Abstract
There are more than 180 different genetic causes of primary immunodeficiencies identified to date. Approaches for identifying causative mutations can be broadly classified into 3 strategies: (1) educated guesses based on known signaling pathways essential for immune cell development and function, (2) similarity of clinical phenotypes to mouse models, and (3) unbiased genetic approaches. Next-generation DNA sequencing permits efficient sequencing of whole genomes or exomes but also requires strategies for filtering vast amounts of data. Recent studies have identified ways to solve difficult cases, such as diseases with autosomal dominant inheritance, incomplete penetrance, or mutations in noncoding regions. This review focuses on recently identified primary immunodeficiencies to illustrate the strategies, technologies, and potential pitfalls in finding novel causes of these diseases.
Keywords: Primary immunodeficiencies, whole-genome sequencing, whole-exome sequencing, linkage analysis, homozygosity mapping
Over the past 4 decades, more than 180 molecular defects causing primary immunodeficiencies (PIDs) have been discovered through advances in immunology and genetics. Because the majority of PIDs are monogenic, whole-exome sequencing (WES)/whole-genome sequencing (WGS) has expedited the discovery of pathogenic mutations, particularly when combined with classical methods of identifying genetic defects (Table I). Although there are many published examples, this review will focus on selected cases to illustrate the spectrum of approaches, which includes (1) educated guesses based on known signaling pathways essential for immune cell development and function, (2) similarity of clinical phenotypes to mouse models, and (3) unbiased genetic approaches (Table II). The second half of the review will address methods of overcoming challenges in identifying molecular causes of PIDs.
TABLE I.
Definitions
| Next-generation sequencing (NGS): Sequencing techniques that simultaneously amplify hundreds of millions of template DNA fragments in parallel. |
| Exome: The protein-coding regions of the genome. |
| Whole-exome sequencing (WES): A technique that uses probes to select the exonic regions from the sample before sequencing. |
| Single nucleotide polymorphism (SNP): Single base variants that constitute the most common type of variation between individual genomes. SNPs can be used as powerful genetic mapping tools through linkage analysis and homozygosity mapping. |
| Linkage analysis: A method used to define the genetic locus associated with a disease based on the principle that the causative mutation and nearby genetic markers will be inherited together. This technique identifies a region, but not the causative mutation, associated with a disease. |
| Homozygosity mapping: A strategy used to define loci with clusters of homozygous SNPs, which thus have a high probability of containing the causative homozygous mutation for an autosomal recessive disease. Like linkage analysis, this technique identifies candidate regions, but not specific mutations, associated with a disease. |
TABLE II.
Summary of pros and cons of strategies for gene hunting
| Strategy | Pros | Cons |
|---|---|---|
| Educated guesses based on known signaling pathways essential for cellular development and function | Efficient Does not require significant computational resources |
Unlikely to identify novel or unexpected genes as causes of PIDs Might not identify the causative gene because of phenotypic variability |
| Similarity of clinical phenotypes to mouse models | Might suggest a relevant gene to human disease Permits in vivo study of a mutation on immune function |
Might be confounded by differences between human and mouse immunity Does not account for environmental factors that affect human immunity |
| Unbiased genetic approaches | Unbiased comprehensive approach for identifying novel genetic causes of PIDs | Requires significant technologic resources to generate and analyze large amounts of data Might be difficult to identify the pathogenic mutation from a large candidate list of benign variations |
EDUCATED GUESS BASED ON KNOWN MOLECULAR PATHWAYS
Knowledge of signaling pathways establishes conceptual frameworks for the identification of molecular defects and their clinical consequences. When a clinical phenotype and disease inheritance pattern suggest candidate genes for a PID, targeted sequencing of these genes is the most efficient approach. This has been instrumental for identifying autosomal causes of diseases that were originally discovered as X-linked disorders. A classic example involves the discovery of the defects underlying hyper-IgM syndrome. The identification of CD40 ligand as a critical signal for class-switching set the stage for the discovery of CD40 ligand deficiency as the cause of X-linked hyper-IgM syndrome.1–6 The subsequent discovery of CD40 deficiency as a cause of autosomal recessive hyper-IgM syndrome emerged from the understanding of the CD40–CD40 ligand interaction in B-cell differentiation and class-switching.7,8 Similarly, the signaling pathways important for T-cell development facilitated the discovery of IL2RG, which encodes the IL-2 receptor γ chain (IL-2Rγ), as the cause of X-linked severe combined immunodeficiency (SCID).9 This enabled the subsequent discovery of defects underlying 2 forms of autosomal recessive SCID. Janus kinase 3 was identified as a kinase downstream of IL-2Rγ,10–12 and the IL-7 receptor was found to be a partner of IL-2Rγ in binding the cytokine IL-7, which is essential for thymocyte development.13,14 More recently, ectodermal dysplasia and immunodeficiency (ED-ID) was identified as an X-linked disorder in male patients with mutations in the IκB kinase γ nuclear factor κB (NF-κB) essential modifier (NEMO).15 Knowledge of the NF-κB signaling pathway was instrumental for identifying mutations in IκBα, a member of the NF-κB inhibitor family, as a novel cause of autosomal dominant ED-ID in female patients.16–18
