Abstract
Tissues can be soft like fat, which bears little stress, or stiff like bone, which sustains high stress, but whether there is a systematic relationship between tissue mechanics and differentiation is unknown. Here, proteomics analyses revealed that levels of the nucleoskeletal protein lamin-A scaled with tissue elasticity, E, as did levels of collagens in the extracellular matrix that determine E. Stem cell differentiation into fat on soft matrix was enhanced by low lamin-A levels, whereas differentiation into bone on stiff matrix was enhanced by high lamin-A levels. Matrix stiffness directly influenced lamin-A protein levels, and, although lamin-A transcription was regulated by the vitamin A/retinoic acid (RA) pathway with broad roles in development, nuclear entry of RA receptors was modulated by lamin-A protein. Tissue stiffness and stress thus increase lamin-A levels, which stabilize the nucleus while also contributing to lineage determination.
Stiffness and strength of a tissue should in principle relate to the physical stress in that tissue. Low stresses in brain and fat may explain why these tissues are soft. High stresses on adult bone, in contrast, are thought to promote its growth and stiffening through a “mechanostat” that functions to match the stress (1). At a microscale, physical stress deforms cells (2) and can alter gene expression profiles (3), but cells in vivo might also directly sense the local tissue stiffness or microelasticity E (in kilopascals, kPa) (table S1), which should relate to the typical stress in that tissue (also in kPa). It is unclear, however, whether any specific proteins function across diverse tissues to not only match stiffness with stress but also impact differentiation processes.
When animal cells are cultured on various gels or elastomeric substrates, cell-generated stress or tension increases as cells spread on matrices with increasing elasticity, E (4, 5). Surprising effects on differentiation (5), as well as cell shape and motility (6), have also been observed. Although some studies have suggested a lack of response to matrix elasticity in two-dimensional (2D) (7) or 3D cultures (8), several other studies have found that gels that mimic the compliance of brain or fat, respectively, maximize neurogenesis or adipogenesis (9–11). Gels that are moderately stiff like muscle are best for myogenesis (12–14), and gels that are firm like precalcified bone optimize osteogenesis in 2D and 3D (5, 15, 16). A 3D hierarchy of soft/stiff/rigid tissue might exist, but the presence of any molecular mechanostats that relate to tissue stiffness and that systematically affect lineage remain unknown. Widely expressed transcriptional regulators that include YAP1 of the Hippo pathway, which promotes growth and regeneration (17), as well as components of the serum response factor (SRF) pathway, which promote cytoskeletal gene expression in differentiation (18), exhibit low nuclear activity in cells on substrates designed to limit cell spreading and cytoskeleton tensions (11, 19). How such factors or completely distinct pathways might relate to matrix elasticity and the stiffness of 3D tissues has yet to be addressed.
Forces on a tissue, as well as those generated by cells within a tissue (Fig. 1A), are sustained in rough proportion to microelasticity E by collagens and lineage-specific cytoskeletal proteins (4, 5). Some forces might also propagate into the nucleus and be resisted by the nuclear lamina. Lamins are intermediate filament proteins found in nearly all cell nuclei and contribute to nuclear stiffness (20, 21) and nuclear stability (22). Although lamins might be viewed as similar in mechanical function to keratin intermediate filament proteins that determine nail and skin structure (23), lamins are also believed to modulate transcription (24) and have been speculated to mechanoregulate the genome (25, 26). Here, initial analyses of proteomes from soft and stiff tissues motivated us to examine, both in vivo and in cultures on soft and stiff gels, whether the nuclear lamina is involved in sensing tissue elasticity in differentiation.
Fig. 1. Lamin-A and collagen levels scale with tissue stiffness, but collagen determines stiffness while lamin-A responds.
(A) Tissue deformation under force is quantified by E and transfers stresses through the extracellular matrix and the cytoskeleton into the nucleus. (B and C) The proteomes of adult mouse tissues were profiled to determine whether scaling of mechanical properties with biopolymer concentration exists across tissues. (D) Quantitative proteomics of multiple human and mouse tissues and cells revealed scaling with E of the absolute ratio or stoichiometry of lamin-A to lamin-B through MS quantification of a pan-lamin peptide. Differences in ratios are significant with brain ≪ liver < fat < heart, lung, and muscle ≪ skull ≪ femur and cartilage, where < indicates P ≤ 0.05 and ≪ indicates P ≤ 0.01. Nuclei with abundant lamin-A are stiff (20). Cultured cells showed the same trend as their primary source tissue. HSCP, human hematopoietic stem cell progenitors from marrow; U251, human glioblastoma cells from brain; A549, human adenocarcinoma epithelial cells from lung; C2C12, mouse myoblast cells from muscle; MSC, osteo-prone human mesenchymal stem cells from marrow. (E) MS trends were validated by immunoblotting (representative blots taken from fig. S5A). (F) Lamin-B1 scales very weakly with E, whereas lamin-B2 is constant on average. (G) Collagen-1 isoforms scale strongly with E. (H) Human glioblastoma cells U251Luc (expressing luciferase for imaging) were xenografted into mouse brain and flank, and 4-week-old tumors were profiled by MS proteomics. (I) Mouse-derived collagens in U251 grown in mouse brain and flank scale with E as observed for adult mouse tissues. (J) Stiffness of flank tumors made with high (A549) or low (U251) lamin-A:B cells was similar to the stiffness of the subcutaneous site (subQ). Tumors were 50% softer after only a brief treatment with collagenase (col’ase). (K) Lamin composition and stiffness of the tumors fit adult tissue scaling. All points are significantly different where indicated (n ≥ 3 MS measurements).
