n = 3.
A significantly higher perturbation in global gene-expression was observed in response to diamide treatment in Mtb as well as the complemented strain, rather than the mutant. Hence, none of the 686 genes whose expression was induced by at least two-fold in both Mtb and the complemented strain 30 min post-diamide treatment (t = 30), exhibited induction in the mutant (Figure 2a). In contrast, a greater degree of overlap existed between Mtb and the complemented strain. Of the 471 and 231 genes whose expression was respectively induced in Mtb and the complemented strain, 26 of the genes exhibited overlapping induction. The expression of only one gene overlapped between Mtb and the mutant and that of only two overlapped between the complemented strain and the mutant at this early time point (Figure 2a). At t = 30, the expression of not a single gene was induced >two-fold in each of the three strains (Figure 2a).
At the t = 60 min post-diamide treatment, the expression of 458 genes was induced >two-fold in either of the strains, Mtb and Mtb:ΔRv2745c (comp). The extent of overlap between Mtb and the complemented strain increased significantly at this time. Of the 213 and 252 genes whose expression was induced >two-fold in Mtb and the complemented strain respectively, 153 (∼72% and 61% respectively) overlapped (Figure 2b). On the other hand, of the 163 genes that exhibited induced expression by least two-fold in the isogenic mutant at t = 60 min post-diamide treatment, only eight overlapped with Mtb as well as the complemented strain. At this time point, the expression of only three genes was induced in a shared manner amongst all three strains (Figure 2b). These patterns are similar to that of t = 90 min post-diamide treatment, with 270 genes out of 362 whose expression was induced >two-fold in Mtb and 360 whose expression was comparably induced in the complemented strain being induced in an overlapping manner (Figure 2c). Of particular note, a majority of the genes that are induced at least two-fold in the isogenic mutant at both 60 and 90 minutes post-diamide treatment were not shared at each time point.
When we only considered genes induced at least four-fold, the expression of 403 genes was induced at the earliest time point, t = 30 min post-diamide, in either of the strains, wild-type and complemented strain, but none of these genes were shared amongst all three (Figure 2d). At this level of higher stringency, >75% of all genes whose expression was induced at both t = 60 and 90 min post-diamide time points overlapped in Mtb as well as the complemented strain. The expression of none of these genes was induced in the Mtb:ΔRv2745c mutant (Figure 2e and 2f). We performed quantitative RT-PCR and were able to validate most of the results obtained from the DNA microarray format (not shown).
Taken together these results implicate that the transcriptional response of the Mtb:ΔRv2745c mutant to diamide is significantly different from Mtb as well as the complemented strain. Hence, Rv2745c plays a role in facilitating the signaling cascades that are required for proper regulation of the redox response. Further, the complemented strain exhibited phenotypic complementation of the response to diamide stress at the t = 60 min and the t = 90 min, but not the t = 30 min time point.
We therefore hypothesized that there may be a delay in the onset of the response of the various regulatory networks in the complemented strain, relative to the wild type strain. We therefore systematically compared the response of Mtb at different times to the complemented strain at not the comparable but the subsequent time point. Consistent with our hypothesis, we observed that >87% of all genes with >two-fold expression in the complemented strain at t = 60 min overlapped with those induced in Mtb at the previous (t = 30 min) time point; similarly >95% of all genes with >four-fold expression in the complemented strain at t = 60 min overlapped with those induced in Mtb at t = 30 min. A high degree of overlap was also present amongst genes whose expression was induced >two- or four-fold in the complemented strain at t = 90 min and Mtb at t = 60 min, again suggesting delayed induction of regulatory networks in the former strain (Figure 2g & i and Figure 2h & j).
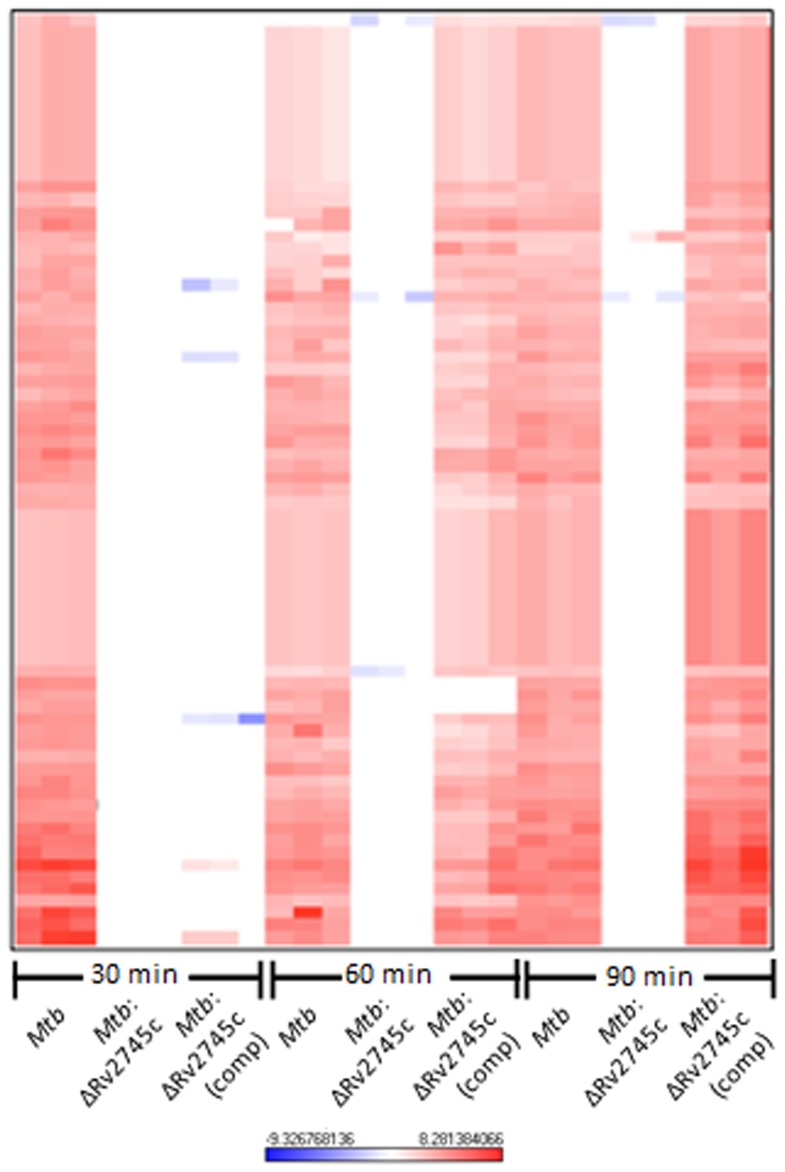
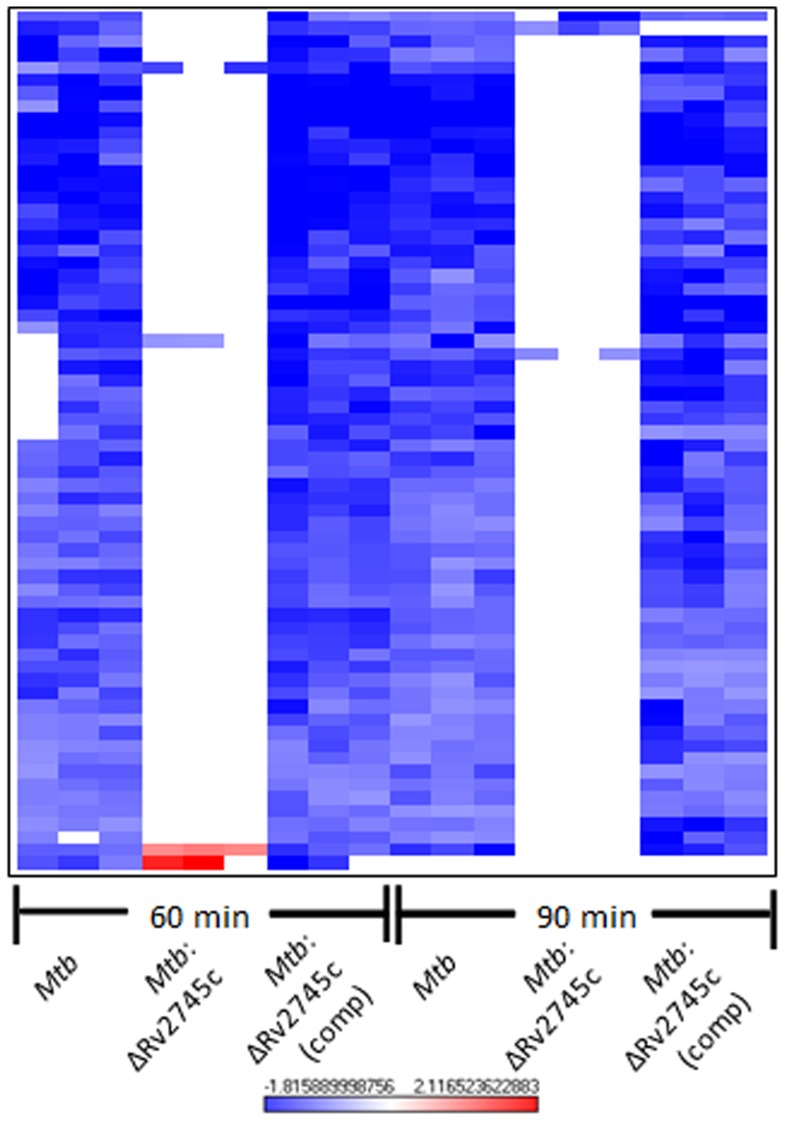
Next, we used hierarchical clustering to identify the specific genes and gene-families with perturbed expression levels in Mtb and the complemented strain, relative to the mutant, in response to redox stress. The transcriptional profiles associated with the isogenic mutant were distinct from those observed for both Mtb and Mtb:ΔRv2745c (comp) (Figure 3, Figure 4a & b). Of the genes induced to the highest levels in the wild type strain, there was no change in expression levels of the isogenic mutant when compared to the untreated control (Figure 3). Again, the delayed response of the complemented strain was reflected in the heat maps, showing that the expression pattern seen in the wildtype strain is restored by 60 min post-diamide treatment in the complemented strain (Figure 3, Figure 4a & b).