Known pathways can also guide a targeted sequencing approach to specific clinical phenotypes, such as chronic mucocutaneous candidiasis (CMC). CMC is a feature of patients with defects in the IL-17 pathway, such as those with signal transducer and activator of transcription 3 deficiency and a resultant lack of TH17 cells or patients with neutralizing autoantibodies against IL-17A and IL-17F.19 This knowledge prompted the discovery of the first human mutations in the genes encoding the IL-17 receptor20 and the identification of gain-of-function mutations in the signal transducer and activator of transcription 1 gene as additional causes of an impaired TH17 response in patients with CMC.21,22
Identifying molecular defects based on known pathways is a focused approach but has a low likelihood of identifying novel genetic causes of PIDs. Additionally, hypomorphic mutations can lead to variable and unexpected phenotypes. For example, mutations in CORO1A, which encodes the actin-binding protein coronin-1A, was initially described as a cause of SCID23 but was subsequently associated with T-cell lymphopenia and EBV-associated lymphoproliferation.24 To overcome these limitations, other methods, such as mouse models and unbiased genetic approaches, have been used to broaden the scope of gene discovery.
SIMILARITY OF CLINICAL PHENOTYPES TO MOUSE MODELS OF DISEASE
Mouse models can demonstrate the physiologic importance of a gene, enabling the identification of pathogenic mutations based on phenotypic similarities between mouse models and patients. This approach was used to identify a mutation in WIPF1,25 which encodes a chaperone protein necessary for stabilizing Wiskott-Aldrich syndrome (WAS) protein,26,27 in a female patient with immunodeficiency, eczema, and thrombocytopenia. Despite phenotypic features suggestive of WAS, her WAS gene sequence was normal, and she had additional immune defects inconsistent with WAS: absence of T-cell proliferation to anti-CD3 stimulation, defective T-cell response to IL-2, and normal platelet size. These features are found in WIP-deficient mice, and targeted Sanger sequencing of WIPF1 identified a homozygous mutation causing a premature stop codon in this patient.
Additionally, mouse models can be used to prioritize a candidate gene list, as was the case in a family with congenital asplenia. WES of 3 affected siblings yielded 32 candidate genes.28 Only 1 gene, NKX2-5, encoded a transcription factor essential for mouse spleen development,29 and in vitro studies demonstrated that the mutation abolished NKX2-5 function.28
A major limitation of this approach is that mouse models do not always recapitulate human disease. For example, TANK-binding kinase 1 (TBK1), a serine/threonine kinase downstream of Toll-like receptor 3 (TLR3), is important for multiple antiviral and antibacterial pathways in mouse models,30,31 suggesting that TBK1 deficiency would manifest as broad susceptibility to viral and bacterial pathogens. However, TBK1 deficiency in human subjects is a risk factor only for HSV encephalitis.32 Therefore the mouse model of TBK1 deficiency did not predict the limited scope of human disease. In another example, mice deficient in the p85α subunit of phosphoinositide 3-kinase have blocked B-cell development, abnormal platelet function, abnormal mast cell development, increased production of IL-12 by dendritic cells, and increased insulin sensitivity.33,34 However, the patient lacking p85α had disease limited to agammaglobulinemia and absent B cells.35 Although mouse models have been indispensible to our understanding of the human immune system, notable differences between the 2 species underscores the need for unbiased approaches for discovery of the causative genes.
UNBIASED GENETIC APPROACHES
When knowledge of signaling pathways and animal models do not suggest candidate genes, genetic techniques have been instrumental for identifying pathogenic mutations. Polymorphic markers are used to map a genetic defect to a specific chromosomal region. These markers include restriction fragment length polymorphisms, microsatellites, and single nucleotide polymorphisms (SNPs). SNPs are single base changes in the human genome that occur with a population frequency of at least 1%. They are widely used in genetic studies because they are ubiquitous throughout the genome,36 adaptable to high-throughput platforms, and catalogued in publically available databases.37,38 Linkage studies use SNPs flanking a pathogenic mutation to define the disease loci shared by the affected subjects (Fig 1).39 Genes within the candidate loci are then sequenced to identify the causative variant.