Results
Lamin-A and Collagen Levels Scale with Tissue Microelasticity
Allometric scaling laws for stress response would be understandable for polymer-based molecular mechanostats, so we examined proteomes for such trends across tissues from brain to bone (Fig. 1A). Nearly 100 of the most abundant structural and nuclear proteins were quantified relative to invariant proteins using label-free mass spectrometry (MS) (Fig. 1B and figs. S1 and S2). Lamin-A was found to increase systematically 30-fold from soft to stiff tissue (Fig. 1D and fig. S3). Lamin-B1 differed by less than threefold, and lamin-B2 varied even less (Fig. 1F), consistent with B-type lamins being constitutively expressed (27). An absolute stoichiometry of the lamin isoforms (lamin-A:B) was directly determined by MS quantitation of a peptide common to all lamins (see Materials and Methods) (fig. S4, A and B) as validated with recombinant protein (fig. S4C), and a power law fit versus tissue microelasticity gave Lamin-A:B ~ E 0.6 (R 2 = 0.88). Combined with findings that B-type lamins were roughly similar in abundance (fig. S4D), the weak scaling of both B-type lamins is consistent with the key result for Lamin-A versus E as a metric of tissue stress: Lamin-A ~ E 0.7.
Primary and immortalized cell types derived from a range of human and mouse tissues follow this scaling in terms of E of the tissue of origin, which helps to generalize the result across species and perhaps ameliorate concerns over tissue heterogeneity. Immunoblotting also validated the A:B scaling and further suggested that the A and C splice-form products of the LMNA gene follow respective scaling exponents of 1.0 and 0.5, so that the 0.7 exponent for total lamin-A is a geometric mean (Fig. 1E and fig. S5). A power law between concentration of a polymer and its stiffness is typical in the physics of biopolymers (28–30). We had previously knocked down lamin-A in human lung–derived A549 cells without affecting lamin-B, and micropipette aspiration showed that knockdown nuclei are softer (20), suggesting that nuclear stiffness increases with A:B stoichiometry. Although the tissue E here provides a metric of the typical stress on a tissue, the power law exponent for lamin-A is midway between the linear response of a simple polymer network (31) and that of a nonlinear, semiflexible meshwork typified by stiffness versus concentration of actin [with exponent 0.4 (29)].
The A:B stoichiometry in Fig. 1D (y axis) indicates that lamin-A dominates over a range of stiff tissues, consistent with LMNA mutations causing lipodystrophies, muscular dystrophies, and premature aging (progeria) that affects heart and large vessels while sparing soft tissues such as brain and marrow (32). Lamin-B dominates in soft tissues, consistent not only with lamin-A appearing low in antibody-staining of neuroendocrine tissues and hematopoietic cells (27) [despite epitope masking (33)] but also with lamin-B knockout mice dying at birth with defects in brain development and tissue innervation (34). In other words, the normal function of cells in stiff tissues is most dependent on lamin-A.
In nuclear-enriched fractions as well as whole tissue lysates, extracellular matrix proteins were the other detected tissue proteins that scaled with E and also showed transcripts scaling with E in both man and mouse (fig. S2). Collagen-1 is the most abundant protein in animals, and its two fiber-coassembling isoforms both gave collagen-1 ~ E 1.5 (Fig. 1G). Gels made with purified collagen-1 scale as ~ E 0.5 (35), but a different exponent for tissue seems consistent with additional matrix or cell components contributing to tissue mechanics. Indeed, collagen-3, -5, -6, -11, and -12 also scaled as ~ E 0.9-1.5. Our tissue profiling was unable to identify any compelling cytoskeletal candidate [particularly in the SRF or YAP1 pathways (fig. S1, A to C)] that could be a universal “mechanostat” similar to lamin-A for the nucleus. A possible reason is tissue-specific isoform usage, such as with the intermediate filament protein vimentin, which is restricted to specific lineages rather than being expressed in all cell types. Similar specialization seems likely to apply to isoforms of actin, myosin, and microtubules.
Matrix Determines Tissue Stiffness and Lamin-A Adjusts in Vivo
To address the relative affect of extracellular matrix and lamins on tissue stiffness, human-derived U251 glioblastoma tumors were grown in the brain and in subcutaneous flank sites of nude mice for label-free MS proteomics (Fig. 1H and fig. S6, A to C). In standard culture, these cells had a low A:B ratio similar to normal mouse brain (Fig. 1D). However, flank tumors of U251s had more matrix and were much stiffer than brain tumors, with scaling of collagen density versus E appearing typical of normal adult tissue (Fig. 1I). Flank tumors of human-derived A549 lung cells (A:B ≈ 2.3) had similar E as U251 tumors and were only slightly stiffer than normal subcutaneous tissue, revealing a response independent of initial lamin levels (Fig. 1J and fig. S6D). Collagenase treatment of fresh tumors reduced E by > 50% in just 10 min, suggesting that collagen is a key determinant of tissue stiffness, unlike lamin-A. Consistent with this interpretation, human matrix or matrix-associated proteins were among the few proteins more than twofold higher in the flank compared with the soft brain site. Moreover, human lamin-A levels proved higher in flank versus brain sites (fig. S6), whereas lamin-B1 and lamin-B2 were only slightly higher in brain. U251 cells thus adjust their lamin-A:B ratio by 1.5-fold, which fits remarkably well to the stiffness-dependent scaling of lamin-A:B found in normal tissues (Fig. 1K).
Two other intermediate filament (IF) proteins exhibited site-dependent differences in U251 cells that were notably similar to lamin-A. Human glial fibrillary acidic protein (GFAP) and vimentin were both lower in the softer brain than in the flank (fig. S6B). GFAP expression is known to be restricted to cells of the central nervous system plus a few nonepithelial lineages, so its up-regulation in flank by human brain–derived U251 cells was not expected. Indeed, mouse GFAP was almost undetectable in flank but abundant in brain (fig. S6C). On the other hand, human nestin (yet another IF protein) in the grafted U251 cells was slightly higher in brain than flank, similar in response to the human B-type lamins. These additional findings for lineage-restricted, cytoplasmic IFs thus reinforced the finding that different IFs exhibit different sensitivities to different microenvironments.