Figure 3. Diamide Induced Genes.
The heat map show results of unsupervised hierarchical clustering focusing on the genes with the highest magnitude of change. A majority of genes induced by diamide treatment in Mtb are also induced in Mtb:ΔRv2745c (comp) by 60 minutes and not induced in Mtb:ΔRv2745c. Genes that are induced in Mtb are also induced in Mtb:ΔRv2745c (comp), whereas there is no change in expression levels in the isogenic mutant indicating that Rv2745c plays a the redox response. n = 3. Red color indicates induction while blue color indicates repression, relative to the control channel. The intensity of each color corresponds to the magnitude.
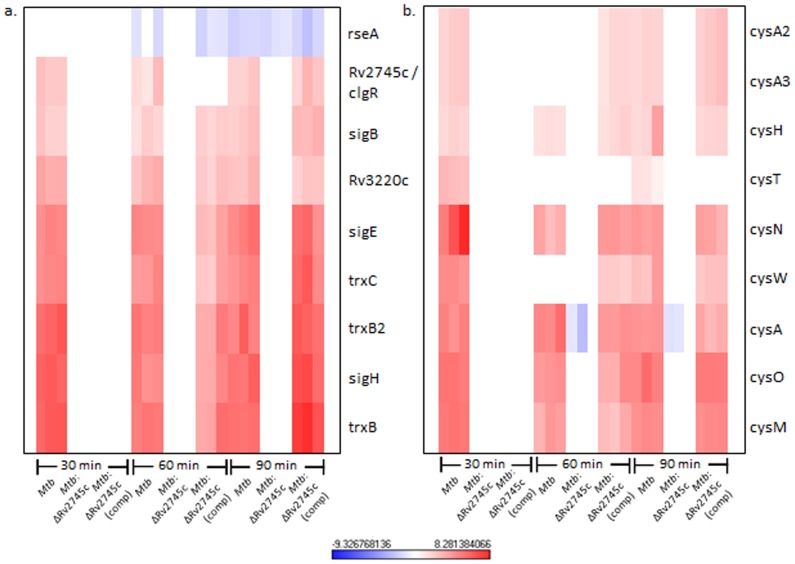
Figure 4. σH Regulon and Cysteine Pathway Induction.
Heat maps show results of unsupervised hierarchical clustering focusing on the genes with the highest magnitude of change. a.) σH regulon and the b.) cysteine pathway are induced in Mtb and Mtb:ΔRv2745c (comp), whereas there is no change in expression levels in the isogenic mutant indicating that Rv2745c plays a role in these regulatory networks, via direct or indirect regulation. n = 3. Red color indicates induction while blue color indicates repression, relative to the control channel. The intensity of each color corresponds to the magnitude.
The regulatory networks that were generally induced in Mtb and the complemented strain, following redox stress, expectedly belonged to the σH-regulon [14]. Surprisingly however, the expression of a large majority of these genes was disrupted in the Mtb:ΔRv2745c mutant (Figure 3, Figure 4a). In addition to positively reinforcing the induction of the σH regulon, these genes play a role in the stress response and detoxification and cysteine biosynthesis (Figure 4a & b) [10], [22]. The cysteine biosynthetic pathway comprises of two arms critical for response to redox stress [23], [24]. The alternative cysteine pathways utilizes thiocarboxylates for cysteine synthesis, which are more resistant to oxidative stress – the type of environment found within the MΦ [23]. Genes encoding CysM and CysO, which are part of the alternative cysteine pathway, exhibit up regulation upon oxidative stress in both Mtb and Mtb:ΔRv2745c (comp). However this expression pattern is disrupted in Mtb:ΔRv2745c (Figure 4b) [23], [25].
While the up regulation of genes within the σH regulon in Mtb and Mtb:ΔRv2745c (comp) is expected, as Rv2745c is activated downstream of σH in response to diamide (Figure 4a) [10], [22], the complete disruption of this regulatory pathway in the mutant is surprising and suggests that the Rv2745c-encoded protein may perform alternative functions in addition to activating ATP-dependent Clp proteases, the expression of which does not occur upon diamide treatment [16].
Additionally, transcription levels of several heat shock protein coding genes were also disrupted in Mtb:ΔRv2745c. Thus, htpX and htrA expression was up regulated in Mtb at all time points, but remain at basal levels in Mtb:ΔRv2745c (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9). The induction of these heat shock genes in both the wild-type and the complemented strains is again not surprising, since their expression is known to be induced in a σH dependent manner [14]. The role of clgR in activation of heat shock proteins is further supported by Mehra, et al., who found that induction of Rv2745c under a tetracycline promoter lead to higher expression levels of heat shock proteins when compared to un-induced samples [10].
Table 1. Mtb 30 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | AverageM Value | ExpressionFold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | 3.135 | 3.814 | 3.920 | 3.623 | 12.320 |
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | 4.291 | 4.419 | 4.201 | 4.304 | 19.749 | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | 1.558 | 1.859 | 1.402 | 1.606 | 3.045 | |
| MT0365 | dnaK | dnaK protein | Rv0350 | 2.964 | 3.292 | 3.109 | 3.122 | 8.703 | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | 2.890 | 2.984 | 2.923 | 2.932 | 7.634 | |
| Transcription | MT2816 | clgR | DNA-binding protein, putative | Rv2745c | 2.137 | 2.537 | 2.170 | 2.281 | 4.862 |
| MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | 1.825 | 2.413 | 1.768 | 2.002 | 4.006 | |
| MT1259 | sigE | RNA polymerase sigma-70 factor, ECF subfamily | Rv1221 | 4.864 | 4.327 | 4.614 | 4.602 | 24.282 | |
| MT3320 | sigH | RNA polymerase sigma-70 factor, ECF subfamily | Rv3223c | 6.455 | 6.134 | 6.033 | 6.208 | 73.904 | |
| MT4030 | sigM | RNA polymerase sigma-70 factor, ECF subfamily | Rv3911 | 1.277 | 2.191 | 1.760 | 1.742 | 3.346 | |
| MT1960 | furA | ferric uptake regulation protein | Rv1909c | 2.369 | 3.518 | 3.244 | 3.044 | 8.247 | |
| MT1260 | rseA | hypothetical protein | Rv1222 | 4.990 | 5.047 | 4.952 | 4.996 | 31.918 | |
| MT3316 | sensor histidine kinase | Rv3220c | 3.158 | 3.583 | 3.231 | 3.324 | 10.014 | ||
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | 4.467 | 4.511 | 3.816 | 4.265 | 19.220 |
| MT2470 | cysT | sulfate ABC transporter, permease protein | Rv2399c | 2.572 | 2.203 | 2.352 | 2.375 | 5.189 | |
| MT2469 | cysW | sulfate ABC transporter, permease protein | Rv2398c | 4.076 | 3.767 | 4.115 | 3.986 | 15.846 | |
| MT1519 | ABC transporter, ATP-binding protein | Rv1473 | 3.819 | 3.999 | 3.820 | 3.879 | 14.716 | ||
| MT2471 | subI | sulfate ABC transporter, sulfate-binding | Rv2400c | 2.003 | 2.246 | 2.398 | 2.216 | 4.645 | |
| Detoxification | MT2719 | cadI | conserved hypothetical protein | Rv2641 | 6.218 | 7.605 | 6.767 | 6.863 | 116.434 |
| MT1517 | trxB1 | thioredoxin | Rv1471 | 6.548 | 5.952 | 6.621 | 6.374 | 82.915 | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | 6.052 | 5.673 | 6.619 | 6.114 | 69.286 | |
| MT4033 | trxC | thioredoxin | Rv3914 | 4.676 | 4.360 | 4.830 | 4.622 | 24.620 | |
| MT1959 | katG | catalase-peroxidase | Rv1908c | 1.313 | 1.553 | 1.786 | 1.551 | 2.929 | |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin-guanine dinucleotide biosynthesis | Rv2453c | 2.806 | 3.000 | 2.753 | 2.853 | 7.226 |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | 4.683 | 4.840 | 4.753 | 4.759 | 27.069 | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | 4.728 | 4.767 | 4.930 | 4.808 | 28.013 |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | 4.620 | 7.578 | 6.087 | 6.095 | 68.357 | |
| MT1323 | cysD | sulfate adenylate transferase, subunit 2 | Rv1285 | 3.022 | 3.410 | 3.629 | 3.354 | 10.224 | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | 4.767 | 4.635 | 4.658 | 4.687 | 25.825 | |
| MT0837 | cysA2 | thiosulfate sulfurtransferase | Rv0815c | 1.511 | 1.891 | 1.856 | 1.753 | 3.370 | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | 1.743 | 1.976 | 1.829 | 1.850 | 3.604 | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | 1.365 | 1.453 | 1.627 | 1.481 | 2.792 | |
| Cell wall associated | MT0856 | lpqQ | hypothetical protein | Rv0835 | 1.244 | 1.123 | 1.351 | 1.239 | 2.361 |
| MT0870 | lpqS | hypothetical protein | Rv0847 | 2.824 | 2.865 | 3.236 | 2.975 | 7.861 | |
| MT1379 | murI | glutamate racemase | Rv1338 | 4.430 | 3.888 | 4.418 | 4.245 | 18.967 |
Genes induced and repressed by functional category at 30 minutes post-diamide treatment in wild-type. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 2. Mtb:ΔRv2745c 30 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | AverageM Value | ExpressionFold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | ND | ND | ND | – | – |
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | ND | ND | ND | – | – | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | ND | ND | ND | – | – | |
| MT0365 | dnaK | dnaK protein | Rv0350 | ND | ND | ND | – | – | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | ND | ND | ND | – | – | |