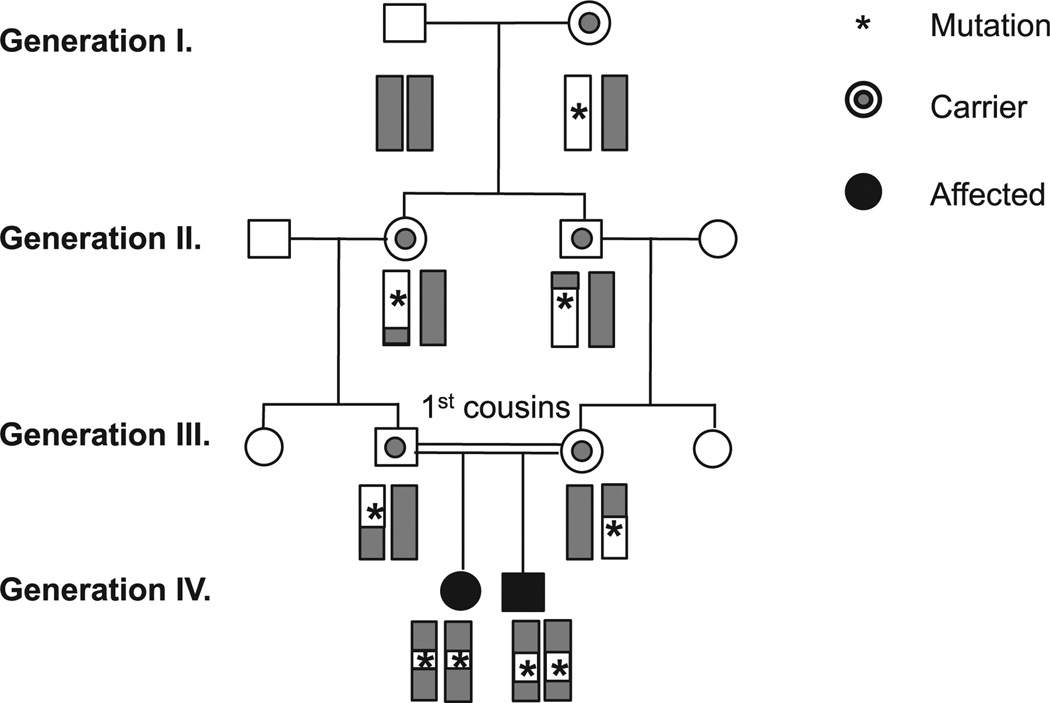
FIG 1.
Linkage analysis in a consanguineous family. The deleterious mutation arose in the patients’ great-grandmother. It is inherited with genetic markers, such as SNPs, which define the genetic locus containing the pathogenic mutation.
Linkage analysis was used to identify homozygous mutations in LRBA, which encodes LPS-responsive beige-like anchor protein (LRBA), in patients with hypogammaglobulinemia, decreased memory B-cell numbers, and autoimmunity.40 Linkage analysis of 5 affected subjects within 4 consanguineous families identified a candidate interval containing 81 genes. Subsequent Sanger sequencing of individual genes revealed homozygous mutations in LRBA in each patient. This approach requires large families with multiple affected members and might identify large candidate regions with many genes requiring expensive and time-consuming Sanger sequencing.
In the mid-2000s, next-generation DNA sequencing (NGS) revolutionized genetics by making it possible to sequence entire human genomes within days. NGS encompasses a variety of methods that simultaneously amplify and sequence millions of DNA fragments.41 Although this technology offers comprehensive sequencing data, it is challenging to distinguish pathogenic variants within the 3.2 billion bases present in the human genome.42 Sequencing can be limited to only the coding region of the genome, which is known as the exome, to further focus NGS data. Although the exome constitutes only 1% of the genome, it harbors approximately 85% of deleterious mutations.43 WES identifies an average of 20,000 to 35,000 single nucleotide variants per exome, depending on the sequencing technology used and the subject’s ethnicity.41,44 The elimination of synonymous variants, which alter DNA but not amino acid sequences, decreases the candidate gene list. For rare diseases, common variants with an allele frequency of greater than 1% in public SNP databases can be eliminated (Fig 2).41,45–47 Bioinformatics algorithms, such as Polyphen-2, SIFT, MutationTaster, and MAPP, can be used to identify missense mutations that can result in altered protein function.48–51 Additionally, programs, such as GERP and Phylo-P SCORE, can be used to assess sequence conservation.52,53 Finally, candidate genes are filtered based on gene function and expression. Genes essential for immune function, such as those important for lymphocyte development, are prioritized; conversely, those unrelated to immune function, such as the genes encoding the olfactory receptors, can be excluded. This approach was used to identify compound heterozygous mutations in IL10R1 as a cause of neonatal-onset Crohn disease in a patient born to nonconsanguineous parents.54
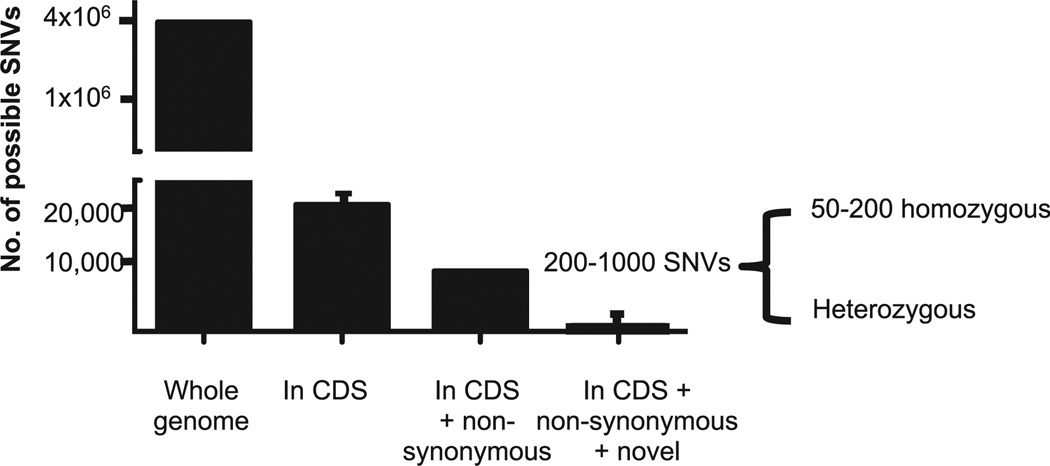
FIG 2.
Any genome will have approximately 4 million variants that differ from the reference genome. Of these, approximately 20,000 will be in the coding and splice site regions (CDS). Approximately 10,000 variants will be nonsynonymous. Finally, approximately 200 to 1000 of these variants will be “novel” (ie, not reported in existing SNP public databases), with the majority of variants existing in heterozygous states (author’s own unpublished data and references45–47).