Lamin-A Conformation and Abundance Are Mechanosensitive in Cultured Cells
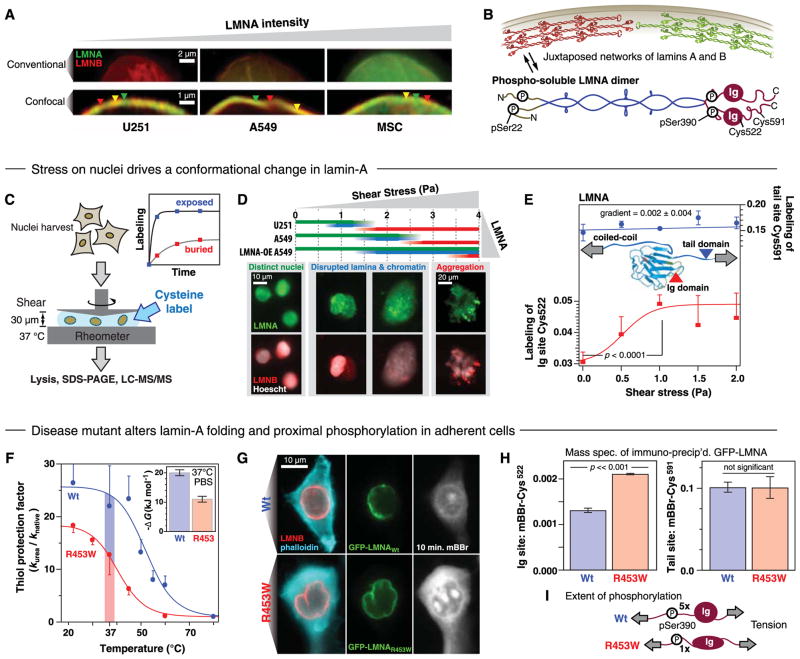
To understand how lamins sustain stress, we focused on cultures of human-derived U251s, A549s, and low-passage mesenchymal stem cells (MSCs), which collectively span the broad range in lamin-A:B (Fig. 1D). Imaging of lamins under constant immunostaining conditions showed the expected increase in lamin-A intensity as well as juxtaposed networks (24) of lamins (Fig. 2, A and B). To dissect molecular responses of lamins to physical stress, we applied cysteine-shotgun MS (CS-MS) which involves using a fluorescent dye to covalently tag cysteines that are conformationally cryptic but exposed by stress (36). When nuclei were isolated from cells and subjected to controlled shear (Fig. 2C), stability against nuclear rupture was seen to increase with lamin-A levels: U251 < A549 < [A549 overexpressing green fluorescent protein (GFP)–lamin-A] (Fig. 2D). Peeling of lamin-A off of stressed nuclei as seen by immuno-fluorescence demonstrated the responsiveness of lamin-A to stress. CS-MS revealed several nuclear proteins in the 60 to 80 kD range as susceptible to stress (fig. S7), with Cys522 in lamin-A’s immuno-globulin (Ig) domain identified as a stress-sensitive site (Fig. 2E). Studies of pure recombinant Ig domain showed the labeling kinetics of Cys522 captured domain unfolding in thermal and solvent denaturation (Fig. 2F and fig. S8, A to E), and this same site in nuclear lamin-A showed 70% more labeling as shear was increased. A nearby Cys591 in the tail was also labeled but was insensitive to stress.
Fig. 2. Nuclear stability is conferred by lamin-A, which unfolds under stress and couples to phosphorylation.
(A) High-resolution images of the nuclear envelope of U251s, A549s, and MSCs show juxtaposed regions of lamins A (green) and B (red), consistent with earlier observations in HeLa cells (24). Triangles highlight domains of lamins A (green), B (red), and overlap (yellow). (B) Higher-order assembly of lamin typical of intermediate filament proteins and the lamin-A dimer solubilized by phosphorylation (38), annotated with MS-detectable phosphorylation and cysteine sites. (C) Shearing of nuclei showed that lamin conformation responds to mechanical stress. A cysteine-reactive label [monobromobimane (mBBr)] was added to nuclei and sheared for 40 min at the indicated stresses in a cone and plate rheometer. All protein was then solubilized and the extent of reaction at each detected cysteine quantified by MS, scaled by the unlabeled protein. (D) A549 nuclei imaged following shear stress. Greater lamin expression confers mechanical robustness to the nuclei, limiting disruption of chromatin. (E) The Ig-like domain of lamin-A has a cryptic cysteine, Cys522, that is buried in the crystal structure (Protein Data Bank accession number 1IFR) but showed 70% more labeling in stressed A549 nuclei. Labeling of Cys591 in the tail of lamin-A did not change with stress (mean ± SEM from curve fit; P ≤ 0.05, n ≥ 3 MS measurements). (F) A point mutant R453W within the lamin Ig domain that is known to cause muscular dystrophy showed decreased domain stability at 37°C as measured by cysteine labeling rates and tryptophan fluorescence (inset). (G) Images of adherent A549 cells transfected with wild-type or mutant GFP-tagged lamin were labeled with 400 μM mBBr for 10 min. (H) Labeling of wild-type and R453W lamin of mBBr measured by MS after immunoprecipitation of GFP (IP-MS) showed greater in vivo labeling of mutant in adherent cells; the tail domain showed no significant difference. (I) In contrast, phosphorylation at Ser390 was fivefold higher in wildtype lamin-A. All points are significantly different, as indicated (n ≥ 3 MS measurements).
