Abstract
Acid ceramidase (encoded by ASAH1) is a lipid hydrolase that catalyzes the conversion of ceramide (cer) into sphingosine (SPH) and a free fatty acid. Adrenocortical steroidogenesis is regulated by the trophic peptide hormone adrenocorticotropin (ACTH), which induces the expression of steroidogenic genes in the human adrenal cortex primarily via a cAMP/protein kinase A (PKA)-dependent pathway. ACTH also stimulates sphingolipid metabolism in H295R adrenocortical cells leading to changes in steroidogenic gene expression. Based on our previous data identifying SPH as an antagonist for the nuclear receptor steroidogenic factor 1 (SF-1) and the role of ACTH-stimulated changes in sphingolipid metabolism on steroidogenic gene transcription, the aim of the current study was to determine the role of ACTH signaling in regulating the expression of the ASAH1 gene in H295R cells. We show that activation of the ACTH signaling pathway induces ASAH1 gene expression by stimulating the binding of the cAMP-responsive element binding protein (CREB) to multiple regions of the ASAH1 promoter. CREB binding promotes the recruitment of the coactivators CREB binding protein (CBP) and p300 to the CREB-responsive regions of the promoter. Consistent with transcriptional activation, we show that cAMP signaling increases the trimethylation of Lys 4 on histone H3 (H3K4) along the ASAH1 promoter. Finally, RNA interference (RNAi) experiments demonstrate that CREB is indispensable for cAMP-induced ASAH1 transcription. These data identify the ACTH/cAMP signaling pathway and CREB as transcriptional regulators of the ASAH1 gene in the human adrenal cortex.
Keywords: Acid ceramidase, cAMP, CREB, Ceramide, Sphingosine, Adrenocorticotropin
1. Introduction
Sphingolipids are a large family of lipids, many of which have bioactive properties. An expanding body of literature has demonstrated roles for sphingolipids in varied cellular processes, including cell proliferation, cell migration, and apoptosis [1–11]. ASAH1 encodes one of three ceramidases (N-acylsphingosine amidohydrolase) expressed in mammalian cells. These lipid hydrolases catalyze the degradation of cer into SPH and a free fatty acid, therefore dynamically regulating the cellular concentrations of these bioactive sphingolipids.
Bioactive sphingolipids such as cer, SPH, and sphingosine-1-phosphate (S1P) have all been implicated in steroidogenesis. Cer suppresses progesterone secretion in ovarian granulosa cells [12] and rat luteal cells [13], decreases testosterone production in rat Leydig cells [9,14], modulates P450c17α enzymatic activity (encoded by CYP17) in rat Leydig cells [9], and inhibits human choriogonadotropin-induced aromatase activity and estradiol production in ovarian granulosa cells [15,16]. We have shown in the H295R human adrenocortical cell line that suppression of ASAH1 expression leads to increased transcription of the CYP17 gene, indicating a role for this ceramidase in adrenocortical steroidogenesis [17]. Further, we have also demonstrated that SPH inhibits CYP17 transcription and cortisol biosynthesis by acting as an antagonistic ligand for SF-1, the nuclear receptor that regulates the transcription of most steroidogenic genes [18,19]. SPH can be rapidly phosphorylated by sphingosine kinases (SKs) to form S1P, which mediates cAMP-stimulated CYP17 transcription in H295R cells [20], increases cortisol secretion in bovine fasciculata cells [10], and stimulates aldosterone secretion in bovine glomerulosa cells [3,21]. In addition to studies demonstrating that sphingolipids regulate steroidogenesis, trophic factors that activate steroid hormone biosynthesis (for example ACTH) have been found to modulate sphingolipid metabolism. In H295R cells, ACTH stimulates sphingolipid metabolism by rapidly promoting the catabolism of sphingomyelin (SM) and cer. ACTH acutely activates SK activity, thus increasing S1P concentrations [17,20,22]. Collectively, these data highlight the intimate, reciprocal relationship between sphingolipid metabolism and steroid hormone biosynthesis.
CREB proteins are leucine zipper-containing transcription factors that regulate the expression of several genes by binding to cAMP-responsive element (CRE) sequences at target promoters [23,24]. In response to cAMP signaling, PKA phosphorylates CREB at Ser133, a post-translational modification that is essential for its transcriptional activity [23,25,26]. CREB binds to the promoter of target genes and facilitates the recruitment of coactivators, including CBP/p300 [27–29] and transducer of regulated CREB binding proteins (TORCs) [30,31] by a mechanism that is either dependent (e.g. CBP/p300) or independent (e.g. TORCs) of Ser133 phosphorylation. In addition to activating target gene transcription, CREB can also mediate transcriptional repression by partnering to repressor proteins. For instance, Kibler and Jeang reported that a CREB/ATF (activating transcription factor)-dependent cyclin A repression occurs through a protein–protein interaction with the human T cell leukemia virus type 1 Tax protein [32]. Further, the transcription factor YY1 represses c-fos transcription by forming a complex with CREB/ATF on the DNA [33].
Based on our previous data identifying SPH as an antagonist for SF-1 and the effect of ACTH-stimulated sphingolipid metabolism on steroidogenic gene transcription and hormone output, we sought to determine the role of ACTH/cAMP signaling in regulating the expression of the ASAH1 gene in H295R adrenocortical cells. We identify ASAH1 as a CREB-responsive gene and show that CREB is essential for cAMP-stimulated ASAH1 transcription. Moreover, CREB enrichment at multiple sites on the ASAH1 promoter facilitates the recruitment of CBP and p300 as well as H3K4 trimethylation. Finally, we demonstrate that cAMP-mediated ASAH1 transcription leads to a significant increase in protein expression and enzymatic activity, thus supporting a role for ASAH1 as an important enzyme in the regulation of cortisol biosynthesis.
2. Materials and methods
2.1. Reagents
Dibutyryl cAMP (Bt2cAMP) was obtained from Sigma (St. Louis, MO).
2.2. Cell culture
H295R adrenocortical cells [34,35] were generously donated by Dr. William E. Rainey (Medical College of Georgia, Augusta, GA) and cultured in Dulbecco’s modified Eagle’s/F12 (DME/F12) medium (Invitrogen, Carlsbad, CA) supplemented with 10% Nu-Serum I (BD Biosciences, Palo Alto, CA), 1% ITS Plus (BD Biosciences, Palo Alto, CA), and antibiotics.
2.3. Real time RT-PCR
Cells were sub-cultured onto 12-well plates and 48 h later treated with 0.4 mM Bt2cAMP for 1–24 h. Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and amplified using a One-Step SYBR Green RT-PCR Kit (Thermo Fisher Scientific Inc, Waltham, MA) and the primers listed in Table 1. ASAH expression was normalized to β-actin content and calculated using the delta–delta cycle threshold (ΔΔCT) method.
Table 1.