An additional filter can be applied to WES data from consanguineous families through the use of homozygous mapping, a special case of linkage analysis. Homozygosity mapping identifies the genomic regions most likely to harbor the pathogenic mutation based on the assumption that the causative mutation for an autosomal recessive disease occurs within a locus containing clusters of homozygous SNPs specific to the affected subjects and inherited from a common ancestor.55,56 The combination of homozygosity mapping and WES is an increasingly common approach for identifying genetic defects underlying PIDs. LRBA deficiency was identified not only through linkage analysis, as described previously, but also through homozygosity mapping and WES in a consanguineous family with chronic inflammatory bowel disease and a combined immunodeficiency.57 Homozygosity mapping of 2 patients and their healthy siblings, combined with WES of 1 patient, identified only 1 novel homozygous variant: a 2-bp deletion in LRBA. In another example 2 siblings from a consanguineous family presented with naive T-cell lymphopenia, defective T-cell activation, and epidermodysplasia verruciformis.58 Homozygosity mapping and WES of 2 patients identified only 2 novel homozygous mutations. One of these mutations resulted in a stop codon in RHOH, which encodes an atypical Rho GTPase important for T-cell development and activation.58,59 As a third example, homozygosity mapping and WGS of 2 consanguineous parents and 1 patient identified a missense mutation in MALT1, which encodes a cysteine protease important for NF-κB activation, as a cause of combined immunodeficiency.60 These examples show that the combination of WES/WGS and homozygosity mapping can generate a shorter list of candidate genes than those identified by means of linkage analysis alone. Although these techniques have greatly expedited the discovery of pathogenic mutations, there are still limitations inherent in WES/WGS. The remainder of this review will highlight some of these difficulties and discuss how recent studies have addressed them.
AUTOSOMAL DOMINANT DISEASES
Autosomal dominant diseases represent a challenge because there are approximately 69% more heterozygous than homozygous variants in any given genome.61 Consequently, effective mapping of an autosomal dominant gene requires large families to narrow the candidate gene list: a pedigree consisting of multiple generations with 6 to 12 affected subjects is often necessary but not always available.39 To circumvent this difficulty, investigators have sequenced affected subjects from unrelated families, as was done in the study identifying heterozygous mutations in GATA2 as a cause of monocytopenia, NK- and B-lymphocytopenia, severe infections with M avium complex (MonoMAC syndrome).62 Assuming that the disease is caused by mutations in a single gene, the authors hypothesized that WES of 4 unrelated patients would identify the gene containing the causative variants while eliminating the majority of nonpathogenic mutations. Only 1 gene, GATA2, which encodes a transcription factor important for stem cell maintenance, contained heterozygous, novel, and deleterious mutations shared by all 4 patients. Another group also identified GATA2 as a cause of MonoMAC syndrome after noting that a small proportion of these patients also have myelodysplasia, acute myeloid leukemia, or chronic myelomonocytic leukemia,63 which have been previously associated with mutations in GATA2.62 Together, these cases illustrate how multiple approaches can be used to successfully identify the genetic cause of autosomal dominant PIDs.