A lamin-A point mutation R453W in the Ig domain that causes muscular dystrophy (37) and that destabilized the purified domain (Fig. 2F) also produced dysmorphic nuclei in A549 cells expressing a GFP-lamin-A with the mutation (Fig. 2G). After labeling the adherent cells with the Cys-reactive fluorescent dye, lamin-A was enriched by immunoprecipitation and analyzed by MS (IP-MS). Labeling of the Ig’s Cys522 increased significantly (Fig. 2H), whereas labeling of the tail’s Cys591 was unaltered. The mutant also showed fivefold less phosphorylation at a proximal Ser390 without differences at head or tail phospho-Ser (Fig. 2I and fig. S8, F and G); synthetic peptides and phosphopeptides were made and confirmed the linearity of quantitation by MS (fig. S8F). Lamin-A phosphorylation is known to promote disassembly (38) and also protein turnover (39).
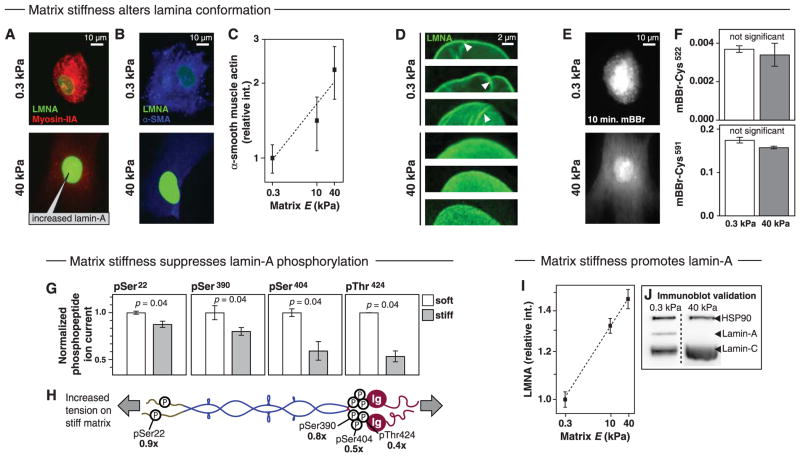
Stresses in the cell are transmitted to the nucleus and lamina through various interactions, and because cytoskeletal tension increases with matrix stiffness (5), CS-MS was used to assess matrix effects on lamin-A. MSCs cultured on either soft gels (0.3 kPa) or stiff gels (40 kPa) exhibited the expected low- and high-tension phenotypes with stiffness-induced increases in (i) cell and nuclear spreading (Fig. 3A and fig. S9A), (ii) stress fiber assembly (Fig. 3A and fig. S9, B and C), and (iii) levels of α-smooth muscle actin (Fig. 3, B and C). On soft matrix, the nuclear envelope appeared highly wrinkled (Fig. 3D), but stiff matrix and high tension “smoothed out” nuclear wrinkles and flattened the nucleus. CS-MS was applied to 3-day cultures, with an anticipation of more labeling of the stress-sensitive Ig domain in the high-tension state, but Cys labeling proved similar in both the Ig and tail sites in cells on soft versus stiff gels (Fig. 3, E and F). On the other hand, phosphorylation proved significantly higher in cells on soft matrices at all four MS-detectable sites (Fig. 3, G and H, and fig. S9D). Because lamin-A phosphorylation promotes disassembly (38) and turnover (39), the results suggested an inverse relationship between phosphorylation and matrix stiffness. Indeed, the total amount of lamin-A increased significantly in both MSCs and A549 cells on stiff substrates (Fig. 3, I and J, and fig. S9, E to G). Lamin-A’s increased levels could thus compensate in part for the increased force per molecule in cells on stiff substrates, and this increased level would tend to maintain stability of the protein and its folded domains. Consistent with an increase in IF assembly with cell tension, MSCs treated with a myosin-II inhibitor to inhibit cell tension have also been found to depolymerize vimentin filaments (36). However, because lamin-B did not change significantly with matrix stiffness (fig. S9G) and no lamin-B phosphopeptides were detected, additional study of this mechanism for lamin-A and other mechanosensitive IFs is needed.
Fig. 3. Cell and nuclei spread on stiff matrix, suppressing lamin-A phosphorylation and increasing lamin-A and cell tension.
Response of MSCs to substrate stiffness was characterized. (A) Cells are more rounded on soft (0.3 kPa) matrix, whereas on stiff (40 kPa) matrix they spread with more pronounced stress fibers, consistent with higher cell tension. (B and C) Levels of α-smooth muscle actin were higher on stiff matrix. (D) Confocal microscopy showed wrinkled nuclei on soft matrix, and smoothed-out and flattened nuclei on stiff matrix. Images are of the middle z-section of different nuclei. (E and F) Cell and nuclei rapidly label with mBBr, but quantitation of lamin-A labeling by IP-MS showed no significant difference in labeling of either the Ig domain or tail sites on soft versus stiff substrate. (G and H) Phosphorylation at Ser390 is ~30% higher on soft substrate, predictive of solubilization. (I and J) Quantitative immunofluorescence and immunoblot show lamin-A increased with substrate stiffness. This tends to reduce the mechanical stress per molecule and maintain the Ig fold. Blots were taken from the same membrane. All points are significantly different (P ≤ 0.05; n ≥ 3 MS and IF measurements).
Lamin-A Enhances Matrix Elasticity-Directed Differentiation
Matrix elasticity directs lineage specification of human bone marrow–derived MSCs in culture toward bone, fat, or other tissue types with mechanisms based in part on myosin-II generated stresses (5). Because lipodystrophy is one of the many diseases involving LMNA and because adipocytes are common in human marrow, the softness of fat was mimicked with a soft gel (E = 0.3 kPa), and precalcified bone or “osteoid” was mimicked with a stiff gel (E = 40 kPa). Bone marrow–derived MSCs typically have a very high A:B ratio (Fig. 1D) that probably reflects their osteogenic niche origins (40), and indeed even with standard adipogenic media only a very small percentage of MSCs on stiff matrix (~1%) (Fig. 4A) showed after 2 weeks of culture the oil-red–positive lipid droplets that are phenotypic of fat. In these cells, stress fibers were displaced by oil droplets that sometimes deformed the nuclear envelope (Fig. 4B and fig. S10A). Soft matrix increased adipogenesis to 8%, but this increased to nearly 20% with partial knockdown of LMNA. MS profiling also revealed an abundant fatty acid ligase (ACSL1) up-regulated nearly 100-fold with knockdown (fig. S11A). Knockdown did not affect adipogenesis of cells on stiff matrix, which invariably showed about 20-fold fewer adipogenic cells than knockdown cells on soft matrix.