Sequences of primers and oligonucleotides.
| Method | Region | Forward primer | Reverse primer |
|---|---|---|---|
| ChIP | −1779/−1596 | TTCCCGGGTTCACGCCAT | GCGCGGTGGCTCACGCCTGTAA |
| −1475/−1364 | CTCCCTTTTCCTCCACTGCATTTGT | ACAATGTGCCAAGCATGTCTCCTGACAC | |
| −1022/−899 | CGGCAGCGTGCTGAGCTTCATCAAAGC | CAGTCGCGCGGGTAGGTGACCGGGTTGG | |
| −325/−214 | ACGGGTGAAGCTCCCGGCCCCACCTA | GAAAAGGGTGGCGTAGAGAAAGAGAGA | |
| −123/+31 | AGTCCCGCCTCCTCCGAGCGTTCCCCCT | GACTAAGGCGACGCAACTCCGGCCCGGC | |
| RT-PCR | β-actin | ACGGCTCCGGCATGTGCAAG | TGACGATGCCGTGCTGCATG |
| ASAH1 | GCACAAGTTATGAAGGAAGCCAAG | TCCAATGATTCCTTTCTGTCTCG | |
| EMSA | −281/−261 | CGAGGGGGATGGATCACGCCAGCCGC | CGGCGGCTGGCGTGATCCATCCCCCT |
| −969/−949 | CGCTGCTTGAGACGTCAGAGGCT | CGAGCCTCTGACGTCTCAAGCAG | |
| −1106/−1086 | CGGAGTGTTGAGTTTTGTAAAGAAATAATACA | CGTGTATTATTTCTTTACAAAACTCAACACTC | |
| −1565/−1545 | CGCCTGTCCCTCTTATTTAAAATTGTAACTCTACCACTTCTGATCTCCACAC | CGGTGTGGAGATCAGAAGTGGTAGAGTTACAATTTTAAATAAGAGGGACAGG | |
| −1776/−1756 | CGTTCCCGGGTTCACGCCATTCTCCTGCCTCA | CGTGAGGCAGGAGAATGGCGTGAACCCGGGAA | |
| −1861/−1841 | CGAGTTTCATTTTTATGTGACGGAGTCTCGCACTGGCGCGCA | CGTGCGCGCCAGTGCGAGACTCCGTCACATAAAAATGAAACT | |
| ASAH1 cloning | −2739 | CGAGCTCTTACGCGTTACCATTTTCTATGAACA | CTTAGATCGCAGATCTGGCGGCAGCCAGGAGGAC |
| −1430 | TCACCGAGAACATACGCCTCAG | CTTAGATCGCAGATCTGGCGGCAGCCAGGAGGAC | |
| −906 | CTTTGAAATCCAACCCGGTCCC | CTTAGATCGCAGATCTGGCGGCAGCCAGGAGGAC | |
| −496 | CGCTTTTCTCAGAGGGCAAAG | CTTAGATCGCAGATCTGGCGGCAGCCAGGAGGAC | |
| −120 | TGGAATGGTGCGGTCCCAGGTC | CTTAGATCGCAGATCTGGCGGCAGCCAGGAGGAC |
2.4. Cloning of the human ASAH1 promoter and site-directed mutagenesis
The human ASAH1 promoter was cloned using LA Taq DNA polymerase (Takara, Madison, WI), 500 ng of human genomic DNA (Promega, Madison, WI) and 300 nM of the primers listed in Table 1. PCR fragments were then cloned into the pGL3 (Promega) reporter gene plasmid at the Mlu I (5′) and Bgl II (3′) sites. The pGL3-ASAH1 constructs were confirmed by sequencing prior to use in transient transfection experiments. In silico analysis using the MatInspector software (Genomatix Software, Ann Arbor, MI) was used to identify putative consensus binding sites for CREB on the ASAH1 promoter.
2.5. Transient transfection and reporter gene analysis
H295R cells were sub-cultured onto 24-well plates and transfected with 150 ng of pGL3-ASAH1, 1.5 ng pRL-TK (Promega), and/or 5 ng of pCMV-CREB (BD Biosciences, Franklin Lakes, NJ) or 5 ng of pCMV-K-CREB (BD Biosciences) using GeneJuice (Novagen, Madison, WI). Twenty-four hours after transfection, the cells were treated with 0.4 mM Bt2cAMP for 16 h and the transcriptional activity of the ASAH1 reporter gene determined using a dual luciferase assay kit (Promega). Firefly (pLG3-ASAH1) luciferase activity was normalized to Renilla luciferase activity (pRL-TK) and expressed as fold change over the mean of the untreated control group.
2.6. Electrophoresis Mobility Shift Assay (EMSA)
Nuclear extracts were isolated from H295R cells that were incubated with 0.4 mM Bt2cAMP for 1 h using the NE-PER kit (Pierce, Rockford, IL). Double-stranded oligonucleotides (Table 1) were labeled with [α-32P] dCTP (MP Biomedicals, Solon, OH) using DNA polymerase I, Klenow fragment (Stratagene, La Jolla, CA). For EMSA reactions, 5 μg of nuclear protein, 0.5 μg poly(dI•dC), 50 μg BSA, and 10,000 CPM of 32P-labeled probe were mixed in 25 μL binding buffer [20 mM HEPES (pH 7.9), 80 mM KCl, 5 mM MgCl2, 2% Ficoll, 5% glycerol, 0.1 mM EDTA, and 0.2 mM dithiothreitol] at room temperature. For supershift assays, 1 μg of anti-CREB antibody (sc-240X, Santa Cruz) was incubated with nuclear extracts, BSA, and poly (dI•dC) for 20 min at room temperature. Labeled probe was added and the reactions were incubated for an additional 15 min at room temperature. In some reactions, 500 ng of recombinant CREB protein (Biomol International, Plymouth Meeting, PA) was used instead of nuclear extracts. DNA–protein complexes were resolved on 5% polyacrylamide/0.5% Ficoll/0.5× TBE gels and the dried gels exposed to a phosphoimager screen. Complexes were visualized by phosphoimager scanning (Phospho/Fluorimager, Fuji Film, Japan).
2.7. Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed as described in Dammer et al. [36]. Briefly, H295R cells were sub-cultured onto 150 mm dishes and treated with 0.4 mM Bt2cAMP for 30 or 60 min. The purified chromatin solutions were immunoprecipitated using 5 μg of anti-phospho-CREB (Ser133), anti-CBP, anti-p300, and anti-trimethyl H3K4. All antibodies were obtained from Millipore. Real-time PCR was carried out using the primer sets indicated in Table 1 and output (immunoprecipitated promoter region) normalized to input DNA. Some reactions were also subjected to agarose (4%) gel electrophoresis.
2.8. RNAi and real time RT-PCR
Cells were sub-cultured onto 12-well plates and 24 h later transfected with 75 nM of nonspecific small interfering RNA (siRNA) oligonucleotides or siRNA oligonucleotides directed against CREB (SI00299894, Qiagen, Valencia, CA) using HiPerfect Transfection Reagent (Qiagen). After 48 h, cells were treated with 0.4 Bt2cAMP for 24 h. Total RNA was extracted using TRIzol (Invitrogen) and ASAH1 mRNA expression was quantified using a One-Step SYBR Green RT-PCR Kit (Thermo Scientific) and normalized to β-actin. Western blotting was performed to confirm reduction of CREB protein expression.