| Transcription | MT2816 | clgR | DNA-binding protein, putative | Rv2745c | ND | ND | ND | – | – |
| MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | ND | ND | ND | – | – | |
| MT1259 | sigE | RNA polymerase sigma-70 factor, ECF subfamily | Rv1221 | ND | ND | ND | – | – | |
| MT3320 | sigH | RNA polymerase sigma-70 factor, ECF subfamily | Rv3223c | ND | ND | ND | – | – | |
| MT4030 | sigM | RNA polymerase sigma-70 factor, ECF subfamily | Rv3911 | ND | ND | ND | – | – | |
| MT1960 | furA | ferric uptake regulation protein | Rv1909c | ND | ND | ND | – | – | |
| MT1260 | rseA | hypothetical protein | Rv1222 | ND | ND | ND | – | – | |
| MT3316 | sensor histidine kinase | Rv3220c | ND | ND | ND | – | – | ||
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | ND | ND | ND | – | – |
| MT2470 | cysT | sulfate ABC transporter, permease protein | Rv2399c | ND | ND | ND | – | – | |
| MT2469 | cysW | sulfate ABC transporter, permease protein | Rv2398c | ND | ND | ND | – | – | |
| MT1519 | ABC transporter, ATP-binding protein | Rv1473 | ND | ND | ND | – | – | ||
| MT2471 | subI | sulfate ABC transporter, sulfate-binding | Rv2400c | ND | ND | ND | – | – | |
| Detoxification | MT2719 | cadI | conserved hypothetical protein | Rv2641 | ND | ND | ND | – | – |
| MT1517 | trxB1 | thioredoxin | Rv1471 | ND | ND | ND | – | – | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | ND | ND | ND | – | – | |
| MT4033 | trxC | thioredoxin | Rv3914 | ND | ND | ND | – | – | |
| MT1959 | katG | catalase-peroxidase | Rv1908c | ND | ND | ND | – | – | |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin-guanine dinucleotide biosynthesis | Rv2453c | ND | ND | ND | – | – |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | ND | ND | ND | – | – | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | ND | ND | ND | – | – |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | ND | ND | ND | – | – | |
| MT1323 | cysD | sulfate adenylate transferase, subunit 2 | Rv1285 | ND | ND | ND | – | – | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | ND | ND | ND | – | – | |
| MT0837 | cysA2 | thiosulfate sulfurtransferase | Rv0815c | ND | ND | ND | – | – | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | ND | ND | ND | – | – | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | ND | ND | ND | – | – | |
| Cell wall associated | MT0856 | lpqQ | hypothetical protein | Rv0835 | ND | ND | ND | – | – |
| MT0870 | lpqS | hypothetical protein | Rv0847 | ND | ND | ND | – | – | |
| MT1379 | murI | glutamate racemase | Rv1338 | ND | ND | ND | – | – |
Genes induced and repressed by functional category at 30 minutes post-diamide treatment in the isogenic mutant. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 3. Mtb:ΔRv2745c (comp) 30 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | AverageM Value | ExpressionFold Change |
| Transcription | MT2784 | ideR | Transcriptional Regulatory Protein (Repressor and activator) | Rv2711 | – | −1.580 | −1.508 | −1.544 | −2.916 |
| MT1685 | purine cyclase-related protein | Rv1647 | 4.080 | 2.325 | – | 3.202 | 9.206 | ||
| Transport | MT1907 | modC | molybdate uptake ABC-transporter | Rv1859 | −2.183 | −1.196 | − | −1.690 | −3.225 |
| Detoxification | MT2967 | fdhD | fdhD protein | Rv2899c | − | 3.234 | 2.899 | 3.066 | 8.377 |
| Intermediary metabolism | MT3719 | ephA | probable epoxide hydrolase | Rv3617 | − | −7.782 | −1.798 | −4.790 | −27.670 |
| Cell wall associated | MT1593 | lprI | lipoprotein | Rv1541c | −1.287 | −1.063 | − | −1.175 | −2.258 |
Genes induced and repressed by functional category at 30 minutes post-diamide treatment in the complemented strain. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 4. Mtb 60 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | 1.302 | 1.850 | 2.051 | 1.734 | 3.328 |
| MT3527 | groES | chaperonin, 10 kDa | Rv3418c | −2.060 | −2.279 | −2.545 | −2.295 | −4.906 | |
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | 3.300 | 3.000 | 3.550 | 3.283 | 9.736 | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | 1.085 | 2.251 | 2.747 | 2.028 | 4.077 | |
| MT0365 | dnaK | dnaK protein | Rv0350 | 2.921 | 2.712 | 2.878 | 2.837 | 7.145 | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | 2.362 | 2.776 | 2.447 | 2.528 | 5.769 | |
| Transcription | MT2816 | clgR | DNA−binding protein, putative | Rv2745c | 1.044 | 1.444 | 2.712 | 1.734 | 3.326 |
| MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | 2.010 | 1.250 | 1.580 | 1.613 | 3.060 | |
| MT1259 | sigE | RNA polymerase sigma−70 factor, ECF subfamily | Rv1221 | 4.580 | 4.760 | 4.450 | 4.597 | 24.195 | |
| MT3320 | sigH | RNA polymerase sigma−70 factor, ECF subfamily | Rv3223c | 4.276 | 5.269 | 4.498 | 4.681 | 25.652 | |
| MT1960 | furA | ferric uptake regulation protein | 1.730 | 3.678 | 2.191 | 2.533 | 5.788 | ||
| MT1009 | mprA | DNA−binding response regulator | Rv0981 | −1.566 | −2.026 | −1.646 | −1.746 | −3.355 | |
| MT1260 | rseA | hypothetical protein | Rv1222 | 4.570 | 4.580 | 4.340 | 4.497 | 22.575 | |
| MT3316 | sensor histidine kinase | Rv3220c | 2.953 | 2.307 | 3.289 | 2.850 | 7.208 | ||
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP−binding protein | Rv2397c | 4.264 | 4.103 | 5.166 | 4.511 | 22.801 |
| MT1519 | ABC transporter, ATP−binding protein | Rv1473 | 3.740 | 3.333 | 3.780 | 3.618 | 12.275 | ||
| Detoxification | MT2719 | cadI | conserved hypothetical protein | Rv2641 | 4.500 | 4.640 | 5.040 | 4.727 | 26.477 |
| MT1517 | trxB1 | thioredoxin | Rv1471 | 5.440 | 5.030 | 5.230 | 5.233 | 37.618 | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | 4.930 | 5.430 | 5.120 | 5.160 | 35.753 | |
| MT4033 | trxC | thioredoxin | Rv3914 | 3.974 | 3.897 | 4.842 | 4.238 | 18.865 | |
| MT1959 | katG | catalase−peroxidase | Rv1908c | 1.840 | 1.220 | 1.610 | 1.557 | 2.942 | |
| MT3174 | fadD13 | substrate–CoA ligase | Rv3089 | 1.805 | 1.641 | 1.262 | 1.569 | 2.968 | |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin−guanine dinucleotide biosynthesis | Rv2453c | 2.190 | 1.800 | 2.740 | 2.243 | 4.735 |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | 4.275 | 4.684 | 5.231 | 4.730 | 26.538 | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | 2.700 | 3.640 | 3.410 | 3.250 | 9.514 |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | 3.236 | 2.299 | 2.838 | 2.791 | 6.921 | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | 3.560 | 3.760 | 3.960 | 3.760 | 13.548 | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | 1.190 | 1.870 | 3.260 | 2.107 | 4.307 | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | 1.078 | 1.219 | 1.122 | 1.140 | 2.204 | |
| Intermediary metabolism | MT3949 | bfrB | ferritin family protein | Rv3841 | −1.295 | −1.422 | −1.739 | −1.486 | −2.800 |
| Cell wall associated | MT0870 | lpqS | hypothetical protein | Rv0847 | 1.477 | 1.871 | 2.347 | 1.898 | 3.728 |
| MT1379 | murI | glutamate racemase | Rv1338 | 2.810 | 4.610 | 3.190 | 3.537 | 11.605 |
Genes induced and repressed by functional category at 60 minutes post-diamide treatment in wild-type. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 5. Mtb:ΔRv2745c 60 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | −2.721 | − | −1.053 | −1.887 | −3.699 |
| MT1907 | modC | ABC transporter, ATP-binding protein | Rv1859 | −1.616 | − | −3.854 | −2.735 | −6.657 | |
| MT0951 | mntH | transport protein, NRAMP family | Rv0924c | 6.500 | 3.995 | 1.122 | 3.872 | 14.645 |
Genes induced and repressed by functional category at 60 minutes post-diamide treatment in the isogenic mutant. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 6. Mtb:ΔRv2745c (comp) 60 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | 2.454 | 1.961 | 2.321 | 2.245 | 4.741 |
| MT3526 | groEL1 | Rv3417c | 1.697 | 1.664 | 1.765 | 1.709 | 3.269 | ||