Fig. 4. Matrix elasticity directs stem cell differentiation, which is enhanced by laminA as it regulates SRF and YAP1.
(A) Partial knockdown (KD) of laminA in MSCs with si-LMNA in combination with soft matrix (0.3 kPa) and an adipo-inducing media-maximized adipogenesis (P ≤ 0.02 knockdown versus control). Stiff matrix (40 kPa) suppressed adipogenesis in parallel cultures, with no significant effect of knockdown. Knockdown of lamin-A was to 35% of wild-type or scrambled-siRNA. (B) Adipogenesis in MSCs on plastic showed that cells with oil droplets (phase contrast microscopy; nucleus indicated by blue arrow with asterisk) had minimal stress fibers (myosin-IIa immunofluorescence) compared with cells without oil droplets (nucleus indicated by blue arrow without asterisk). (C) Overexpression (OE) of lamin-A in MSCs in combination with stiff matrix and an osteo-inducing media-maximized osteogenesis (P ≤ 0.0001). Soft matrix suppressed osteogenesis in parallel cultures, with no significant effect of overexpression. (D) Alkaline phosphatase (ALP) staining was done after 1 week as a measure of osteogenic signal, together with the fraction of cells with staining. (E) Correlation between nuclear area and lamin-A level with treatments on soft and stiff matrix (normalized to Hoechst stain). NT, non-treated control. Inset cartoons highlight the relationship between cell and nuclear spread area as well as cell tension. (F) Pathway analyses after knockdown of LMNA in three different MSCs. Gene symbols are colored according to mRNA abundance in MSCs (green, low; red, high) from microarray data for 11 soft tissues in human and 10 soft tissues in adult mouse (of 14,985 gene annotations common to mouse and human), and genes are ranked based on Pearson correlations with lamin-A. SRF and related transcription factors and target genes all show reduced levels with lamin-A knockdown, whereas neither YAP1 nor its target genes were affected. TEAD1 has been implicated in both YAP1 and SRF pathways, but lamin-A knockdown suppresses TEAD1 similar to SRF, suggesting that it is in the SRF pathway. A transcription factor predicted to regulate lamin-A (RARG) was not affected by lamin-A knockdown, and few RA pathway transcripts changed with LMNA knockdown except CRABP2 (89), which decreased. CRABP2 is up-regulated in osteoarthritis models where COL1A1 increases in osteogenic-like processes (90). The SREBF1-regulated gene, FABP5, increases to give an average ratio for message of CRABP2/FABP5 ~ 0.3 relative to untreated cells; both CRABP2 and FABP5 are known to bind RA, and the change in the RA signaling ratio (CRABP2/FABP5) was consistent with switching of differentiation pathways (91). (G) The decrease in ACTA2, downstream of SRF, was confirmed at the protein level in MSCs by immunofluorescence. (H) High-resolution confocal microscopy of YAP1 in MSCs cultured on substrates of increasing stiffness show increasing nuclear localization, as reported previously (11). Insets highlight observation of enrichment at the nuclear envelope, which was especially evident with lamin-A overexpression. (I) Plot shows a fourfold increase in nuclear to cytoplasmic ratio of YAP1 with increasing matrix stiffness in MSCs, except that lamin-A overexpression decreases nuclear YAP1. (J) YAP1 was also bimodally distributed on substrates of intermediate stiffness (10 kPa). (K) YAP1 protein and mRNA levels in tissues of increasing stiffness showed nonmonotonic trends, with the mRNA data averaged from human and mouse microarrays. All points are significantly different, as indicated (n ≥ 3 imaging, IF, and immunoblot experiments).
Osteogenesis of MSCs was modulated over a 20-fold range through a combination of matrix elasticity and controlled expression of lamin-A (Fig. 4, C and D). Soft matrix always repressed osteogenesis, but stiff matrix plus lamin-A over-expression led to 80% of cells being positive for a standard marker of osteogenesis. Optimal osteogenic conditions also increased endogenous lamin-A expression (by twofold), whereas adipogenic conditions slightly suppressed lamin-A (Fig. 4E). Thus, insoluble and soluble factors combined to promote lamina remodeling consistent in trend with soft and stiff tissue lineages (Fig. 1D). Although traditional cultures of MSCs on rigid plastic or glass (with unknown matrix) also showed that lamin-A knockdown favored adipogenesis (41) and that lamin-A overexpression favored osteogenesis (42), cultures here on controlled matrix suggest that matrix is upstream, consistent with tissue studies (Fig. 1, H to J).
Matrix elasticity–directed lineage specification of MSCs is based in part on myosin-II–generated cell tension and the accompanying cell spreading (5, 11), which roughly paralleled nuclear shape changes (Figs. 3A and 4E, and fig. S9, A and B). Myosin-IIAwas indeed increasingly active and assembled with stiffness-dependent decreases in phosphorylation near myosin-IIA’s coiled-coil tail (fig. S9C), as we reported recently (43). Thus, phosphorylation of Ser/Thr residues just beyond the coiled coils (e.g., Fig. 3H) inhibits assembly of both myosin-IIA and lamin-A homodimers into the respective functional higher-order filaments, and such phosphorylation appeared consistently higher in cells on soft matrices compared with stiff matrices for both proteins. On the other hand, overexpression of lamin-A in cells on stiff matrix did not increase tail phosphorylation of myosin-IIA, suggesting a nonlinear relationship between cytoskeleton tension and lamin-A at the highest levels.