2.9. Western blotting (WB)
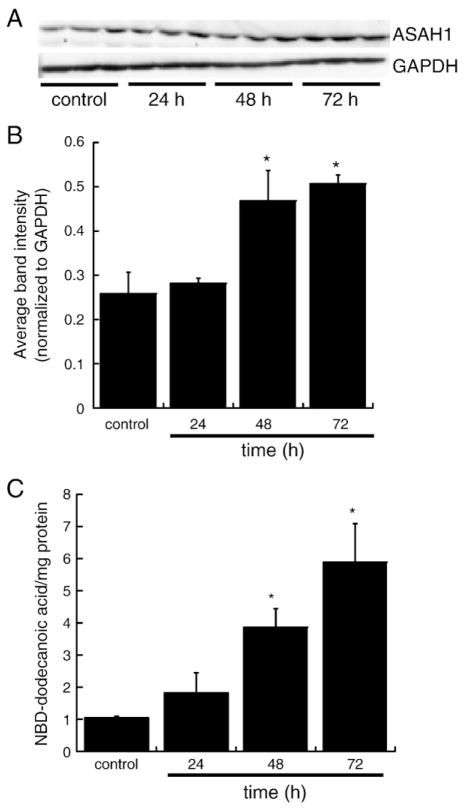
H295R cells were treated with 0.4 mM Bt2cAMP for 24, 48, or 72 h and harvested into RIPA buffer. Cells were then lysed by sonication (one 5 s burst) followed by incubation on ice for 30 min. Lysates were centrifuged for 10 min at 4 °C and the supernatant collected for analysis by SDS-PAGE. Aliquots of each sample (25 μg of protein) were run on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Thermo Scientific, Rockford, IL). Blots were probed with an anti-ASAH1 antibody (HPA005468, Sigma, St. Louis, MO) and expression was detected using an ECF western blotting kit (GE Healthcare, Piscataway, NJ) and visualized by scanning blots on a Typhoon Trio Scanner (GE Healthcare). Protein concentrations were determined by bicinchoninic acid (BCA) Protein Assay (Pierce).
2.10. In vitro Ceramidase Activity Assay
Activity assays were performed as described in Nikolova-Karakashian and Merrill [37]. H295R cells were sub-cultured in 100-mm dishes as treated for 24, 48, or 72 h with 0.4 mM Bt2cAMP. After treatment, cells were harvested into lysis buffer (0.2% Triton X-100, 10 mM Tris–Cl, pH 7.4, 1 mM 2-mercaptoethanol, 1 mM EDTA, 15 mM NaCl) with protease inhibitors (EMD Chemicals, Gibbstown, NJ) and sonicated 5 times for 2 s burst. Protein concentrations were determined by BCA Protein Assay (Pierce). Acid ceramidase activity was assayed by incubating 100 μL of cell lysate (at least 15 μg/μL of proteins) with 2 μL of a 1 mM NBD-12-cer stock in 0.5 M acetate buffer (pH 4.5) for 2 h at 37 °C. Reactions were terminated by the addition of 10 μL oleic acid (10 mg/mL), 1 mL chloroform:methanol (2:1, v/v), and 1 mL Dole’s solution (isopropanol:heptane:2 N H2SO4, 40:10:1, v/v/v) followed by vortexing and a 10 min incubation at room temperature. Four hundred microliters heptane and 600 μL distilled H2O were added, the mixtures were vortexed for 2 min, and then centrifuged for 10 min at 4000 rpm. The lower organic phase was dried under a stream of nitrogen and spotted on Silica Gel 60 thin-layer chromatography (TLC) plates (EMD Chemicals). Plates were developed in chloroform:methanol:25% NH4OH (90:20:0.5, v/v/v) and visualized by fluorescence scanning on a Typhoon Trio Scanner (GE Healthcare). NBD-dodecanoic acid formation was quantified by densitometry and normalized to the protein content of each sample.
2.11. Statistical analysis
One-way ANOVA, Tukey–Kramer multiple comparison, and unpaired student t-tests were performed using GraphPad InStat software (GraphPad Software Inc., San Diego, CA). Significant differences from a compared value were defined as p<0.05 and denoted by asterisks (*) or carats (^).
3. Results
3.1. cAMP regulates ASAH1 mRNA expression
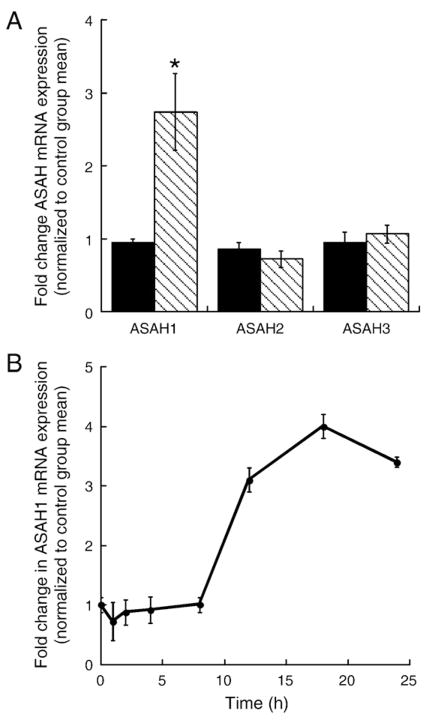
As discussed earlier, steroidogenesis in adrenocortical cells is activated by a cAMP-dependent pathway. There are 3 genes that encode ceramidase enzymes (ASAH1, ASAH2, ASAH3). Therefore, we determined the effect of increased intracellular cAMP on ASAH gene expression in H295R cells by treating with 0.4 mM Bt2cAMP for 24 h. ASAH1 mRNA expression is increased by 2.8-fold after 24 h treatment with Bt2cAMP (Fig. 1A). Conversely, no increase in expression in response to Bt2cAMP was observed for both ASAH2 and ASAH3 (Fig. 1A), indicating that the cAMP signaling pathway differentially regulates the expression of these genes. Of note, similar results were obtained for mouse Y1 adrenocortical cells (data not shown). Next, we assessed the kinetics of the ASAH1 response to Bt2cAMP by treating H295R cells for 1 to 24 h. These temporal experiments revealed that Bt2cAMP evoked a rapid and transient decrease in ASAH1 mRNA expression within 1 h, followed by a chronic increase in mRNA expression (Fig. 1B). A maximal 4-fold increase in ASAH1 mRNA expression was observed at the 18 h time point.
Fig. 1.
Bt2cAMP increases ASAH1 mRNA expression. (A) H295R cells were cultured onto 12-well plates and treated for 24 h with 0.4 mM Bt2cAMP. Total RNA was isolated for analysis of ASAH1, ASAH2, or ASAH3, and β-actin mRNA expression by qRT-PCR. Data is graphed as fold change in ASAH mRNA expression and normalized to the mRNA expression of β-actin. Data graphed represent the mean±SEM of three separate experiments, each performed in triplicate. Statistically different from untreated control group, p<0.05. (B) H285R cells were treated for 1 to 24 h with 0.4 mM Bt2cAMP and ASAH1 and β-actin mRNA expression quantified by real time RT-PCR. Data is graphed as fold change in ASAH1 mRNA content and is normalized to the mRNA expression of β-actin. Data graphed represent the mean±SEM of three separate experiments each performed in triplicate. Statistically different from untreated control group, p<0.05.