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | 2.973 | 2.698 | 3.026 | 2.899 | 7.459 | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | 1.759 | 1.651 | 1.804 | 1.738 | 3.336 | |
| MT0365 | dnaK | dnaK protein | Rv0350 | 2.158 | 2.336 | 3.394 | 2.629 | 6.187 | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | 2.377 | 2.301 | 3.632 | 2.770 | 6.821 | |
| Transcription | MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | 1.894 | 1.589 | 2.014 | 1.832 | 3.560 |
| MT1259 | sigE | RNA polymerase sigma-70 factor, ECF subfamily | Rv1221 | 2.737 | 2.447 | 3.679 | 2.954 | 7.750 | |
| MT3320 | sigH | RNA polymerase sigma-70 factor, ECF subfamily | Rv3223c | 3.223 | 3.345 | 4.739 | 3.769 | 13.635 | |
| MT1960 | furA | Rv1909c | 2.465 | 2.646 | 2.811 | 2.641 | 6.236 | ||
| MT1009 | mprA | Rv0981 | −1.426 | −1.742 | −1.638 | −1.602 | −3.035 | ||
| MT1260 | rseA | hypothetical protein | Rv1222 | 2.657 | 2.646 | 3.716 | 3.006 | 8.034 | |
| MT3316 | sensor histidine kinase | Rv3220c | 2.100 | 1.708 | 2.563 | 2.124 | 4.358 | ||
| Transport | MT2468 | cysA1 | Rv2397c | 3.588 | 3.563 | 3.985 | 3.712 | 13.107 | |
| MT2469 | cysW | sulfate ABC transporter, permease protein | Rv2398c | 1.845 | 1.869 | 1.635 | 1.783 | 3.442 | |
| MT1519 | Rv1473 | 3.022 | 2.958 | 3.803 | 3.261 | 9.588 | |||
| MT2471 | subI | sulfate ABC transporter, sulfate−binding | Rv2400c | 1.098 | 1.467 | 1.011 | 1.192 | 2.285 | |
| Detoxification | MT2719 | cadI | Rv2641 | 4.175 | 4.322 | 6.555 | 5.017 | 32.389 | |
| MT1517 | trxB1 | thioredoxin | Rv1471 | 3.349 | 3.605 | 5.726 | 4.227 | 18.725 | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | 3.218 | 3.290 | 5.195 | 3.901 | 14.936 | |
| MT4033 | trxC | thioredoxin | Rv3914 | 2.160 | 2.018 | 3.730 | 2.636 | 6.216 | |
| MT1959 | katG | catalase−peroxidase | Rv1908c | 1.713 | 2.331 | 2.757 | 2.267 | 4.814 | |
| Lipid metabolism | MT0882 | fadA | Rv0859 | −2.372 | −1.416 | −1.240 | −1.676 | −3.195 | |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin−guanine dinucleotide biosynthesis | Rv2453c | 2.375 | 2.000 | 2.914 | 2.430 | 5.388 |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | 2.481 | 2.625 | 3.363 | 2.823 | 7.075 | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | 2.346 | 2.104 | 2.969 | 2.473 | 5.552 |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | 3.611 | 3.698 | 3.389 | 3.566 | 11.841 | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | 2.797 | 2.808 | 4.142 | 3.249 | 9.507 | |
| MT0837 | cysA2 | thiosulfate sulfurtransferase | Rv0815c | 1.008 | 1.467 | 1.478 | 1.318 | 2.493 | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | 1.516 | 1.358 | 1.686 | 1.520 | 2.868 | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | 1.108 | 1.405 | 1.696 | 1.403 | 2.645 | |
| Intermediary metabolism | MT3949 | bfrB | Rv3841 | −1.440 | −1.332 | −1.177 | −1.316 | −2.490 | |
| MT3969 | ethA | Rv3854c | − | −1.173 | −1.226 | −1.200 | −2.297 | ||
| Cell wall associated | MT0870 | lpqS | hypothetical protein | Rv0847 | 1.258 | 1.157 | 1.404 | 1.273 | 2.416 |
| MT1379 | murI | glutamate racemase | Rv1338 | 3.322 | 2.855 | 3.729 | 3.302 | 9.863 |
Genes induced and repressed by functional category at 60 minutes post-diamide treatment in the complemented strain. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 7. Mtb 90 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | 3.003 | 3.121 | 3.426 | 3.183 | 9.083 |
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | 5.176 | 4.723 | 5.038 | 4.979 | 31.534 | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | 1.806 | 2.009 | 2.109 | 1.975 | 3.930 | |
| MT0365 | dnaK | dnaK protein | Rv0350 | 3.616 | 3.287 | 3.971 | 3.625 | 12.336 | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | 2.993 | 2.707 | 3.254 | 2.985 | 7.916 | |
| Transcription | MT2816 | clgR | DNA-binding protein, putative | Rv2745c | 1.737 | 1.731 | 2.349 | 1.939 | 3.835 |
| MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | 2.162 | 1.906 | 2.612 | 2.227 | 4.680 | |
| MT1259 | sigE | RNA polymerase sigma-70 factor, ECF subfamily | Rv1221 | 5.082 | 4.539 | 5.660 | 5.094 | 34.147 | |
| MT3320 | sigH | RNA polymerase sigma-70 factor, ECF subfamily | Rv3223c | 5.271 | 5.486 | 6.261 | 5.673 | 51.011 | |
| MT4030 | sigM | RNA polymerase sigma-70 factor, ECF subfamily | Rv3911 | 1.286 | 0.768 | 1.015 | 1.023 | 2.032 | |
| MT1960 | furA | ferric uptake regulation protein | Rv1909c | 2.279 | 2.663 | 2.635 | 2.525 | 5.757 | |
| MT2784 | ideR | iron-dependent repressor IdeR | Rv2711 | 1.185 | 1.088 | 0.911 | 1.061 | 2.087 | |
| MT1009 | mprA | DNA-binding response regulator | Rv0981 | −1.807 | −1.601 | −1.578 | −1.662 | −3.164 | |
| MT1260 | rseA | hypothetical protein | Rv1222 | 5.400 | 5.001 | 5.968 | 5.456 | 43.910 | |
| MT3316 | sensor histidine kinase | Rv3220c | 2.304 | 2.344 | 2.997 | 2.548 | 5.850 | ||
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | 3.785 | 3.824 | 3.643 | 3.751 | 13.461 |
| MT2469 | cysW | sulfate ABC transporter, permease protein | Rv2398c | 2.201 | 3.545 | 2.075 | 2.607 | 6.092 | |
| MT1519 | ABC transporter, ATP-binding protein | Rv1473 | 3.679 | 3.609 | 3.936 | 3.741 | 13.371 | ||
| MT2471 | subI | sulfate ABC transporter, sulfate-binding | Rv2400c | 1.399 | 1.475 | 1.462 | 1.446 | 2.724 | |
| Detoxification | MT2719 | cadI | conserved hypothetical protein | Rv2641 | 6.100 | 5.606 | 6.200 | 5.969 | 62.619 |
| MT1517 | trxB1 | thioredoxin | Rv1471 | 5.489 | 5.535 | 5.724 | 5.583 | 47.931 | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | 6.348 | 4.685 | 4.867 | 5.300 | 39.399 | |
| MT4033 | trxC | thioredoxin | Rv3914 | 4.689 | 4.151 | 5.029 | 4.623 | 24.643 | |
| MT1959 | katG | catalase-peroxidase | Rv1908c | 1.760 | 2.116 | 2.419 | 2.098 | 4.282 | |
| MT0179 | mce1B | virulence factor mce family protein | Rv0170 | −1.355 | −1.366 | −1.269 | −1.330 | −2.514 | |
| MT3960 | sodA | superoxide dismutase | Rv3846 | − | −1.528 | −1.841 | −1.684 | −3.214 | |
| Lipid metabolism | MT2303 | fabD | malonyl CoA-acyl carrier protein transacylase | Rv2243 | 1.512 | 1.672 | 1.971 | 1.718 | 3.290 |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin-guanine dinucleotide biosynthesis | Rv2453c | 2.598 | 2.386 | 2.693 | 2.559 | 5.893 |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | 4.691 | 4.579 | 5.222 | 4.831 | 28.457 | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | 3.860 | 4.007 | 4.132 | 4.000 | 15.996 |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | 3.646 | 3.630 | 3.324 | 3.533 | 11.579 | |
| MT1323 | cysD | sulfate adenylate transferase, subunit 2 | Rv1285 | 3.377 | 2.964 | 3.064 | 3.135 | 8.784 | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | 4.195 | 4.459 | 5.209 | 4.621 | 24.606 | |
| MT0837 | cysA2 | thiosulfate sulfurtransferase | Rv0815c | 1.307 | 1.543 | 1.616 | 1.489 | 2.806 | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | 1.338 | 1.350 | 1.649 | 1.446 | 2.724 | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | 1.275 | 3.358 | 1.492 | 2.042 | 4.117 | |
| Cell wall associated | MT0870 | lpqS | hypothetical protein | Rv0847 | 2.474 | 2.392 | 2.852 | 2.573 | 5.950 |
| MT1379 | murI | glutamate racemase | Rv1338 | 3.520 | 3.437 | 3.730 | 3.562 | 11.814 |
Genes induced and repressed by functional category at 90 minutes post-diamide treatment in wild-type. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 8. Mtb:ΔRv2745c 90 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Transcription | MT4030 | sigM | RNA polymerase sigma-70 factor, ECF subfamily | Rv3911 | −1.090 | − | −1.495 | −1.292 | −2.449 |
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | −1.043 | −1.266 | − | −1.154 | −2.226 |
Genes induced and repressed by functional category 90 minutes post-diamide treatment in the isogenic mutant. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Table 9. Mtb:ΔRv2745c (comp) 90 min post-diamide treatment.