Additional indicators of cell tension tended to increase in vitro with matrix elasticity and/or lamin-A levels in MSCs. Not only was α-smooth muscle actin suppressed on soft matrix where lamin-A was low (Fig. 3, B and C), but knockdown of lamin-A also suppressed α-smooth muscle actin (ACTA2) transcript and protein (Fig. 4, F and G and fig. S12, A and B), together with many other key targets and components of the SRF pathway that regulates expression of ACTA2 as well as many cytoskeletal genes (18) involved in differentiation to both soft and stiff tissue lineages [neurogenesis (44), myogenesis (18), and osteogenesis (45)]. SRF is regulated in part by nuclear actin (18, 46), and lamin-A binds nuclear actin (47) as well as other proteins that also bind nuclear actin (48); this provided a mechanism for SRF regulation by lamin-A, as also suggested by overexpression studies of one protein (emerin) that binds both lamin-A and actin (49). Tissue analyses showed that SRF target proteins did not generally scale with tissue E (figs. S2B and S3D) and that SRF and ACTA2 transcripts increased nontrivially with E (fig. S2C). Because high SRF activity can inhibit differentiation of some lineages [e.g., epithelial cells (11)], mechanosensitive lamin-A is likely just one coregulator of the SRF pathway.
One transcription factor implicated in lipodystrophy, SREBP1 (SREBF1), is known to bind laminA in distributing between nucleus and cytoplasm (50). SREBP1 is not only an early response factor in adipogenesis (51) but, according to chromatin-IP (fig. S12C), it also regulates ACSL1 and another adipogenic survival factor FABP5 (52), both of which increased with LMNA knockdown (Fig. 4F).
YAP1 has been reported to be excluded from the nucleus in a functionally important manner during adipogenesis of MSCs and also functionally localized to the nucleus during osteogenesis of MSCs (11), but neither YAP1 transcript levels nor its binding partners or target genes changed with lamin-A knockdown (Fig. 4F). Although YAP1 protein levels did decrease with lamin-A knockdown (fig. S12, A and B), and YAP1 did tend to translocate as expected into the nucleus with increased matrix E (Fig. 4, H to J, and fig. S12D), lamin-A overexpression in cells on stiff matrix also produced decreases in both total YAP1 levels and nuclear localization (Fig. 4H). Fluorescence intensity profiles through the nucleus further showed many of the overexpressing cells as well as a fraction of wild-type cells on stiff matrix with YAP1 enriched at the nuclear envelope. The non-monotonic response of YAP1 versus lamin-A levels in vitro was also found for YAP1 protein and transcript levels versus tissue stiffness (Fig. 4K and fig. S12E). Consistent with this, neither YAP1 nor SRF were predicted to directly drive LMNA expression (fig. S13A), and LMNA has not been found to be a direct target of these factors as detected by chromatin-IP (18, 53). We thus sought a pathway that could directly regulate LMNA and thereby impact lineage.
Retinoic Acid Pathway Regulates Lamin-A Transcription, but Lamin-A Protein Regulates an RA Receptor
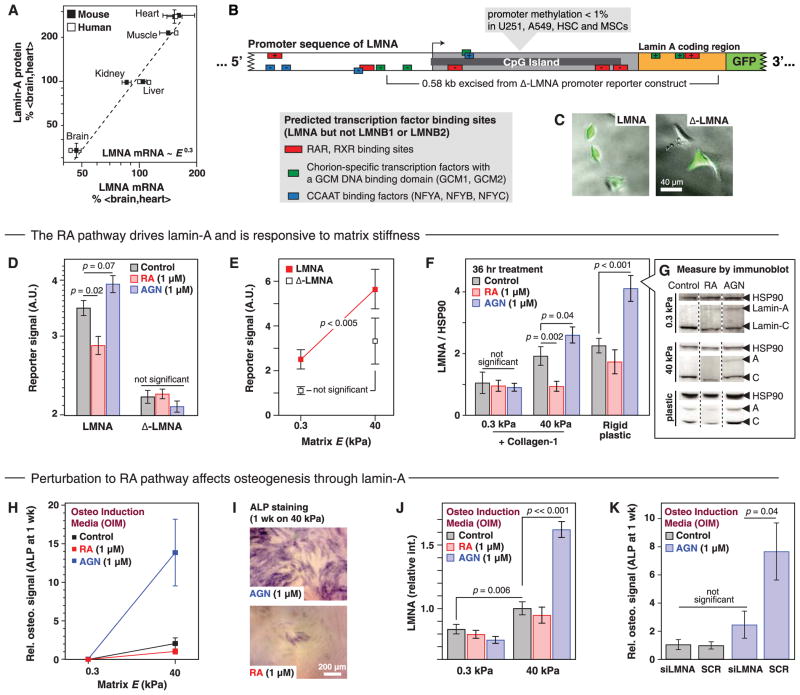
LMNA level is transcriptionally regulated, with both message and protein fitting the same power law scaling in mouse and man (R2 = 0.95) (Fig. 5A). Promoter methylation was minimal in LMNA across a range of cell types (fig. S13B). Bioinformatics analyses of promoters for LMNA, LMNB1, and LMNB2 predict retinoic acid (RA) transcription factor sites only in LMNA (Fig. 5B and fig. S14), and chromatin-IP has confirmed binding of RA nuclear receptors to LMNA (fig. S12C) consistent with experiments on RA-responsive elements(RARE) in LMNA (54). Neither RA factors nor collagens were greatly affected by lamin-A knockdown (Fig. 4F and fig. S13C), except for RARB, which is a downstream target of the RA pathway. This placed extra-cellular matrix upstream of lamin-A together with the level of transcription factors that likely regulate lamin-A, while also suggesting that the RA pathway might be modulated by lamin-A.