3.2. CREB increases ASAH1 reporter gene activity
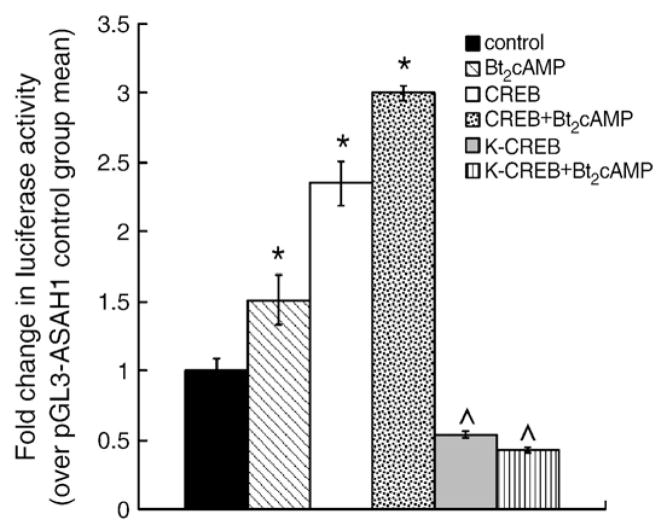
To further define the mechanism by which Bt2cAMP modulates ASAH1 mRNA expression, we cloned 2.7 kb of the human ASAH1 promoter and ligated the amplified product into the pGL3 vector. H295R cells were transfected with this reporter construct for 24 h followed by exposure to 0.4 mM Bt2cAMP for 16 h. As shown in Fig. 2, Bt2cAMP treatment significantly increased the transcriptional activity of the 2.7 kb reporter gene by 1.5-fold. Because we observed a significant increase in ASAH1 transcription in response to Bt2cAMP treatment, we performed in silico analysis of the 2.7 kb ASAH1 promoter and found 6 putative CREs (Fig. 2). Overexpression of CREB increased luciferase activity of the 2.7 kb construct by 2.3 fold when compared to cells transfected with only pGL3-ASAH1(−2740). Bt2-cAMP stimulation resulted in a further 30% increase in reporter gene activity over the CREB-mediated activation in luciferase expression.
Fig. 2.
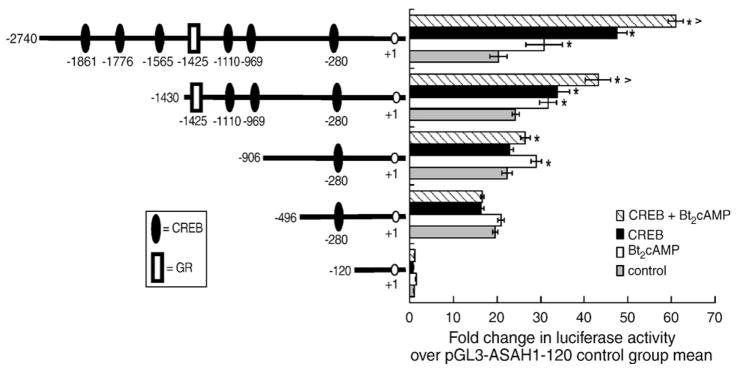
Localization of the cAMP- and CREB-responsive region(s) of the ASAH1 promoter. H295R cells were transfected with reporter gene plasmids (pGL3-ASAH1) containing varying regions of the ASAH1 promoter and a CREB expression plasmid using Gene Juice as described in Materials and methods. Twenty-four hours after transfection, cells were treated with 0.4 mM Bt2cAMP for 16 h and luciferase activity in the cell lysates quantified by luminometry. Statistical difference from untreated control within each transfection group or between control and Bt2cAMP-treated cells transfected with CREB is denoted by * and ^, respectively; p<0.05.
To define the cAMP-responsive region(s) of the ASAH1 promoter, deletion constructs were generated and assayed in transfection assays. While deletion of region −2740 to −906 had no significant effect on the ability of Bt2cAMP to stimulate reporter gene activity, removal of 410 base pairs (−496 construct) completely attenuated the cAMP response (Fig. 2). Deletion of the region encompassing −2740 to −1430 decreased the stimulatory effect of CREB overexpression by approximately 33% in both control and Bt2cAMP-treated cells and deletion of the region encompassing −1430 to −906 completed ablated the CREB response. The integral role of CREB was confirmed in reporter gene studies using a dominant-negative CREB (pCMV-K-CREB) [38] which completely attenuated the stimulatory effects of both CREB and Bt2cAMP (Fig. 3). Notably, dominant-negative CREB also reduced basal luciferase activity by 50%, suggesting that CREB may play a role in regulating the constitutive expression of ASAH1.
Fig. 3.
Dominant-negative mutant CREB abrogates ASAH1 reporter gene activity. H295R cells were transfected with pGL3-ASAH1(−2740), wild type (pCMV-CREB) or dominant-negative mutant (pCMV-K-CREB) CREB expression plasmids, and pRL-TK. Luciferase activity in lysates isolated from control and Bt2cAMP-treated cells was quantified by luminometry. Data are expressed as the fold change in pGL3-ASAH1 (−2740) reporter gene activity over the untreated control group mean and represent the mean±SEM of three separate experiments, each performed in triplicate. Asterisks (*) and carats (^) denote a statistically significant difference (p<0.05) from the untreated control group and the untreated CREB-transfected group, respectively.
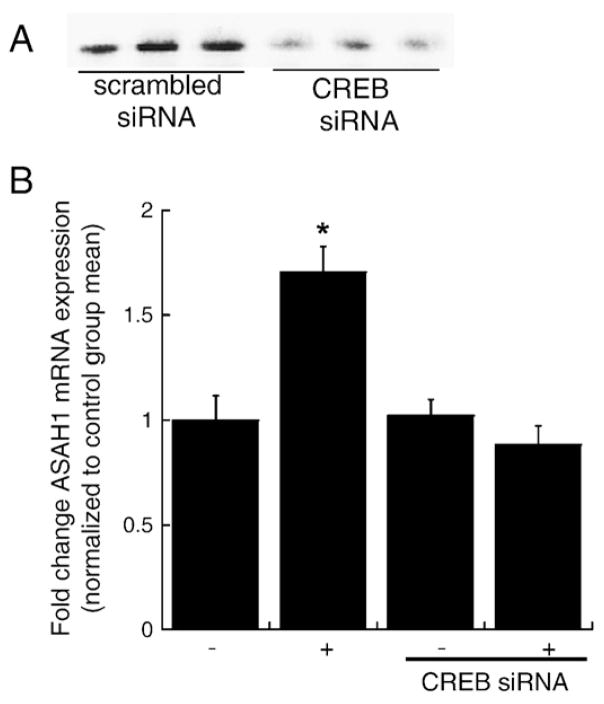
3.3. CREB is required for cAMP-stimulated ASAH1 mRNA expression
In order to investigate if CREB was required for ASAH1 transcription, we used RNAi to suppress CREB translation (Fig. 4A) and assessed the effect of reduced CREB expression on cAMP-stimulated ASAH1 transcription. As shown in Fig. 4B, H295R cells transfected with CREB siRNA oligonucleotides lose the ability to respond to Bt2cAMP.
Fig. 4.
Silencing CREB decreases cAMP-stimulated ASAH1 transcription. (A) H295R cells were transfected with 75 nM CREB or scrambled siRNA oligonucleotides for 72 h and total cell lysates were isolated for SDS-PAGE and western blotting. Blots were hybridized to an anti-CREB antibody. (B) RNA isolated from untreated or Bt2cAMP-stimulated cells that were transfected with CREB siRNA were subjected to qRT-PCR. Data is graphed as fold change in ASAH1 mRNA expression and is normalized to the mRNA expression of β-actin and represent the mean±SEM of three separate experiments, each performed in triplicate. −, Control; +, Bt2cAMP. (*) indicates a statistically significant difference compared to untreated controls (p<0.05).