| Function | MT # | Symbol | Description | Rv# | M1 | M2 | M3 | Average M Value | Expression Fold Change |
| Heat Shock | MT0397 | clpB | ATP-dependent Clp protease, ATP-binding subunit | Rv0384c | 3.987 | 4.158 | 3.667 | 3.937 | 15.320 |
| MT3526 | groEL1 | chaperonin, 60 kDa | Rv3417c | 1.868 | 1.944 | 2.967 | 2.260 | 4.789 | |
| MT3527 | groES | chaperonin, 10 kDa | Rv3418c | 1.298 | 2.344 | 3.283 | 2.308 | 4.952 | |
| MT0265 | hsp | heat shock protein, HSP20 family | Rv0251c | 5.430 | 5.139 | 5.495 | 5.355 | 40.917 | |
| MT0589 | htpX | heat shock protein HtpX | Rv0563 | 2.416 | 2.120 | 2.321 | 2.286 | 4.876 | |
| MT0365 | dnaK | dnaK protein | Rv0350 | 5.646 | 5.429 | 4.661 | 5.245 | 37.927 | |
| MT0367 | dnaJ1 | dnaJ protein | Rv0352 | 4.054 | 4.365 | 4.101 | 4.173 | 18.042 | |
| Transcription | MT2816 | clgR | DNA-binding protein, putative | Rv2745c | 2.978 | 1.678 | 2.261 | 2.306 | 4.944 |
| MT2783 | sigB | RNA polymerase principal sigma factor SigB | Rv2710 | 2.667 | 2.720 | 3.112 | 2.833 | 7.125 | |
| MT1259 | sigE | RNA polymerase sigma-70 factor, ECF subfamily | Rv1221 | 5.964 | 5.573 | 4.292 | 5.276 | 38.753 | |
| MT3320 | sigH | RNA polymerase sigma-70 factor, ECF subfamily | Rv3223c | 7.174 | 6.878 | 5.741 | 6.597 | 96.836 | |
| MT4030 | sigM | RNA polymerase sigma-70 factor, ECF subfamily | Rv3911 | 1.592 | 1.395 | 1.143 | 1.377 | 2.597 | |
| MT0017 | pknB | serine/threonine protein kinase | Rv0014c | −1.240 | −2.333 | −1.012 | −1.528 | −2.884 | |
| MT1960 | furA | ferric uptake regulation protein | Rv1909c | 3.102 | 2.845 | 3.261 | 3.069 | 8.393 | |
| MT2784 | ideR | iron-dependent repressor IdeR | Rv2711 | 1.199 | 1.113 | 1.074 | 1.129 | 2.187 | |
| MT1009 | mprA | DNA-binding response regulator | Rv0981 | −2.224 | −1.402 | −1.606 | −1.744 | −3.349 | |
| MT1260 | rseA | hypothetical protein | Rv1222 | 6.595 | 6.524 | 5.785 | 6.301 | 78.872 | |
| MT3316 | sensor histidine kinase | Rv3220c | 2.331 | 1.731 | 2.241 | 2.101 | 4.290 | ||
| Transport | MT2468 | cysA1 | sulfate ABC transporter, ATP-binding protein | Rv2397c | 2.478 | 2.905 | 3.187 | 2.857 | 7.243 |
| MT2469 | cysW | sulfate ABC transporter, permease protein | Rv2398c | 2.201 | 2.229 | 1.920 | 2.117 | 4.337 | |
| MT1519 | ABC transporter, ATP-binding protein | Rv1473 | 5.628 | 4.510 | 3.948 | 4.695 | 25.911 | ||
| MT2471 | subI | sulfate ABC transporter, sulfate-binding | Rv2400c | 1.735 | 1.457 | 1.438 | 1.543 | 2.915 | |
| Detoxification | MT2719 | cadI | conserved hypothetical protein | Rv2641 | 7.428 | 6.957 | 6.975 | 7.120 | 139.088 |
| MT1517 | trxB1 | thioredoxin | Rv1471 | 8.175 | 7.631 | 6.737 | 7.514 | 182.799 | |
| MT4032 | trxB2 | thioredoxin reductase | Rv3913 | 6.124 | 6.487 | 5.555 | 6.055 | 66.508 | |
| MT4033 | trxC | thioredoxin | Rv3914 | 6.542 | 5.853 | 4.581 | 5.659 | 50.522 | |
| MT1959 | katG | catalase-peroxidase | Rv1908c | 3.273 | 3.200 | 3.162 | 3.212 | 9.265 | |
| Lipid metabolism | MT1001 | accA2 | acetyl/propionyl-CoA carboxylase, alpha subunit | Rv0973c | −1.270 | −1.101 | −1.222 | −1.197 | −2.293 |
| MT3350 | alkB | alkane-1 monooxygenase | Rv3252c | −1.885 | −1.538 | −1.399 | −1.607 | −3.047 | |
| MT0882 | fadA | thiolase | Rv0859 | −1.413 | −1.627 | −1.280 | −1.440 | −2.712 | |
| Molybdopterin biosynthesis | MT2528 | mobA | molybdopterin-guanine dinucleotide biosynthesis | Rv2453c | 3.241 | 3.688 | 3.189 | 3.373 | 10.360 |
| MT3301 | moeB1 | HesA/MoeB/ThiF family protein | Rv3206c | 4.233 | 4.164 | 4.196 | 4.198 | 18.349 | |
| Sulfate metabolism | MT1377 | cysM | cysteine synthase | Rv1336 | 4.163 | 4.227 | 3.935 | 4.108 | 17.246 |
| MT1324 | cysN | sulfate adenylate transferase, subunit 1 | Rv1286 | 3.263 | 2.650 | 3.421 | 3.111 | 8.641 | |
| MT1323 | cysD | sulfate adenylate transferase, subunit 2 | Rv1285 | 2.797 | 2.243 | 3.263 | 2.768 | 6.811 | |
| MT1376.1 | cysO | conserved hypothetical protein | Rv1335 | 4.680 | 4.595 | 4.668 | 4.648 | 25.068 | |
| MT0837 | cysA2 | thiosulfate sulfurtransferase | Rv0815c | 1.922 | 2.300 | 1.637 | 1.953 | 3.872 | |
| MT3199 | cysA3 | thiosulfate sulfurtransferase | Rv3117 | 2.059 | 2.400 | 1.490 | 1.983 | 3.953 | |
| MT2462 | cysH | phosphoadenosine phosphosulfate reductase | Rv2392 | 1.536 | 1.563 | 1.349 | 1.483 | 2.794 | |
| Intermediary metabolism | MT3949 | bfrB | ferritin family protein | Rv3841 | −3.178 | −3.401 | −1.809 | −2.796 | −6.944 |
| MT3969 | ethA | monooxygenase, flavin-binding family | Rv3854c | −3.630 | −2.818 | −3.209 | −3.219 | −9.310 | |
| MT3349 | rubA | rubredoxin | Rv3251c | −2.793 | −2.330 | −2.688 | −2.604 | −6.078 | |
| MT3348 | rubB | rubredoxin | Rv3250c | −2.064 | −1.773 | −2.262 | −2.033 | −4.093 | |
| Cell wall associated | MT3169 | lipR | acetyl−hydrolase | Rv3084 | −2.807 | −1.934 | −1.114 | −1.951 | −3.867 |
| MT2912 | efpA | efflux protein | Rv2846c | −1.691 | −1.269 | −1.243 | −1.401 | −2.641 | |
| MT0870 | lpqS | hypothetical protein | Rv0847 | 2.801 | 2.422 | 2.643 | 2.622 | 6.156 | |
| MT1379 | murI | glutamate racemase | Rv1338 | 3.229 | 3.500 | 3.325 | 3.351 | 10.205 |
Genes induced and repressed by functional category 90 minutes post-diamide treatment in the complemented strain. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
Profiles that exhibit repression, i.e. lower expression levels relative to baseline following redox stress were apparent at t = 60 min and t = 90 min rather than t = 30 min. Thus, of the 274 genes that are down regulated >2-fold at t = 30 min, there was no overlap in all three strains (Figure 5a). However, at t = 60 min, a significantly higher number of genes (506) showed repression >2-fold (Figure 5b). Of the 252 and 258 genes repressed >2-fold in Mtb and Mtb:ΔRv2745c (comp), respectively, 155 (62% and 60% respectively) were commonly repressed, whereas of the 170 genes repressed >2-fold in Mtb:ΔRv2745c only 3 were shared amongst all three strains (Figure 5b). This pattern was reflected at t = 90 min, wherein, of the 518 genes that are repressed >2-fold, 126 were shared between Mtb and the complemented strain (Figure 5c). A total of 220 and 343 genes were repressed >2-fold in wild-type and complemented strains, respectively, while only 92 genes were repressed >2-fold in the mutant (Figure 5c).
Figure 5. Venn Diagrams of Diamide Repressed Genes.
Venn diagrams describe the extent of overlap between gene-expression upon diamide treatment in Mtb (blue circles), Mtb:ΔRv2745c (red circles) and Mtb:ΔRv2745c (comp) (green circles). Genes repressed at least two-fold at: a.) 30, b.) 60, and c.) 90 min post-diamide treatment are shown. Genes repressed at least four-fold at: d.) 30, e.) 60, and f.) 90 min post-diamide treatment. g–j.): Delayed response of Mtb:ΔRv2745c (comp). g.) Genes repressed at least two-fold comparing Mtb at 30 min to Mtb:ΔRv2745c (comp) at 60 min and h.) Genes repressed at least two-fold comparing Mtb at 60 min to Mtb:ΔRv2745c (comp) at 90 min. i.) Genes repressed at least four-fold comparing Mtb at 30 min to Mtb:ΔRv2745c (comp) at 60 min. j.) Genes repressed at least four-fold comparing Mtb at 60 min to Mtb:ΔRv2745c (comp) at 90 min. n = 3.
This delayed response amongst genes that are repressed by >2-fold in the wild-type and complemented strains indicates that the repression of gene-expression is a secondary effect rather than a primary function mediated by the Rv2745c-encoded transcription factor (Figures 5 a–c, Figure 6, Tables 4, 6, 7, and 9). This implicates that the main role of clgR is to induce a subset of genes upon oxidative stress, and that some of these induced genes may eventually mediate or result in the repression of downstream genes. At both t = 60 min and t = 90 min, a majority of the genes that were repressed by at least two-fold were down regulated in both Mtb and Mtb:ΔRv2745c (comp) (Figure 5b & 4c).
Figure 6. Diamide Repressed Genes.