DISEASES CAUSED BY MUTATIONS IN NONCODING REGIONS
Mutations in noncoding regions typically will not be detected by using WES, which primarily captures only exonic regions. Noncoding regions comprise approximately 99% of the genome, and the effect of a variant in a noncoding region is often indeterminate. Therefore identification of deleterious noncoding mutations typically occurs only after an excellent candidate gene is identified. Exonic mutations in UNC13D, which encodes Munc13-4, were known to cause familial hemophagocytic lymphohistiocytosis type 3, prompting the discovery of mutations in conserved intronic regions that abolish protein expression and result in familial hemophagocytic lymphohistiocytosis type 3.64,65 Additional examples include intronic GATA2 mutations in patients with MonoMAC syndrome66; an intronic mutation in SH2D1A, which encodes SAP, resulting in X-linked lymphoproliferative disorder67; and a mutation in the 5′ untranslated region of NEMO as a cause of X-linked ED-ID.68 In all these examples, identification of a causative intronic mutation was possible because a candidate gene was identified based on the clinical phenotype and the mutation severely impaired protein expression. In the future, the decreasing cost of WGS will facilitate efficient identification of intronic mutations. Recent research has been directed at improving both bioinformatic and laboratory tools for predicting the effect of intronic mutations,69 but anticipating the effect of intronic mutations remains a nascent and challenging field.
DISEASES CAUSED BY STRUCTURAL VARIATIONS OF THE GENOME
Identifying large structural variations, such as deletions, inversions, and translocations, by using WES/WGS can be problematic for multiple reasons. It can be difficult to differentiate a bona fide deletion from a genomic interval with poor exome capture or sequencing. Because of its large size, the human genome is fragmented before NGS. Biotinylated oligonucleotide probes complementary to the exome bind to DNA fragments encoding the exome and are collected with magnetic streptavidin beads. The fragments are amplified and sequenced in parallel. Although sequencing is performed to achieve an overall average sequencing depth, or coverage of typically 30× to 150×, a fragment might be sequenced from 1 to more than 200 times because of genomic variations, such as repetitive regions or differences in guanosine-cytosine content. As a result, WES might not adequately sequence as much as 10% of genes.44 The sequences are then aligned to the reference human genome. Fragments containing a large deletion or inversion will not align properly. Instead, the fragment must be analyzed as series of smaller subunits to identify the best-fit alignment for each subunit until the structural variations are pinpointed, a process that requires significant bioinformatics resources.70
Yet even WGS, which avoids the step of exome capture, does not guarantee easy detection of structural variations and copy number variations, which include deletions and duplications. This was illustrated by a study of patients from 3 families with dominantly inherited cold-induced urticaria, antibody deficiency, and autoimmunity.71 Linkage analysis of 2 families identified 24 candidate genes, but WGS did not reveal novel variants in this region. PLCG2, which encodes phospholipase Cγ2, was the primary candidate because of its known role in B-cell function and signaling. Sanger sequencing of PLCG2 identified a heterozygous deletion in all the patients, which was confirmed by using retrospective reanalysis of the WGS data. It is difficult to distinguish a heterozygous deletion from an area of low coverage because of the significant variability in coverage inherent in WES.
Array comparative genomic hybridization is an alternative method of identifying copy number variations in which DNA is fragmented, fluorescently labeled, and hybridized to microarrays containing oligonucleotide probes complementary to the reference genome. Increased signal intensity corresponds to a duplication, whereas decreased or absent signal intensity would correspond to a heterozygous or homozygous deletion.72 This technique identified novel deletions in dedicator of cytokinesis 8 (DOCK8) in patients with autosomal recessive hyper-IgE syndrome.73 Array comparative genomic hybridization has also identified large deletions in CYBB, which encodes the gp91phox subunit of the NADPH oxidase, resulting in chronic granulomatous disease.74 These arrays cannot identify structural variations that do not lead to changes in copy number, such as inversions and translocations. Detection of such variants remains an area of active research in the development of NGS technology.