Fig. 5. Matrix stiffness is upstream of RA regulation of lamin-A transcription.
(A) LMNA message correlates with protein (R2 = 0.95) across tissue. (B) Promoter-reporter construct for LMNA is annotated with six predicted binding sites of transcription factors in RA pathway and a deletion construct (Δ-LMNA) lacking four RA factor binding sites. (C) A549 cells transfected with GFP reporter constructs. (D) Antagonist (AGN) and agonist (RA) increase and decrease, respectively, expression from the LMNA promoter-reporter and not Δ-LMNA; lamin-A protein shows the same response. (E) LMNA reporter activity increased significantly in MSCs grown on stiff (40 kPa) versus soft (0.3 kPa) matrix, but Δ-LMNA showed no significant difference. (F) RA and AGN regulate lamin-A in MSCs only on stiff matrix. Gels were coated with collagen-1 for comparison to cultures on untreated plastic, and immunoblotting (G) was performed after 36 hours culture (mean ± SEM from titration; all blots from same membrane; P ≤ 0.05; n ≥ 3 immunoblots). (H) AGN coupled with stiff matrix to increase osteogenesis, determined by ALP staining (I). (J) Increased osteogenic potential was coincident with increased lamin-A levels, measured by immunofluorescence. (K) Lamin-A was necessary for the increased osteogenic potential of MSCs treated with AGN as coincident treatment with siRNA against LMNA-abrogated osteogenesis.
Enzymatically derived from vitamin A, RA regulates development and regeneration and is a normal component of serum (~10 nM). It enhances lamin-A expression in embryonic carcinoma cells (54) while repressing lamins in adult granulocyte differentiation (55). Here, a lamin-A promoter driving GFP in A549 cells (Fig. 5C and fig. S14B) showed that RA was repressive, whereas an antagonist (AGN-193109, denoted AGN) enhanced expression (Fig. 5D). RA nuclear receptors are the major RA effectors and were likely involved; indeed, a mutated promoter construct (Δ-LMNA) lacking four of six RAREs (Fig. 5B and fig. S14B) showed no significant response to RA or AGN (Fig. 5D). Immunoblots of endogenous lamin-A confirmed RA responsiveness (fig. S15, A to E), and a pharmacodynamics study demonstrated nM activity (56) as well as a twofold dynamic range in lamin-A:B expression (fig. S15, F and G). MSCs transfected with the promoter constructs showed an increase in expression in cells on stiff matrix compared with soft matrix for the full promoter, whereas Δ-LMNA showed no significant difference (Fig. 5E). Mechanical and chemical cues were combined to assess lamin-A protein in MSCs cultured on gels (Fig. 5, F and G), with the cells on stiff matrix (or on plastic) showing the expected lamin-A increase with AGN treatment and decrease with RA, but the effects were entirely suppressed on soft matrix. Matrix elasticity is thus upstream of the RA pathway, which is, in turn, upstream of lamin-A transcription.
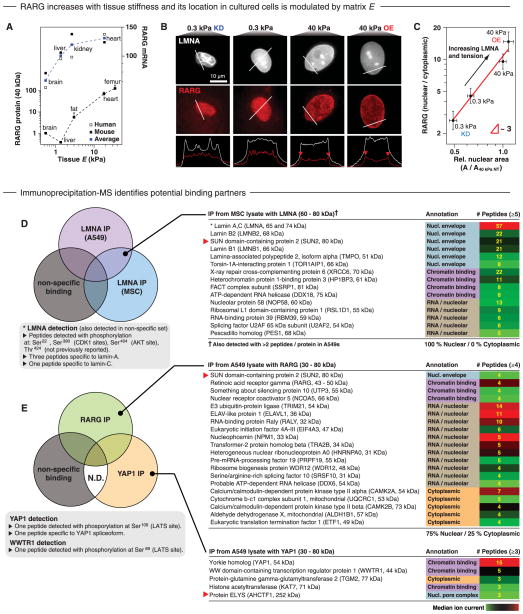
Based on RA pathway effects on lamin-A expression in cells on stiff matrix (Fig. 5F), we hypothesized measurable effects on osteogenesis. AGN indeed enhanced osteogenesis of MSCs on stiff matrix, consistent with the AGN-driven increase in lamin-A level, whereas RA suppressed osteogenesis on stiff matrix, and neither drug had an effect on MSCs on soft matrix (Fig. 5, H and I). The increased osteogenic potential of AGN-treated MSCs on stiff matrix was reflected in higher lamin-A levels (Fig. 5J), and the effect was nullified by simultaneous treatment with small interfering RNA (siRNA) against LMNA (Fig. 5K). Ectopic bone with MSC grafts has been found to be inhibited by RA and a RARG-specific agonist (57, 58), and RARG knockout mice also exhibit osteochondral defects and live only weeks longer than the few weeks that LMNA-deficient mice survive (59–61). Chromatin-IP has thus far identified the RUNX2 gene to be a target of RA transcription factors but not yet SRF (fig. S12C). RARG message generally increased in mouse and man versus tissue E, and RARG protein likewise increased in mouse tissue (Fig. 6A and fig. S12E). Fat showed low levels of RARG, and lamin-A knockdown did appear to switch RA pathways toward one that should favor adipo-genesis (Fig. 4F). In MSCs, RARG was mostly nuclear in immunostaining (Fig. 6B), and its levels were relatively unperturbed by knockdown of lamin-A (fig. S12, A and B). However, RARG’s nuclear-to-cytoplasmic ratio increased fourfold from soft to stiff matrix and was further suppressed by lamin-A knockdown (Fig. 6C). This effect of the lamina on nuclear translocation of RARG was similar in magnitude to the translocation of mechanosensitive YAP1 (Fig. 4I). Moreover, in cells on stiff matrix, RARG also localized to the nuclear envelope (Fig. 6B).