The heat map shows results of unsupervised hierarchial clustering focusing on genes with the greatest level of repression compared to the control channel. A majority of genes repressed in Mtb are also repressed in Mtb:ΔRv2745c (comp) at both 60 and 90 minutes post-diamide treatment, whereas there is no change in expression levels in Mtb:ΔRv2745c. n = 3. Red color indicates induction while blue color indicates repression, relative to the control channel. The intensity of each color corresponds to the magnitude.
To enhance the stringency of our analysis, we assessed the repression of genes in the three strains by >4-fold post-diamide treatment. A total of only 90 genes fulfilled this criterion at t = 30 min (Figure 5d). Of these, a majority (83) was repressed in the complemented strain and there was no overlap of shared repressed genes amongst the three strains (Figure 5d). The total number of repressed genes >4-fold increased to 163 at t = 60 min (Figure 5e). Of these, 31 were repressed in both wild-type and the complemented strain, whereas no genes in the mutant strain were also similarly repressed >4-fold in Mtb or the complemented strain at this time. (Figure 5e). At t = 90 min, the expression of a total of 94 genes was repressed >4-fold, of which 19 were commonly repressed in both wild-type and the complemented strain (Figure 5f). Of the 75 genes repressed >4-fold in the complemented strain, one was repressed in the mutant, as well (Figure 5f). The delayed response of the complemented strain that was seen in induced genes was not reflected in the repressed set of genes both in genes repressed >2-fold and >4-fold (Figures 5 g–j).
At 60 and 90 min post-diamide treatment the expression of ESAT-6 was repressed >2-fold in the wild-type and complemented strain, relative to its basal level of expression in the mutant strain (Tables 4, 5, 6, 7, 8, 9) as well as in all three strains at t = 30 min (Tables 1, 2, 3). This repression is most likely a secondary effect in response to the action of clgR induced genes that are upstream of ESAT-6 in the signaling cascade. Additionally, the levels of mprA were also repressed in both the wild-type and the complemented strain strains at both 60 and 90 min post-diamide treatment while remaining at basal levels in the isogenic mutant (Tables 4, 5, 6, 7, 8, 9).
When comparing repressed genes within Mtb and Mtb:ΔRv2745c (comp), a high degree of similarity emerges 60 and 90 min post-diamide treatment (Figure 6). However, this pattern of repression is ablated in the isogenic mutant, implicating that a secondary response occurs in which clgR activation of a subset of genes leads to repression of an additional subset of genes (Figure 6).
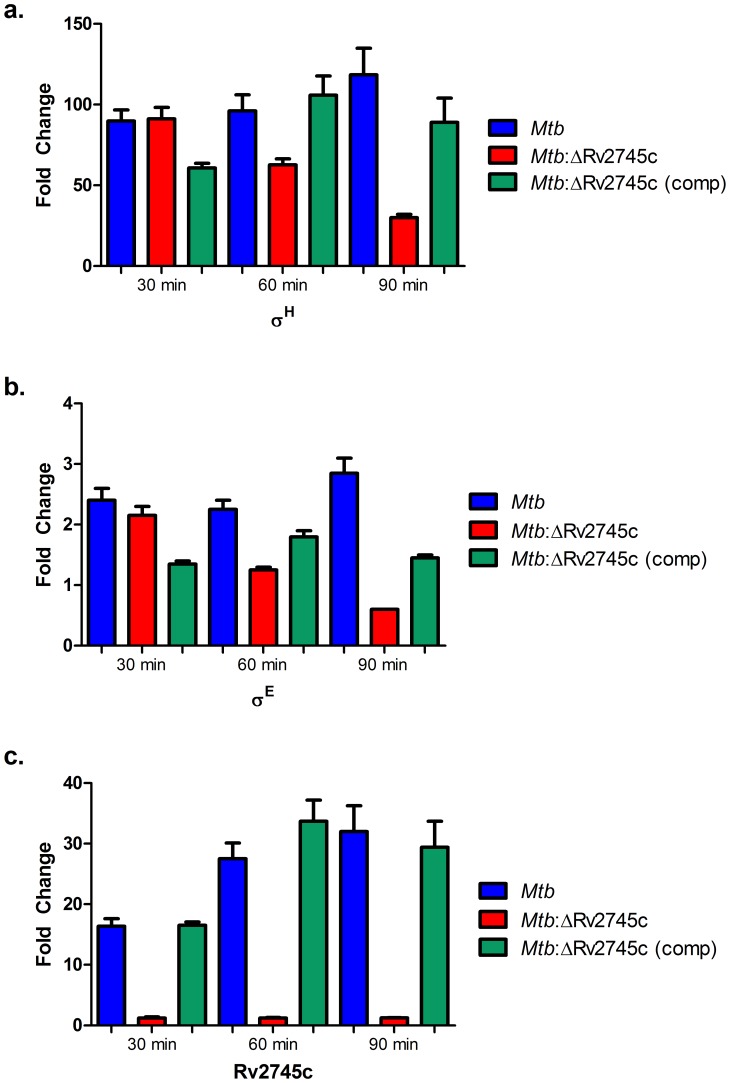
Due to the extensive use of microarray technology in order to understand the transcriptomic networks that involve Rv2745c, we performed quantitative RT-PCR in order to confirm many of the salient results obtained using the microarray platform (Figure 7). RT-PCR confirmed that the expression of Rv2745c was not induced at any time point in the mutant, but was highly induced in both Mtb and the complemented stain at all time points, with levels of the transcript increasing in the complemented strain (Figure 7c). Furthermore, RT-PCR detected very low levels of σH induction in the isogenic mutant at the 30-minute time point (Figure 7a) similar to those observed in microarray experiments. Furthermore, the levels of σH transcript decreased throughout the time course in the isogenic mutant. RT-PCR revealed low levels of σE induction in each of the strains but the levels of this transcript significantly increased in Mtb and the complemented strains, relative to the mutant, over the course of time (Figure 7b).
Figure 7. RT-PCR Confirmation.
a.) The data shows that there is a small increase in σH levels of the isogenic mutant, relative to that of the wild type and complemented strain. However, σH levels are not sustained in the isogenic mutant, whereas they increase in the wild-type and complemented strain. b.) σE levels remain low in all three strains. c.) There is Rv2745c activation in the wild-type and the complemented strain, whereas there is no Rv2745c expression in the isogenic mutant, as expected.
Discussion
The ability of Mtb to persist within host tissues for extended period of time indicates that this pathogen has developed unique mechanisms for its survival. Undoubtedly, some of these mechanisms involve coping with the various host and environmental stresses. Mtb is uniquely positioned to respond effectively to numerous stress conditions at it encodes numerous stress response transcription factors including sigma factors, two-component systems, eukaryotic like kinases etc. [26]. The proteins encoded by these diverse transcriptional and post-transcriptional regulators form specific stress-response regulons, which respond to a variety of environmental changes. The complexity of this system is enhanced by the interaction and interdependence of different stress regulons.
A key Mtb stress regulon is controlled by the expression of sigma factors, σH, primarily in response to oxidative stress [16], [27]. In turn σH results in the induced expression of σE, σB and a number of other transcription factors, thus directly and indirectly shaping the oxidative-stress dependent expression of over 500 Mtb genes [14]. Loss of σH ameliorates pathology in mice [27], reduces Mtb cfu levels in primate macrophages [28] and results in the complete lack of virulence in primates [29]. Thus, σH appears to be critical for Mtb to survive the host phagocyte oxidative burst. Expression of σE can be induced by σH as well as independent of it. During redox stress, modeled by diamide, the expression of both σH and σE is induced. Loss of σE also results in diminished virulence and pathology in the mouse model [30]. Rv2745c is predicted to encode a transcription factor with homology to Clp gene regulator (ClgR) in related gram-positive organisms [31]. The expression of Rv2745c is induced in response to numerous conditions that result in the up regulation of either σH or σE e.g. redox stress by diamide [14], cell envelope damage by thioridazine, vancomycin or SDS [19], [20], [32], low pH [6], enduring hypoxia and reaeration [18], etc. Interestingly however, this protein appears to perform different functions in many of these situations. Thus, Personne, et al. report that the expression of Clp protease genes clpP1, clpP2, clpC1 and clpX, known to be induced by ClgR upon subjecting Mtb to hypoxia, are not induced during redox stress [21]. Our group has similarly reported that while the expression of Rv2745c is induced >50-fold in Mtb following thioridazine treatment, the expression of the downstream clp genes was not induced [20]. Similarly, the expression of clp genes was also not induced by elevated levels of Rv2745c following vancomycin treatment of Mtb [32]. These results raise the possibility that the Rv2745c-encoded protein may indeed perform diverse functions in response to the different stress conditions, which result in the induction of its expression in Mtb. If this is true, then it may be possible that Mtb has adapted a stress response pathway present in related, saprophytic actinomyces to benefit its specialized pathogenic needs. It is possible that while retaining its function of inducing the expression of Clp proteases in response to hypoxia, this protein additionally also performs a moonlighting function in response to oxidative and envelope damaging stress. To specifically address this issue we have begun to modularly study the phenotype of the Mtb:ΔRv2745c mutant in response to these conditions. The current manuscript serves as the initial report in this regard. Here we demonstrate that the mutant is exquisitely sensitive to redox stress by diamide, which leads to rapid and strong induction of Rv2745c expression. We further show that the induction of Rv2745c does not result in the up regulation of Clp genes during redox stress. Instead, the expression of the σH/σE network is dysregulated in the ΔRv2745c mutant.