DISEASES WITH INCOMPLETE PENETRANCE
All of the cases discussed thus far are Mendelian diseases with full penetrance. This assumption governs the filtering strategies used to eliminate candidate mutations. For example, in an autosomal recessive disease, all candidate mutations must be homozygous in the patient and heterozygous in at least 1 parent because the patient might have a de novo mutation in the second affected allele. However, the possibility of incomplete penetrance should be considered when developing a filtering strategy so that causative variants are not inadvertently eliminated. This was demonstrated in the study identifying an autosomal dominant mutation in TRIF, which encodes an adaptor protein important for TLR3 signaling, as a risk factor for herpes virus encephalitis (HSE).75 A heterozygous missense mutation in TRIF was identified in a patient with HSE, but the patient’s mother and maternal grandfather had the same mutation and HSV-1 antibodies without a history of HSE. The authors demonstrated the deleterious effect of this mutation by showing that fibroblasts from the patient and her mother had impaired cytokine production to TLR3 stimulation, which was restored by transfection with wild-type TRIF. This study indicates the importance of functional assays in proving the effect of a variant on host defense because a strictly Mendelian filtering strategy would have excluded the causative variant. Furthermore, because only one of the 3 subjects with the mutation had HSE, this case also suggests how other modifier genes, epigenetic factors, or environmental exposures can affect the clinical phenotype. This phenomenon occurs with autosomal recessive PIDs as well. Even patients with the same homozygous mutation in RAG2 can present with different phenotypes, such as Omenn syndrome and hyper-IgM syndrome.76 Identification of these modifier genes represents a challenging frontier for research.
CONCLUSIONS AND FUTURE DIRECTIONS
Advances in immunology and genetics have facilitated the discovery of novel defects underlying PIDs. However, there is still much progress to be made. Epigenetic modifications regulating gene expression, such as DNA methylation, histone modification, and noncoding RNAs, modulate the immune system,77 and defects in these mechanisms might contribute to PIDs. NGS can be used to investigate the transcriptome to detect disease-causing splice variants, leading to exon skipping, alternative splicing, and alternative start and polyadenylation sites.78 For our patients, the identification of the defects underlying PIDs enables genetic counseling and preimplantation diagnosis. Lastly, pinpointing these genetic defects is the foundation for the development of gene therapy as a cure.
Acknowledgments
Supported by grants from the National Institutes of Health (AI-076210 and AI094017 to R.S.G. and 1 K12 HD052896-01A1 to J.C.), the Dubai Harvard Foundation for Medical Research (R.S.G.), the Jeffrey Modell Foundation (R.S.G.), and a Manton Foundation Pilot Award (J.C.).
Disclosure of potential conflict of interest: R. S. Geha has received research support from the National Institutes of Health (NIH), the Jeffrey Modell Foundation, and the Dubai Harvard Foundation. J. Chou has received research support from the Manton Foundation and the NIH. C. Platt declares that he has no relevant conflicts of interest.
Abbreviations used
- CMC
Chronic mucocutaneous candidiasis
- ED-ID
Ectodermal dysplasia and immunodeficiency
- HSE
Herpes virus encephalitis
- IL-2Rγ
IL-2 receptor γ chain
- LRBA
LPS-responsive beige-like anchor protein
- MonoMAC
Monocytopenia NK- and B-lymphocytopenia, severe infections with M avium complex
- NF-κB
Nuclear factor κB
- NGS
Next-generation DNA sequencing
- PID
Primary immunodeficiency
- SCID
Severe combined immunodeficiency
- SNP
Single nucleotide polymorphism
- TBK1
TANK-binding kinase 1
- TLR3
Toll-like receptor 3
- WAS
Wiskott-Aldrich syndrome
- WES
Whole-exome sequencing
- WGS
Whole-genome sequencing
REFERENCES
- 1.Fuleihan R, Ramesh N, Loh R, Jabara H, Rosen RS, Chatila T, et al. Defective expression of the CD40 ligand in X chromosome-linked immunoglobulin deficiency with normal or elevated IgM. Proc Natl Acad Sci U S A. 1993;90:2170–2173. doi: 10.1073/pnas.90.6.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen RC, Armitage RJ, Conley ME, Rosenblatt H, Jenkins NA, Copeland NG, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259:990–993. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 3.Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 4.DiSanto JP, Bonnefoy JY, Gauchat JF, Fischer A, de Saint Basile G. CD40 ligand mutations in x-linked immunodeficiency with hyper-IgM. Nature. 1993;361:541–543. doi: 10.1038/361541a0. [DOI] [PubMed] [Google Scholar]
- 5.Korthauer U, Graf D, Mages HW, Briere F, Padayachee M, Malcolm S, et al. Defective expression of T-cell CD40 ligand causes X-linked immunodeficiency with hyper-IgM. Nature. 1993;361:539–541. doi: 10.1038/361539a0. [DOI] [PubMed] [Google Scholar]