3.4. CREB binds to the ASAH1 promoter in vitro and cAMP strengthens this binding
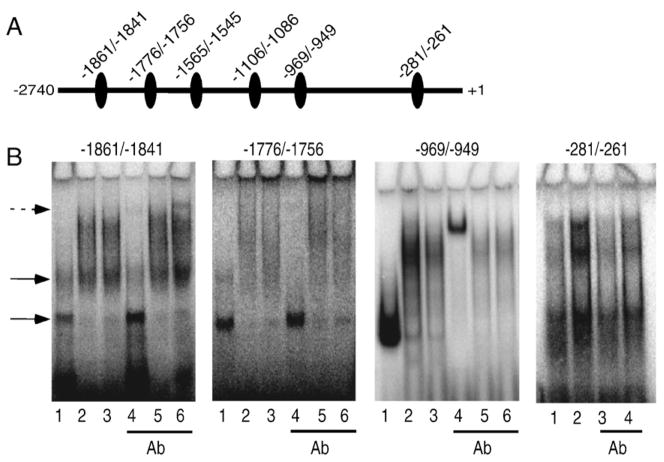
Our reporter gene assay with ASAH1 deletion constructs indicated that CREs located between −2750 and −906 confer CREB responsiveness (Fig. 2). Therefore, we carried out EMSA to determine which of the putative CREs interacted with CREB. Radiolabeled double-stranded oligonucleotides corresponding to the 6 regions depicted in Fig. 5A (sequences in Table 1) were incubated with either recombinant CREB or nuclear extracts isolated from control or Bt2cAMP-treated cells. As shown in Fig. 5B, specific DNA–protein complexes were found with 4 of the 6 probes tested (−1861/−1841, −1776/−1756, −969/−949, −281/−261). The putative CREs at −1861/−1841 (panel 1, lane 1) and −1776/−1756 (panel 2, lane 1) exhibited weak interaction with recombinant CREB when compared to the −969/−949 (panel 3, lane 1) probe.
Fig. 5.
CREB binds to the ASAH1 promoter in vitro. (A) Depiction of the regions of the ASAH1 promoter corresponding to each oligonucleotide probe. Ovals indicate putative CREB binding sites. (B) Nuclear extracts isolated from cells treated with 0.4 mM Bt2cAMP for 1 h or recombinant CREB (0.5 μg) were incubated with 32P radiolabeled oligonucleotides (10,000 cpm) corresponding to the following regions of the ASAH1 promoter: −1861/−1841, −1776/−1756, −969/−949, and −281/−261. Lanes in panels 1–3: 1) recombinant CREB, 2) control nuclear extracts, 3) Bt2cAMP-treated nuclear extracts, 4) recombinant CREB+anti-CREB antibody, 5) control nuclear extracts+anti-CREB antibody, and 6) Bt2cAMP-treated nuclear extracts+anti-CREB antibody. Panel 4: untreated nuclear extracts (lane 1), Bt2cAMP-treated nuclear extracts (lane 2), untreated nuclear extracts+anti-CREB antibody (lane 3), and Bt2cAMP-treated nuclear extracts+anti-CREB antibody (lane 4). Solid arrows indicate CREB binding and dashed arrow denotes shifted bands.
A weak upper and more intense lower band was revealed in reactions containing recombinant CREB and −1861/−1841 or −1776/−1756 (lane 1 in panels 1 and 2), suggesting the interaction of monomeric (lower band) and dimeric (upper band) CREB with these regions of the ASAH1 promoter. In contrast, the −969/−949 probe exhibited one major DNA–protein complex when incubated with recombinant CREB (panel 3, lane 1). A supershift was observed when anti-CREB antibody was added to the reaction containing recombinant CREB and the −969/−949 probe (panel 3, lane 4). Further, when nuclear extracts were incubated with the −969/−949 probe in the presence of anti-CREB antibody, a significant reduction in DNA–protein complex formation was observed (panel 3, lanes 5 and 6). Notably, the mobility of DNA–protein complexes formed when the −969/−949 region was incubated with nuclear proteins (panel 3, lanes 3 and 4) was significantly retarded when compared to reactions containing recombinant CREB (panel 3, lane 1). This difference in mobility suggests that CREB may be a component of a multi-protein complex.
A weak supershift was also found when the antibody was added to reactions containing the −1861/−1841 probe and nuclear extracts isolated from Bt2cAMP-treated cells (panel 1, lane 6). Bt2cAMP treatment increased the affinity of nuclear proteins for the −281/−261 region (compare lanes 1 and 2 in panel 4) and the CREB antibody decreased cAMP-stimulated complex formation (compare lanes 2 and 4 in panel 4). No significant binding to −1565/−1545 and −1110/−1086 oligonucleotides was observed (data not shown). Additionally, we did not observe changes in the formation of DNA–protein complexes when reactions contained antibodies against ATF-1 or ATF-2 (data not shown).
3.5. cAMP promotes the recruitment of CREB and other coregulatory proteins to the ASAH1 promoter
We next examined the effect of cAMP stimulation on the recruitment of CREB and coactivator proteins to the endogenous ASAH1 promoter (Fig. 6A) by performing ChIP assays using chromatin isolated from H295R cells that were treated with 0.4 mM Bt2cAMP for 30 or 60 min. Our results indicate that CREB is enriched by 2.6-fold at region A (−1779/−1596) and 15-fold at region D (−325/−214) in cells treated with Bt2cAMP for 30 min (Fig. 6B). One h exposure to Bt2cAMP increased CREB recruitment to region A by 4.9-fold. Region A is also shown in the agarose gel in the panel B inset. Bt2cAMP had no significant effect on CREB binding in the other three regions (B, C, and E) of the ASAH1 promoter that were examined.
Fig. 6.
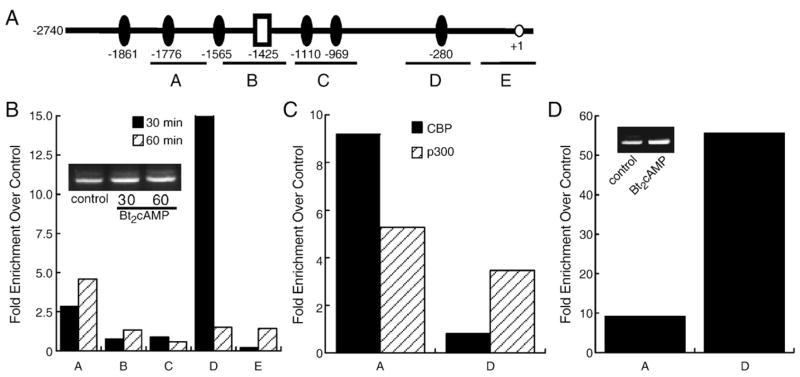
CREB, CBP, and p300 bind to the ASAH1 promoter in vivo. (A) Diagram of the promoter regions amplified by each primer set used for ChIP assay. (B) H295R cells were treated for 30 or 60 min with 0.4 mM Bt2cAMP, cross-linked with 1% formaldehyde, and the sheared chromatin immunoprecipitated with antibodies against anti-phospho-CREB (Ser133). (C) Chromatin isolated from cells treated for 60 min with 0.4 mM Bt2cAMP was immunoprecipitated with antibodies against anti-CBP or anti-p300 and recruitment to region A or D of the ASAH1 promoter assessed by qPCR. (D) H295R cells were treated for 30 min with 0.4 mM Bt2cAMP and the isolated chromatin immunoprecipitated with an antibody against trimethyl-H3K4. In panels B, C, and D, DNA purified was quantified by real time PCR and normalized to the ΔCt values of input DNA. Data is expressed as fold change over untreated control. Representative agarose gels of the PCR products obtained for region A are shown as insets to panel B (control, 30, and 60 min Bt2cAMP treatment) and panel D (control and 30 min Bt2cAMP treatment).