Our results indicate that at least during redox stress due to diamide, the major function of Rv2745c induction in the absence of downstream clp gene induction may be to supplement the σH/σE regulon. Hence, the Mtb:ΔRv2745c exhibits an in-vitro phenotypic susceptibility to diamide mediated oxidative stress, which is not observed for either Mtb or Mtb:ΔRv2745c (comp), implicating that clgR plays a role in mediated signaling cascades involved in response to redox stress. This altered phenotype is further supported by the differential expression patterns seen in the isogenic mutant when compared to both the wild type and complemented strain. Thus, the transcriptional phenotype of the Mtb:ΔRv2745c mutant in response to diamide stress in-vitro closely resembles those of the Mtb:Δ-σH and Mtb:Δ-σE mutants. Hence, the expression pathways, expression of σH, σE, σB, the thioredoxin/thioredoxin reductase and the cysteine metabolic pathways was disrupted in the Mtb:ΔRv2745c mutant. It is conceivable, based on our data, that the Rv2745c-encoded protein is somehow involved in the reinforcement of the σH/σE response to redox stress (Figure 6), perhaps by playing a regulatory role in the positive feedback loop that maintains σH or σE regulation [20], [22]. This role is independent of clp protease activation, indicating that Rv2745c may also directly activate σE and/or σH so that σH can mediate other downstream signaling events. This indicates that the positive feedback loop of Rv2745c with the downstream gene of the σH regulon, σE, is more influential and plays a more prominent role in inducing the σH regulon than previously thought.
The acute dysregulation of the σH/σE regulon, which involves genes upstream of clgR, as well as downstream genes in the sulfate assimilation pathway, occurred in Mtb:ΔRv2745c, implicates a strong alternative role for Rv2745c in signaling cascades responsible for responding to redox stress. This may be mediated through a direct binding of ClgR to either (or both) σH or σE promoter elements to reinforce the transcriptional signal or through an indirect effect of Rv2745c activation of clp proteases. This could be independent of the Clp protease complex. Accordingly, we did not detect an increase in clp protease (clpP1 as well as clpP2) transcriptional levels compared to t = 0 min, which is supported by the studies of Personne, et al., indicating that clpP activation is condition dependent [21]. However, we must acknowledge the possibility that Clp protease complex is indeed activated by ClgR during redox stress, albeit at lower levels such that the transcriptional induction of clp genes by either microarrays or RT-PCR is virtually undetectable. In this scenario, the enhanced activity of the protease complex could result in a higher turnover of RshA and RseA, the cognate anti-sigma factors for σH or σE. In this regard, Sureka, et al. have shown that RseA is indeed a biological substrate for the Mtb clp protease system during stringent response [33]. Decreased availability of either anti-sigma factor could also potentially result in an over-exuberant expression of the σH/σE network. Similarly, it is possible that the expression of a negative regulator of clp protease genes is induced during redox stress but not during hypoxia. Such an arrangement could result in the availability of ClgR during both conditions but the expression of clp protease system only during the latter situation. The expression profiles of a few known transcriptional repressors were found to be up regulated in Mtb vis-à-vis the mutant, during diamide stress (Table 10). However, unraveling the exact mechanism of this discrepancy will require more work.
Table 10. Negative Regulators Expressed in Mtb post-diamide treatment.
| Expression Fold-Change | |||||
| MT # | Description | Rv # | 30 min | 60 min | 90 min |
| MT0073 | transcriptional regulator, TetR family | Rv0067c | 10.573 | 6.246 | 6.246 |
| MT0206 | transcriptional regulator, putative | Rv0196 | 2.364 | 5.363 | 5.363 |
| MT0343 | transcriptional regulator, TetR family | Rv0328 | 14.405 | 4.157 | 4.157 |
| MT1079 | transcriptional regulator, MarR family | Rv1049 | 20.578 | 8.545 | 8.545 |
| MT3262 | transcriptional regulator, TetR family | Rv3173c | 3.198 | 2.802 | 2.802 |
| MT3938 | transcriptional regulator, TetR family | Rv3830c | 16.089 | 3.924 | 8.074 |
| MT3948 | hypothetical protein | Rv3840 | 16.382 | 4.902 | 8.711 |
Probable negative transcriptional regulators induced in Mtb post-diamide at each time point. n = 3. MT # and Rv # denote the CDC1551 and the H37Rv gene IDs, respectively.
We also report higher expression levels of katG in Mtb at all time points upon redox stress, whereas the levels of katG remained unchanged in Mtb:ΔRv2745c (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9). katG is a catalase peroxidase that is required for both countering peroxide products generated by NADPH oxidase of the phagocyte and activation of isoniazid, which is a key drug in TB treatment [34], [35]. However, in a study conducted by Mehra, et al., Rv2745c induction led to increased clp proteases and decreased levels of katG [10]. Hence, it is conceivable that katG may be a direct target of clpP proteases that are under the transcriptional control of clgR.
While the binding region of Mtb clgR has not yet been characterized, the consensus sequence for the clgR operator in C. glutamicum, S. lividans, and Bif. breve is well studied [11]. According to Estorninho, et al., there are several genes with a promoter-binding region specific to clgR when comparing consensus sequences, which support our result that Rv0384c (clpB), Rv3269 and Rv0251c were up regulated in the Mtb and not Mtb:ΔRv2745c at all time points (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9) [11]. Rv0250c, which also has a clgR consensus sequence, was also up regulated in Mtb at two of the three time-points post-diamide treatment when compared to the untreated control (Tables 1 & 7) [11]. The Rv0251c – Rv0249c operon encodes for Acr2 (hsp), which is a chaperone that functions as an oxidoreductase and succinate dehydrogenase, whose regulation has been shown to be σE dependent upon SDS stress [11], [36]. Up regulation of acr2 also occurs upon activation of σH and σE during oxidative stress, consequently clgR may also be responsible for the activation of acr2, as promoter binding sites for σH and σE are also found upstream of acr2 [37].
A critical observation in our study was that while the complemented strain resulted in comparable expression of ClgR at the transcript as well as the protein level, and was able to rescue the mutant phenotype, at the molecular level, a significant delay existed in the complete complementation of the stress response. Thus, gene-expression at t = 60 min in Mtb mirrored that observed at t = 90 min for the complemented strain (Figure 2 h–j). These results reinforce the importance of using complemented strains in gene-functional evaluation experiments. Further, our results suggest that trans-complementation of bacterial genes could sometimes result in either a partial or, as observed in this instance, a delayed complementation of the phenotype. What effect this delay would have on the phenotype of the complemented strain in-vivo, remains to be seen. Such experiments are currently underway in our laboratory.
We also observed that genes whose expression was repressed in Mtb and the complemented strain (at either >2- or >4-fold limit) occurred only at later (t = 60 min & t = 90 min, but not t = 30 min) stages, indicating that secondary effect(s) are at play. It is likely that some of the genes induced directly in Mtb due to diamide stress cause the repression of other genes by either transcriptional or post-transcriptional mechanisms. Hence, down-regulation of mprA occurred in both wild-type and complemented strains while remaining at basal levels in the mutant at all time points, implicating an additional role for Rv2745c-encoded ClgR protein (Tables 4, 5, 6, 7, 8, 9). MprA can function as both an activator and a repressor of acr2, depending on condition [36]. However, the expression of mprA was repressed at both 60 and 90 min in the wild type and the complemented strain, indicating, that in the absence of MprA, Rv2745c may induce acr2 [36]. MprA also induces the expression of σE independent of σH [38]. Thus, the induction of σE via σH due to redox stress apparently shuts down mprA transcription via feedback and it appears that Rv2745c plays a role in this process. The delay in repression is more than likely due a secondary response of repression, in which Rv2745c activated genes may be responsible for repression of downstream genes, which also explains why there is a general delay in down-regulation in the wild-type and complemented strains (Figure 6).
RT-PCR data revealed that there are low levels of σH induction in the isogenic mutant (Figure 7a). However, these levels continued to decrease throughout the course of diamide treatment, implicating that Rv2745c helps facilitate maintenance of the σH regulon upon application of redox stress. Additionally, the complemented strain and wild type Mtb had similar σH induction, which further supports that Rv2745c plays a role in the σH positive feedback loop. RT-PCR also confirms that Rv2745c was not induced in the isogenic mutant, but was induced to similar levels in both the wild-type and complemented strain (Figure 7c).
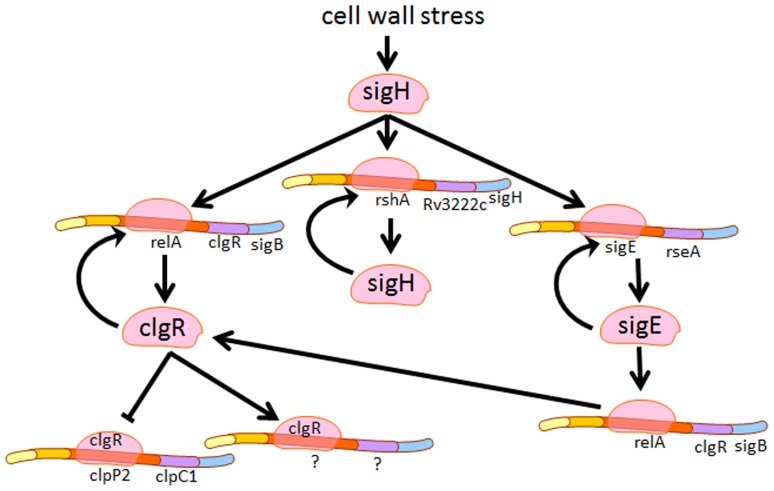
Our results show that the Rv2745c-encoded protein facilitates key down-stream signaling in response to redox stress. Deletion of Rv2745c leads to disruption of key regulatory networks, such as the σH regulon, the cysteine biosynthetic pathway, and the thioredoxin pathways. Disruption of upstream genes, such as σH and σE, implicate that Rv2745c may facilitate the positive feedback loop of this regulatory network either via direct transcriptional or an indirect post-transcriptional mechanism (Figure 8). Thus, further studies are required to clarify role of Rv2745c in the pathogenesis of Mtb. Further understanding of the function of Rv2745c in response to various environmental pressures may help lead to a better understanding as to how Mtb is able to survive and persist within the AMΦ.
Figure 8. clgR activation schematic.