- 6.Noelle RJ, Roy M, Shepherd DM, Stamenkovic I, Ledbetter JA, Aruffo A. A 39-kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells. Proc Natl Acad Sci U S A. 1992;89:6550–6554. doi: 10.1073/pnas.89.14.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari S, Giliani S, Insalaco A, Al-Ghonaium A, Soresina AR, Loubser M, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci U S A. 2001;98:12614–12619. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutukculer N, Moratto D, Aydinok Y, Lougaris V, Aksoylar S, Plebani A, et al. Disseminated cryptosporidium infection in an infant with hyper-IgM syndrome caused by CD40 deficiency. J Pediatr. 2003;142:194–196. doi: 10.1067/mpd.2003.41. [DOI] [PubMed] [Google Scholar]
- 9.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu ZJ, et al. Functional activation of Jak1 and Jak3 by selective association with IL-2 receptor subunits. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 11.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 12.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 13.Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T(−)B(+)NK(+) severe combined immunodeficiency. Nat Genet. 1998;20:394–397. doi: 10.1038/3877. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 15.Puel A, Picard C, Ku CL, Smahi A, Casanova JL. Inherited disorders of NF-kappaB-mediated immunity in man. Curr Opin Immunol. 2004;16:34–41. doi: 10.1016/j.coi.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 16.McDonald DR, Mooster JL, Reddy M, Bawle E, Secord E, Geha RS. Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency. J Allergy Clin Immunol. 2007;120:900–907. doi: 10.1016/j.jaci.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 17.Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, Bonnet M, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112:1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen R, van Wengen A, Hoeve MA, ten Dam M, van der Burg M, van Dongen J, et al. The same IkappaBalpha mutation in two related individuals leads to completely different clinical syndromes. J Exp Med. 2004;200:559–568. doi: 10.1084/jem.20040773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna S, Etzioni A. New host defense mechanisms against Candida species clarify the basis of clinical phenotypes. J Allergy Clin Immunol. 2011;127:1433–1437. doi: 10.1016/j.jaci.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 23.Shiow LR, Roadcap DW, Paris K, Watson SR, Grigorova IL, Lebet T, et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat Immunol. 2008;9:1307–1315. doi: 10.1038/ni.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moshous D, Martin E, Carpentier W, Lim A, Callebaut I, Canioni D, et al. Whole-exome sequencing identifies coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J Allergy Clin Immunol. 2013;131:1594–1603. doi: 10.1182/blood-2012-07-440339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanzi G, Moratto D, Vairo D, Masneri S, Delmonte O, Paganini T, et al. A novel primary human immunodeficiency due to deficiency in the WASP-interacting protein WIP. J Exp Med. 2012;209:29–34. doi: 10.1084/jem.20110896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anton IM, de la Fuente MA, Sims TN, Freeman S, Ramesh N, Hartwig JH, et al. WIP deficiency reveals a differential role for WIP and the actin cytoskeleton in T and B cell activation. Immunity. 2002;16:193–204. doi: 10.1016/s1074-7613(02)00268-6. [DOI] [PubMed] [Google Scholar]
- 27.de la Fuente MA, Sasahara Y, Calamito M, Anton IM, Elkhal A, Gallego MD, et al. WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP) Proc Natl Acad Sci U S A. 2007;104:926–931. doi: 10.1073/pnas.0610275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koss M, Bolze A, Brendolan A, Saggese M, Capellini TD, Bojilova E, et al. Congenital asplenia in mice and humans with mutations in a Pbx/Nkx2-5/p15 module. Dev Cell. 2012;22:913–926. doi: 10.1016/j.devcel.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 30.O’Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Weidberg H, Elazar Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci Signal. 2011;4 doi: 10.1126/scisignal.2002355. pe39. [DOI] [PubMed] [Google Scholar]
- 32.Herman M, Ciancanelli M, Ou YH, Lorenzo L, Klaudel-Dreszler M, Pauwels E, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fruman DA. Regulatory subunits of class IA PI3K. Curr Top Microbiol Immunol. 2010;346:225–244. doi: 10.1007/82_2010_39. [DOI] [PubMed] [Google Scholar]
- 34.Fukao T, Yamada T, Tanabe M, Terauchi Y, Ota T, Takayama T, et al. Selective loss of gastrointestinal mast cells and impaired immunity in PI3K-deficient mice. Nat Immunol. 2002;3:295–304. doi: 10.1038/ni768. [DOI] [PubMed] [Google Scholar]
- 35.Conley ME, Dobbs AK, Quintana AM, Bosompem A, Wang YD, Coustan-Smith E, et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. J Exp Med. 2012;209:463–470. doi: 10.1084/jem.20112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhlenbaumer G, Hullmann J, Appenzeller S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat. 2011;32:144–151. doi: 10.1002/humu.21400. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Herrera G, Tampella G, Pan-Hammarstrom Q, Herholz P, Trujillo-Vargas CM, Phadwal K, et al. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet. 2012;90:986–1001. doi: 10.1016/j.ajhg.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 42.Chou J, Ohsumi TK, Geha RS. Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies. Curr Opin Allergy Clin Immunol. 2012;12:623–628. doi: 10.1097/ACI.0b013e3283588ca6. [DOI] [PubMed] [Google Scholar]