We also assessed the effect of cAMP signaling on coactivator association with the regions (A and D) of the ASAH1 promoter that exhibited enriched CREB binding. Both CBP and p300 were recruited to region A in cells exposed to Bt2cAMP for 60 min (Fig. 6C). Finally, since activated gene transcription is associated with the increased presence of specific histone modifications, we determined the effect of Bt2cAMP on the amount of trimethylated H3K4, a modification associated with active transcription [39], in regions A and D of the ASAH1 promoter. As shown in Fig. 6D (region A shown in agarose gel inset), Bt2cAMP evoked an increase in the trimethylation of H3K4, indicating that cAMP-stimulated CREB binding occurs concomitantly with histone H3 modification.
3.6. cAMP increases ASAH1 protein expression and enzymatic activity
Because Bt2cAMP increased ASAH1 mRNA levels, we sought to investigate if this effect translates into an increase in protein expression. We carried out western blotting analysis in H295R cells treated for 24, 48, or 72 h with Bt2cAMP. As shown in Fig. 7A and B, Bt2cAMP significantly increased ASAH1 protein levels by 1.81- and 1.96-fold after 48 and 72 h treatment, respectively. In addition, to determine if the increase in protein expression was functionally significant, we quantified ASAH activity in Bt2cAMP-treated H295R cells. As shown in Fig. 7C, Bt2cAMP significantly increased total ceramidase activity by 3.7- and 5.6-folds after 48 and 72 h.
Fig. 7.
Bt2cAMP increases ASAH1 protein expression and enzymatic activity. (A) Representative western blot of H295R cells treated for 24, 48, or 72 h with 0.4 mM Bt2cAMP. Cell lysates were harvested and separated in a 10% SDS-PAGE gel followed by western blotting using anti-ASAH1 or anti-GAPDH antibodies. (B) Graphical analysis of data obtained in western blotting studies of ASAH1 protein expression in cells treated for 24 to 72 h with 0.4 mM Bt2cAMP. Data graphed represent the mean±SEM of two separate experiments, each carried out in triplicate. Asterisks indicate a statistically significant difference compared to untreated controls (p<0.05). (C) H295R cells were treated for 24 to 72 h with 0.4 mM Bt2cAMP and cell lysates were isolated and for ceramidase assays as described in the Materials and methods section. Plates were developed in chloroform:methanol:25% NH4OH (90:20:0.5, v/v/v) and visualized by fluorescence scanning on a Typhoon Trio Scanner. NBD-dodecanoic acid formation was quantified and normalized to the protein content of each sample. Data graphed are the mean±STD of two experiments each performed in duplicate. Asterisks indicate a statistically significant difference compared to untreated controls (p<0.05).
4. Discussion
ACTH/cAMP signaling regulates cortisol biosynthesis in the human adrenal cortex by chronically stimulating the transcription of multiple genes required for cholesterol metabolism and transport [18]. In addition, ACTH acutely increases sphingolipid metabolism in H295R cells, resulting in changes in the cellular concentrations of bioactive sphingolipids that regulate steroidogenic gene transcription [22]. Herein we show that ACTH/cAMP induces ASAH1 gene transcription by promoting the binding of CREB to multiple regions on the ASAH1 promoter.
We show that cAMP differentially regulates ASAH genes (Fig. 1A) and that the transcription of the ASAH1 gene is modulated by cAMP in a time-dependent manner with maximal induction of the gene at the 18 h time point (Fig. 1B). Interestingly, cAMP acutely reduces ASAH1 mRNA expression within 1 h. These data indicate that ACTH/cAMP signaling evokes a biphasic pattern of ASAH1 mRNA expression, with an initial decrease followed by transcriptional induction. The rapid decrease in the mRNA expression ASAH1 suggests that cAMP may regulate the stability of ASAH1 transcripts and possibly promote the acute degradation of ASAH1 mRNAs. Regulation of RNA stability is a fundamental mechanism of gene regulation.
MicroRNAs (miRNA) and 3′UTR AU-rich elements (AURE) are two types of regulatory molecules capable of destabilizing transcripts [40–42]. miRNAs are single-stranded RNA molecules that down-regulate gene expression by binding to complementary regions of mRNA molecules and either targeting them for degradation or blocking the assembly of the translational machinery [42]. AUREs contain clustered or distributed AUUUA pentameric motifs or an U-rich sequence where interacting proteins bind to either stabilize (e.g. HuR) or destabilize (e.g. TTP, KSRP) the transcript by signal transduction pathways [43,44]. Of note, these regulatory elements have been shown to regulate the expression of genes involved in steroidogenesis including SK1 [45] and steroidogenic acute regulatory protein (StAR) [46]. We have found multiple miRNA target sites and 3′UTR AU-rich regions on the ASAH1 mRNA (Lucki and Sewer, unpublished observation). Thus, it is possible that these regulatory mechanisms are involved in the observed ASAH1 mRNA decrease induced by cAMP. Interestingly, a similar acute decrease in mRNA expression was also observed for ASAH2 and ASAH3 in response to ACTH and Bt2cAMP, suggesting that a similar regulatory mechanism may be involved (Lucki and Sewer, unpublished observation). Studies aimed at defining the mechanism by which cAMP promotes the acute decrease in mRNA expression of ASAH transcripts are underway.
Luciferase assays using deletion constructs of the ASAH1 promoter identified a role for CREB in increased reporter gene activity that was localized to the region encompassing −2740 and −906 base pairs upstream of the transcription start site (Fig. 2). ChIP (Fig. 6) and EMSA (Fig. 5B) studies support a role for cAMP in stimulating the association of CREB with the ASAH1 promoter. Our ChIP experiments (Fig. 6) revealed that CREB is enriched at region −325/−214 (region D) of the ASAH1 promoter, however we did not observe a stimulatory effect of CREB overexpression on reporter gene activity when cells were transfected with the pGL3-ASAH1(−496) plasmid that contains the −325/−214 region (Fig. 2). It is possible that although cAMP promotes CREB recruitment to the endogenous promoter, an increase in transcription requires the coordinate binding of CREB to multiple regions of the ASAH1 promoter. It is equally likely that cAMP signaling increases the binding of additional transcription factors in conjunction with CREB to the promoter. Nonetheless, RNAi (Fig. 4) and the use of a dominant-negative CREB expression plasmid (Fig. 3) confirm the integral role of CREB in conferring increased gene expression in response to cAMP. Trimethylation of H3K4 and the recruitment of CBP and p300 to both the distal (region A) and proximal (region D) regions of the promoter further support a role for CREB in activating ASAH1 gene transcription.
Functional promoter characterization has been reported for some of the genes encoding sphingolipid metabolizing enzymes, including S1P phosphatase 2 [47], subunit 2 of serine palmitoyltransferase [48], ganglioside GM3 synthase [49], ceramide glucosyltransferase [50], and ASAH2 [51]. However, few studies have identified the transcription factors involved in the regulation of these genes. Sobue et al. reported a nerve growth factor-mediated induction of the SK1 gene via binding of Sp1 (specificity protein 1) to a specific 5′ region of the promoter [52] and Mechtcheriakova et al. demonstrated that NF-κB is necessary for induction of the S1P phosphatase 2 gene in response to inflammatory stimuli [47].