Proposed overview of possible clgR activation upon cell wall stress. Induction of the σH regulon requires clgR activation. However, it is unclear if induction of the σH regulon is through direct or indirect effects of clgR activation.
Supporting Information
PCR Screening for Mtb :ΔRv2745c. a. PCR using Rv2745c primers. Amplification of ∼339 base pairs in lane 4 shows the presence of Rv2745c in Mtb wild type, while its absence in lane 5 indicates deletion of Rv2745c from Mtb:ΔRv2745c. b. PCR amplification of hygr. The absence of a band in lane 4 confirms the expected finding that hygr is not present within the Mtb genome. The presence of a band from hygr carrying plasmid as well as from genomic DNA derived from Mtb:ΔRv2745c is indicative of the replacement of Rv2745c by hygr (Lane 5). Lane 1–5(L–R): 0.1–12 kbp Ladder; Neg. Ctrl. (No DNA);ΔRv2745c::HygR phasmid; Mtb and Mtb:ΔRv2745c.
(TIF)
Western Blot of Rv2745c levels. Rv2745c protein levels after 60 minutes post-diamide treatment were detected via Western Blot. Whole cell lysates from several isolated colonies from the complementation were used. Rv2745c levels were restored to similar levels in the complemented strain relative to wild-type. From left to right, lane order: Mtb, Mtb:ΔRv2745c (comp, 2), Mtb:ΔRv2745c (comp, 4), Mtb:ΔRv2745c (comp, 10), Mtb:ΔRv2745c. Rv2745c levels were absent in the isogenic mutant 60-minutes post-diamide treatment.
(TIF)
Primers for PCR, Southern Blot, and RT-PCR.
(PDF)
Acknowledgments
We would like to thank Prof. William R Jacobs, Jr., PhD, Howard Hughes Medical Institute Investigator, Albert Einstein College of Medicine, Bronx, NY for providing us with bacteriophage lysate.
Funding Statement
This work was supported by the NIH grants AI089323, with additional support from NIH grants HL106790, AI091457, RR026006, RR020159, RR000164/OD011104 and C06AI058609. Additionally, the authors also acknowledge the support from the Louisiana Board of Regents (LEQSF (2007-12)-ENHPKSFI- PRS), the Tulane Research Enhancement Fund, the Tulane Center for Infectious Diseases, the Tulane National Primate Research Center Office of the Director and a Bridge Award from the Tulane Office of the Vice-President for Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. (2012) Global tuberculosis report 2012 (in IRIS). Geneva: World Health Organization. viii, 272 p. [Google Scholar]
- 2. Dye C (2013) Making wider use of the world’s most widely used vaccine: Bacille Calmette-Guerin revaccination reconsidered. J R Soc Interface 10: 20130365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeena PM, Chhagan MK, Topley J, Coovadia HM (2001) Safety of the intradermal Copenhagen 1331 BCG vaccine in neonates in Durban, South Africa. Bull World Health Organ 79: 337–343. [PMC free article] [PubMed] [Google Scholar]
- 4. Harding CV, Boom WH (2010) Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat Rev Microbiol 8: 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arko-Mensah J, Julian E, Singh M, Fernandez C (2007) TLR2 but not TLR4 signalling is critically involved in the inhibition of IFN-gamma-induced killing of mycobacteria by murine macrophages. Scand J Immunol 65: 148–157. [DOI] [PubMed] [Google Scholar]
- 6. Rohde KH, Abramovitch RB, Russell DG (2007) Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2: 352–364. [DOI] [PubMed] [Google Scholar]
- 7. Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG (2012) Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog 8: e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A, Farhana A, Guidry L, Saini V, Hondalus M, et al. (2011) Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev Mol Med 13: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vandal OH, Nathan CF, Ehrt S (2009) Acid resistance in Mycobacterium tuberculosis. J Bacteriol 191: 4714–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mehra S, Dutta NK, Mollenkopf HJ, Kaushal D (2010) Mycobacterium tuberculosis MT2816 encodes a key stress-response regulator. J Infect Dis 202: 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Estorninho M, Smith H, Thole J, Harders-Westerveen J, Kierzek A, et al. (2010) ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology 156: 3445–3455. [DOI] [PubMed] [Google Scholar]
- 12. Sherrid AM, Rustad TR, Cangelosi GA, Sherman DR (2010) Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS One 5: e11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR, Jr. (2007) Genetic Manipulation of Mycobacterium tuberculosis. Curr Protoc Microbiol Chapter 10: Unit 10A 12. [DOI] [PubMed]
- 14. Mehra S, Kaushal D (2009) Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J Bacteriol 191: 3965–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hernandez-Abanto SM, Woolwine SC, Jain SK, Bishai WR (2006) Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements. Arch Microbiol 186: 459–464. [DOI] [PubMed] [Google Scholar]
- 16. Manganelli R, Voskuil MI (2002) Schoolnik GK, Dubnau E, Gomez M, et al (2002) Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol 45: 365–374. [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Huang Y, Xue C, He Y, He ZG (2011) ClpR protein-like regulator specifically recognizes RecA protein-independent promoter motif and broadly regulates expression of DNA damage-inducible genes in mycobacteria. J Biol Chem 286: 31159–31167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rustad TR, Harrell MI, Liao R, Sherman DR (2008) The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3: e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, et al. (2009) The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191: 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dutta NK, Mehra S, Kaushal D (2010) A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PLoS One 5: e10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Personne Y, Brown AC, Schuessler DL, Parish T (2013) Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities. PLoS One 8: e60228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhat SA, Singh N, Trivedi A, Kansal P, Gupta P, et al. (2012) The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic Biol Med 53: 1625–1641. [DOI] [PubMed] [Google Scholar]
- 23. Agren D, Schnell R, Oehlmann W, Singh M, Schneider G (2008) Cysteine synthase (CysM) of Mycobacterium tuberculosis is an O-phosphoserine sulfhydrylase: evidence for an alternative cysteine biosynthesis pathway in mycobacteria. J Biol Chem 283: 31567–31574. [DOI] [PubMed] [Google Scholar]
- 24. Jurgenson CT, Burns KE, Begley TP, Ealick SE (2008) Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis. Biochemistry 47: 10354–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatzios SK, Bertozzi CR (2011) The regulation of sulfur metabolism in Mycobacterium tuberculosis. PLoS Pathog 7: e1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sachdeva P, Misra R, Tyagi AK, Singh Y (2010) The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. Febs j 277: 605–626. [DOI] [PubMed] [Google Scholar]
- 27. Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, et al. (2002) Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A 99: 8330–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dutta NK, Mehra S, Martinez AN, Alvarez X, Renner NA, et al. (2012) The stress-response factor SigH modulates the interaction between Mycobacterium tuberculosis and host phagocytes. PLoS One 7: e28958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehra S, Golden NA, Stuckey K, Didier PJ, Doyle LA, et al. (2012) The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J Infect Dis 205: 1203–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manganelli R, Fattorini L, Tan D, Iona E, Orefici G, et al. (2004) The extra cytoplasmic function sigma factor sigma(E) is essential for Mycobacterium tuberculosis virulence in mice. Infect Immun 72: 3038–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bellier A, Mazodier P (2004) ClgR, a novel regulator of clp and lon expression in Streptomyces. J Bacteriol 186: 3238–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Provvedi R, Boldrin F, Falciani F, Palu G, Manganelli R (2009) Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155: 1093–1102. [DOI] [PubMed] [Google Scholar]
- 33. Barik S, Sureka K, Mukherjee P, Basu J, Kundu M (2010) RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol Microbiol 75: 592–606. [DOI] [PubMed] [Google Scholar]
- 34. Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD (2004) Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol 52: 1291–1302. [DOI] [PubMed] [Google Scholar]
- 35. Ascenzi P, Coletta A, Cao Y, Trezza V, Leboffe L, et al. (2013) Isoniazid inhibits the heme-based reactivity of Mycobacterium tuberculosis truncated hemoglobin N. PLoS One. 8: e69762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pang X, Howard ST (2007) Regulation of the alpha-crystallin gene acr2 by the MprAB two-component system of Mycobacterium tuberculosis. J Bacteriol 189: 6213–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stewart GR, Newton SM, Wilkinson KA, Humphreys IR, Murphy HN, et al. (2005) The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol Microbiol 55: 1127–1137. [DOI] [PubMed] [Google Scholar]
- 38. He H, Hovey R, Kane J, Singh V, Zahrt TC (2006) MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol 188: 2134–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR Screening for Mtb :ΔRv2745c. a. PCR using Rv2745c primers. Amplification of ∼339 base pairs in lane 4 shows the presence of Rv2745c in Mtb wild type, while its absence in lane 5 indicates deletion of Rv2745c from Mtb:ΔRv2745c. b. PCR amplification of hygr. The absence of a band in lane 4 confirms the expected finding that hygr is not present within the Mtb genome. The presence of a band from hygr carrying plasmid as well as from genomic DNA derived from Mtb:ΔRv2745c is indicative of the replacement of Rv2745c by hygr (Lane 5). Lane 1–5(L–R): 0.1–12 kbp Ladder; Neg. Ctrl. (No DNA);ΔRv2745c::HygR phasmid; Mtb and Mtb:ΔRv2745c.
(TIF)
Western Blot of Rv2745c levels. Rv2745c protein levels after 60 minutes post-diamide treatment were detected via Western Blot. Whole cell lysates from several isolated colonies from the complementation were used. Rv2745c levels were restored to similar levels in the complemented strain relative to wild-type. From left to right, lane order: Mtb, Mtb:ΔRv2745c (comp, 2), Mtb:ΔRv2745c (comp, 4), Mtb:ΔRv2745c (comp, 10), Mtb:ΔRv2745c. Rv2745c levels were absent in the isogenic mutant 60-minutes post-diamide treatment.
(TIF)
Primers for PCR, Southern Blot, and RT-PCR.
(PDF)