- 43.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 44.Clark MJ, Chen R, Lam HY, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng SB, Nickerson DA, Bamshad MJ, Shendure J. Massively parallel sequencing and rare disease. Hum Mol Genet. 2010;19:R119–R124. doi: 10.1093/hmg/ddq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 51.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao H, Yang W, Lee PP, Ho MH, Yang J, Zeng S, et al. Exome sequencing identifies novel compound heterozygous mutations of IL-10 receptor 1 in neonatal-onset Crohn’s disease. Genes Immun. 2012;13:437–442. doi: 10.1038/gene.2012.8. [DOI] [PubMed] [Google Scholar]
- 55.Gibbs JR, Singleton A. Application of genome-wide single nucleotide polymorphism typing: simple association and beyond. PLoS Genet. 2006;2:e150. doi: 10.1371/journal.pgen.0020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alkuraya FS. Homozygosity mapping: one more tool in the clinical geneticist’s toolbox. Genet Med. 2010;12:236–239. doi: 10.1097/GIM.0b013e3181ceb95d. [DOI] [PubMed] [Google Scholar]
- 57.Alangari A, Alsultan A, Adly N, Massaad MJ, Kiani IS, Aljebreen A, et al. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J Allergy Clin Immunol. 2012;130:481–488.e2. doi: 10.1016/j.jaci.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crequer A, Troeger A, Patin E, Ma CS, Picard C, Pedergnana V, et al. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J Clin Invest. 2012;122:3239–3247. doi: 10.1172/JCI62949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Y, Chae HD, Siefring JE, Jasti AC, Hildeman DA, Williams DA. RhoH GTPase recruits and activates Zap70 required for T cell receptor signaling and thymocyte development. Nat Immunol. 2006;7:1182–1190. doi: 10.1038/ni1396. [DOI] [PubMed] [Google Scholar]
- 60.Jabara HH, Ohsumi T, Chou J, Massaad MJ, Benson H, Megarbane A, et al. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J Allergy Clin Immunol. 2013;132:151–158. doi: 10.1016/j.jaci.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meeths M, Chiang SC, Wood SM, Entesarian M, Schlums H, Bang B, et al. Familial hemophagocytic lymphohistiocytosis type 3 (FHL3) caused by deep intronic mutation and inversion in UNC13D. Blood. 2011;118:5783–5793. doi: 10.1182/blood-2011-07-369090. [DOI] [PubMed] [Google Scholar]
- 65.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115:461–473. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 66.Hsu AP, Johnson KD, Falcone EL, Sanalkumar R, Sanchez L, Hickstein DD, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121:3830–3837. S1–S7. doi: 10.1182/blood-2012-08-452763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Recher M, Fried AJ, Massaad MJ, Kim HY, Rizzini M, Frugoni F, et al. Intronic SH2D1A mutation with impaired SAP expression and agammaglobulinemia. Clin Immunol. 2013;146:84–89. doi: 10.1016/j.clim.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mooster JL, Cancrini C, Simonetti A, Rossi P, Di Matteo G, Romiti ML, et al. Immune deficiency caused by impaired expression of nuclear factor-kappaB essential modifier (NEMO) because of a mutation in the 5’ untranslated region of the NEMO gene. J Allergy Clin Immunol. 2010;126:127–132.e7. doi: 10.1016/j.jaci.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baralle D, Lucassen A, Buratti E. Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep. 2009;10:810–816. doi: 10.1038/embor.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ombrello MJ, Remmers EF, Sun G, Freeman AF, Datta S, Torabi-Parizi P, et al. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N Engl J Med. 2012;366:330–338. doi: 10.1056/NEJMoa1102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361:2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arai T, Oh-ishi T, Yamamoto H, Nunoi H, Kamizono J, Uehara M, et al. Copy number variations due to large genomic deletion in X-linked chronic granulomatous disease. PLoS One. 2012;7:e27782. doi: 10.1371/journal.pone.0027782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sancho-Shimizu V, Perez de Diego R, Lorenzo L, Halwani R, Alangari A, Israelsson E, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chou J, Hanna-Wakim R, Tirosh I, Kane J, Fraulino D, Lee YN, et al. A novel homozygous mutation in recombination activating gene 2 in 2 relatives with different clinical phenotypes: Omenn syndrome and hyper-IgM syndrome. J Allergy Clin Immunol. 2012;130:1414–1416. doi: 10.1016/j.jaci.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Knight JC. Genomicmodulators of the immune response. Trends Genet. 2013;29:74–83. doi: 10.1016/j.tig.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kassahn KS, Waddell N, Grimmond SM. Sequencing transcriptomes in toto. Integr Biol (Camb) 2011;3:522–528. doi: 10.1039/c0ib00062k. [DOI] [PubMed] [Google Scholar]