Although previous studies have defined the structural units of the ASAH1 gene [53,54], to our knowledge there is only one report that investigates the transcriptional regulation of an ASAH gene. Park et al. characterized a 1931 base pair region of the murine ASAH1 promoter and demonstrated that Kruppel like factor 6, Sp1, and AP2 (activator protein 2) can bind the promoter in vitro [55]. Our findings herein provide functional characterization of the ASAH1 promoter and establish the factors that facilitate ACTH/cAMP-dependent ASAH1 gene expression.
We also show that the effect of ACTH/cAMP signaling on ASAH1 transcription results in a significant increase in protein expression (Fig. 7). Increased protein concentration is, in turn, concomitant with an upregulation in ceramidase enzymatic activity. These data herein, coupled with our previous findings establishing the role of ACTH/ cAMP signaling in acutely modulating sphingolipid metabolism [56], demonstrate that activation of the ACTH signaling pathway elicits two temporally distinct effects on sphingolipid metabolism; a rapid response and a chronic, transcriptional response. In addition, these data supports a role for ASAH1 as an important enzyme for the regulation of sphingolipid metabolism in response to ACTH/cAMP signaling that consequently modulates cortisol biosynthesis.
We have previously demonstrated that ACTH promotes rapid changes in sphingolipid intracellular concentrations, including a decrease in cer [22]. Thus, our data indicating an ACTH-regulated transcription and activity of ASAH1 is in agreement with such previously finding and suggests that ACTH signaling is directly regulating intracellular SPH and S1P concentrations. Given that SPH is an antagonist ligand for SF-1 and S1P promotes CYP17 expression, it is tempting to speculate that the transcriptional regulation of the ASAH1 gene by ACTH/cAMP signaling is part of a regulatory mechanism through which ACTH modulates cortisol production. In summary, we identify CREB as a central regulator of ASAH1 gene transcription and demonstrate that activation of the cAMP signaling pathway promotes the coordinate interaction of CREB and the coactivators CBP and p300 to multiple regions of the ASAH1 promoter, concomitant with the trimethylation of H3K4.
References
- 1.Gomez-Munoz A. Ceramide 1-phosphate/ceramide, a switch between life and death. Biochim Biophys Acta. 2006;1758:2049–2056. doi: 10.1016/j.bbamem.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Gomez-Munoz A, Kong J, Salh B, Steinbrecher UP. Sphingosine-1-phosphate inhibits acid sphingomyelinase and blocks apoptosis in macrophages. FEBS Lett. 2003;539:56–60. doi: 10.1016/s0014-5793(03)00197-2. [DOI] [PubMed] [Google Scholar]
- 3.Brizuela L, Rabano M, Pena A, Gangoiti P, Macarulla JM, Trueba M, Gomez-Munoz A. Sphingosine 1-phosphate: a novel stimulator of aldosterone secretion. J Lipid Res. 2006;47:1238–1249. doi: 10.1194/jlr.M500510-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Budnik LT, Jahner D, Mukhopadhyay AK. Inhibitory effects of TNF alpha on mouse tumor Leydig cells: possible role of ceramide in the mechanism of action. Mol Cell Endocrinol. 1999;150:39–46. doi: 10.1016/s0303-7207(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 5.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 6.Degnan BM, Bourdelat-Parks B, Daniel A, Salata K, Francis GL. Sphingomyelinase inhibits in vitro Leydig cell function. Ann Clin Lab Sci. 1996;26:234–242. [PubMed] [Google Scholar]
- 7.Hannun YA. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 8.Kihara A, Mitsutake S, Mizutani Y, Igarashi Y. Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog Lipid Res. 2007;46:126–144. doi: 10.1016/j.plipres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Meroni SB, Pellizzari EH, Canepa DF, Cigorraga SB. Possible involvement of ceramide in the regulation of rat Leydig cell function. J Steroid Biochem Mol Biol. 2000;75:307–313. doi: 10.1016/s0960-0760(00)00188-6. [DOI] [PubMed] [Google Scholar]
- 10.Rabano M, Pena A, Brizuela L, Marino A, Macarulla JM, Trueba M, Gomez-Munoz A. Sphingosine-1-phosphate stimulates cortisol secretion. FEBS Lett. 2003;535:101–105. doi: 10.1016/s0014-5793(02)03882-6. [DOI] [PubMed] [Google Scholar]
- 11.Thon L, Mohlig H, Mathieu S, Lange A, Bulanova E, Winoto-Morbach S, Schutze S, Bulfone-Paus S, Adam D. Ceramide mediates caspase-independent programmed cell death. FASEB J. 2005;19:1945–1956. doi: 10.1096/fj.05-3726com. [DOI] [PubMed] [Google Scholar]
- 12.Santana P, Llanes L, Hernandez I, Gonzalez-Robayna I, Tabraue C, Gonzalez-Reyes J, Quintana J, Estevez F, Ruiz de Galarreta CM, Fanjul LF. Interleukin-1 beta stimulates sphingomyelin hydrolysis in cultured granulosa cells: evidence for a regulatory role of ceramide on progesterone and prostaglandin biosynthesis. Endocrinology. 1996;137:2480–2489. doi: 10.1210/endo.137.6.8641202. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Ni J, Bian S, Yao L, Zhu H, Zhang W. Inhibition of steroidogenesis and induction of apoptosis in rat luteal cells by cell-permeable ceramide in vitro. ShengLi XueBao. 2001;53:142–146. [PubMed] [Google Scholar]
- 14.Morales V, Santana P, Diaz R, Tabraue C, Gallardo G, Blanco FL, Hernandez I, Fanjul LF, Ruiz de Galarreta CM. Intratesticular delivery of tumor necrosis factor-{alpha} and ceramide directly abrogates steroidogenic acute regulatory protein expression and Leydig cell steroidogenesis in adult rats. Endocrinology. 2003;144:4763–4772. doi: 10.1210/en.2003-0569. [DOI] [PubMed] [Google Scholar]
- 15.Santana P, Llanes L, Hernandez I, Gallardo G, Quintana J, Gonzalez J, Estevez F, Ruiz de Galarreta C, Fanjul LF. Ceramide mediates tumor necrosis factor effects on P450-aromatase activity in cultured granulosa cells. Endocrinology. 1995;136:2345–2348. doi: 10.1210/endo.136.5.7720683. [DOI] [PubMed] [Google Scholar]
- 16.Son DS, Arai KY, Roby KF, Terranova PF. Tumor necrosis factor alpha (TNF) increases granulosa cell proliferation: dependence on c-Jun and TNF receptor type 1. Endocrinology. 2004;145:1218–1226. doi: 10.1210/en.2003-0860. [DOI] [PubMed] [Google Scholar]
- 17.Urs AN, Dammer E, Sewer MB. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology. 2006;147:5249–5258. doi: 10.1210/en.2006-0355. [DOI] [PubMed] [Google Scholar]
- 18.Sewer MB, Waterman MR. ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc Res Tech. 2003;61:300–307. doi: 10.1002/jemt.10339. [DOI] [PubMed] [Google Scholar]
- 19.Bakke M, Zhao L, Hanley NA, Parker KL. SF-1: a critical mediator of steroidogenesis. Mol Cell Endocrinol. 2001;171:5–7. doi: 10.1016/s0303-7207(00)00384-1. [DOI] [PubMed] [Google Scholar]
- 20.Ozbay T, Rowan A, Leon A, Patel P, Sewer MB. Cyclic adenosine 5′-monophosphate-dependent sphingosine-1-phosphate biosynthesis induces human CYP17 gene transcription by activating cleavage of sterol regulatory element binding protein 1. Endocrinology. 2006;147:1427–1437. doi: 10.1210/en.2005-1091. [DOI] [PubMed] [Google Scholar]
- 21.Brizuela L, Rabano M, Gangoiti P, Narbona N, Macarulla JM, Trueba M, Gomez-Munoz A. Sphingosine-1-phosphate stimulates aldosterone secretion through a mechanism involving the PI3K/PKB and MEK/ERK 1/2 pathways. J Lipid Res. 2007;48:2264–2274. doi: 10.1194/jlr.M700291-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Ozbay T, Merrill AH, Jr, Sewer MB. ACTH regulates steroidogenic gene expression and cortisol biosynthesis in the human adrenal cortex via sphingolipid metabolism. Endocr Res. 2004;30:787–794. doi: 10.1081/erc-200044040. [DOI] [PubMed] [Google Scholar]
- 23.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev, Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 24.Sands WA, Palmer TM. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008;20:460–466. doi: 10.1016/j.cellsig.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 26.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 27.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 28.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 29.Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, Chirn GW, McWhinnie E, Cohen D, Skelton J, Terry R, Yu Y, Bodian D, Buxton FP, Zhu J, Song C, Labow MA. From the cover: identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Kibler KV, Jeang KT. CREB/ATF-dependent repression of cyclin a by human T-cell leukemia virus type 1 Tax protein. J Virol. 2001;75:2161–2173. doi: 10.1128/JVI.75.5.2161-2173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Gedrich RW, Engel DA. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staels B, Hum DW, Miller WL. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol Endocrinol. 1993;7:423–433. doi: 10.1210/mend.7.3.8387159. [DOI] [PubMed] [Google Scholar]
- 35.Rainey WE, Bird IM, Mason JI. the NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol Cell Endocrinol. 1994;99:R17–R20. doi: 10.1016/0303-7207(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 36.Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol. 2007;21:415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- 37.Nikolova-Karakashian M, Merrill AH., Jr Ceramidases. Methods Enzymol. 2000;311:194–201. doi: 10.1016/s0076-6879(00)11081-x. [DOI] [PubMed] [Google Scholar]
- 38.Walton KM, Rehfuss RP, Chrivia JC, Lochner JE, Goodman RH. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)- regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol Endocrinol. 1992;6:647–655. doi: 10.1210/mend.6.4.1350057. [DOI] [PubMed] [Google Scholar]
- 39.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 40.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 41.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 43.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 44.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Sobue S, Nemoto S, Murakami M, Ito H, Kimura A, Gao S, Furuhata A, Takagi A, Kojima T, Nakamura M, Ito Y, Suzuki M, Banno Y, Nozawa Y, Murate T. Implications of sphingosine kinase 1 expression level for the cellular sphingolipid rheostat: relevance as a marker for daunorubicin sensitivity of leukemia cells. Int J Hematol. 2008;87:266–275. doi: 10.1007/s12185-008-0052-0. [DOI] [PubMed] [Google Scholar]
- 46.Duan H, Jefcoate CR. The predominant cAMP-stimulated 3×5 kb StAR mRNA contains specific sequence elements in the extended 3′UTR that confer high basal instability. J Mol Endocrinol. 2007;38:159–179. doi: 10.1677/jme.1.02153. [DOI] [PubMed] [Google Scholar]
- 47.Mechtcheriakova D, Wlachos A, Sobanov J, Kopp T, Reuschel R, Bornancin F, Cai R, Zemann B, Urtz N, Stingl G, Zlabinger G, Woisetschlager M, Baumruker T, Billich A. Sphingosine 1-phosphate phosphatase 2 is induced during inflammatory responses. Cell Signal. 2007;19:748–760. doi: 10.1016/j.cellsig.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Linn SC, Andras LM, Kim HS, Wei J, Nagiec MM, Dickson RC, Merrill AH., Jr Functional characterization of the promoter for the mouse SPTLC2 gene, which encodes subunit 2 of serine palmitoyltransferase. FEBS Lett. 2006;580:6217–6223. doi: 10.1016/j.febslet.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SW, Lee SH, Kim KS, Kim CH, Choo YK, Lee YC. Isolation and characterization of the promoter region of the human GM3 synthase gene. Biochim Biophys Acta. 2002;1578:84–89. doi: 10.1016/s0167-4781(02)00505-5. [DOI] [PubMed] [Google Scholar]
- 50.Ichikawa S, Ozawa K, Hirabayashi Y. Molecular cloning and characterization of the mouse ceramide glucosyltransferase gene. Biochem Biophys Res Commun. 1998;253:707–711. doi: 10.1006/bbrc.1998.9855. [DOI] [PubMed] [Google Scholar]
- 51.Okino N, Mori K, Ito M. Genomic structure and promoter analysis of the mouse neutral ceramidase gene. Biochem Biophys Res Commun. 2002;299:160–166. doi: 10.1016/s0006-291x(02)02540-8. [DOI] [PubMed] [Google Scholar]
- 52.Sobue S, Hagiwara K, Banno Y, Tamiya-Koizumi K, Suzuki M, Takagi A, Kojima T, Asano H, Nozawa Y, Murate T. Transcription factor specificity protein 1 (Sp1) is the main regulator of nerve growth factor-induced sphingosine kinase 1 gene expression of the rat pheochromocytoma cell line, PC12. J Neurochem. 2005;95:940–949. doi: 10.1111/j.1471-4159.2005.03399.x. [DOI] [PubMed] [Google Scholar]
- 53.Li CM, Park JH, He X, Levy B, Chen F, Arai K, Adler DA, Disteche CM, Koch J, Sandhoff K, Schuchman EH. The human acid ceramidase gene (ASAH): structure, chromosomal location, mutation analysis, and expression. Genomics. 1999;62:223–231. doi: 10.1006/geno.1999.5940. [DOI] [PubMed] [Google Scholar]
- 54.Li CM, Hong SB, Kopal G, He X, Linke T, Hou WS, Koch J, Gatt S, Sandhoff K, Schuchman EH. Cloning and characterization of the full-length cDNA and genomic sequences encoding murine acid ceramidase. Genomics. 1998;50:267–274. doi: 10.1006/geno.1998.5334. [DOI] [PubMed] [Google Scholar]
- 55.Park JH, Eliyahu E, Narla G, DiFeo A, Martignetti JA, Schuchman EH. KLF6 is one transcription factor involved in regulating acid ceramidase gene expression. Biochim Biophys Acta. 2005;1732:82–87. doi: 10.1016/j.bbaexp.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Ozbay T, Merrill AH, Jr, Sewer MB. ACTH regulates steroidogenic gene expression and cortisol biosynthesis in the human adrenal cortex via sphingolipid metabolism. Endocr Res. 2004;30:787–794. doi: 10.1081/erc-200044040. [DOI] [PubMed] [Google Scholar]