Fig. 6. Lamin-A protein regulates nuclear translocation of RA receptor.
(A) RARG protein and message (mouse and human average) increased in tissues of increasing stiffness. (B) High-resolution confocal microscope images of RARG in MSCs on matrices of various stiffnesses and with knockdown or overexpression of lamin-A. Nuclear midsections showed cytoplasmic RARG on soft matrix and increasing localization of RARG to the nuclear periphery with increasing lamin level. (C) Nuclear-to-cytoplasmic ratio of RARG scales with matrix elasticity and was directly affected by lamin-A knockdown or overexpression. All points are significantly different, as indicated (n ≥ 3 IF experiments). (D) Proteins coimmunoprecipitated with lamin-A, common to both A549s and MSCs but not found in a nonspecific control (against GFP, which was not present in the sample), were analyzed by MS. (E) Proteins associated with immunoprecipitated RARG or YAP1, but not with control samples (a combined list of proteins precipitating with antibody to GFP, not present in the sample, and proteins binding to antibody-free beads). Protein lists were compiled by combining hits from duplicate experiments. The nuclear membrane protein SUN2 was common to both lamin-A and RARG immunoprecipitation experiments. N.D., not detected.
One of the very few other factors found in the nuclear proteome that correlated well with tissue E was polymerase I and transcript release factor (PTRF) (fig. S1). PTRF transcript also correlated strongly with both LMNA and COL1A1 across mouse and man transcriptomes (fig. S2D), and lamin-A knockdown strongly decreased PTRF levels in MSCs (fig. S13C). Consistent with PTRF being downstream of lamin-A, chromatin-IP identifies PTRF to be a target gene for both RA and SRF transcription factors (fig. S12C).
To directly perturb the nuclear envelope by means other than knockdown or overexpression of lamin-A, we overexpressed the membrane protein SUN2, which shuttles from the endoplasmic reticulum (ER) to the inner nuclear envelope, where it cross-links the nuclear lamina to the cytoskeleton (48). We hypothesized that SUN2 overexpression would saturate cross-linking sites and effectively decouple the nucleus from the cytoskeleton. SUN2 overexpression indeed produced nuclear rounding and decreased lamin-A levels (figs. S16, A and B), and it also increased cytoplasmic RARG (fig. S16C). SUN2 interacted with lamin-A based on MS analyses of proteins that coimmunoprecipitated with lamin-A (Fig. 6D) [as seen in other assays (62)]. Consistent with such an interaction, nucleus-enriched tissue proteomics indicated SUN2 ~ E0.9, even though total SUN2 showed no trend with E (fig. S1B). SUN2 was also found in one co-IP with RARG (Fig. 6E), which might explain RARG’s enrichment at the envelope (Fig. 6B); however, interactions are likely to be indirect and/or weak at least because cytoplasmic RARG appeared more diffuse than SUN2 in the ER (fig. S16C). In the same set of experiments, a similar IP-MS approach was taken to identify possible binding partners for YAP1, particularly any factors involved in enrichment at the envelope (Fig. 4H). IP-MS (Fig. 6E) identified the YAP1 paralog TAZ (WWTR1), which has been reported to form a heterodimer with YAP1 (63), and YAP1 phosphorylation at a serine indicated interactions with a key kinase in the Hippo pathway (64). In addition, the co-IP included ELYS (AHCTF1 and MEL-28), which is known to be enriched at the nuclear envelope (65), but any YAP1-ELYS interactions are again likely to be indirect and/or weak and in need of further study together with RARG and SUN2. Nonetheless, our finding that lamin-A protein indirectly regulated lamin-A transcription (through factors that might also bind RARG) means that the apparent mechanoregulation of message could simply be a consequence of feedback from mechanoregulated protein.
High Lamin-A Impedes Nuclear Remodeling Under Stress
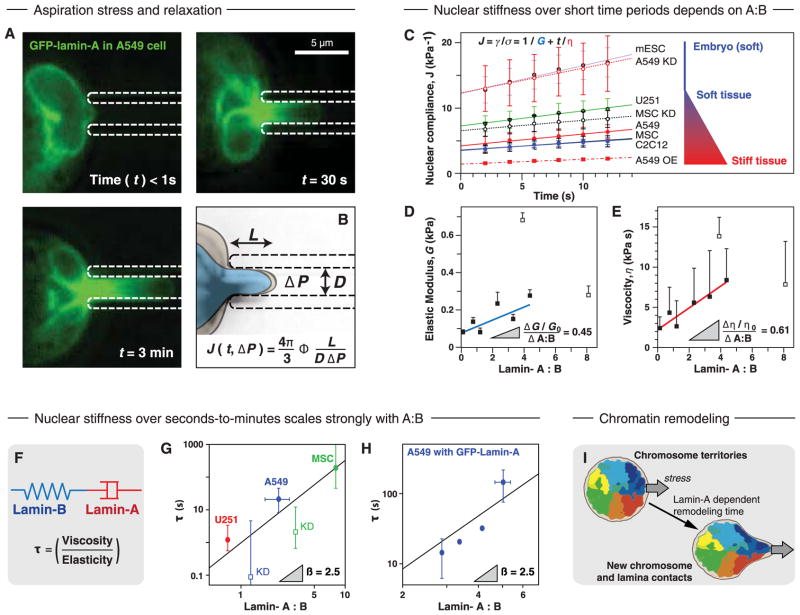
Because tissue stress and matrix stiffness stabilize expression of lamin-A, which clearly conferred protection against stress-driven rupture of isolated nuclei (Fig. 2D), we sought a real-time analysis of single-cell nuclear responses to stress. Nuclei in diverse cell types—including embryonic stem cells with very low lamin-A (20)—were aspirated into micropipettes at stresses of kPa, which is similar to tensions in cells (5) and also similar in magnitude to tissue stiffnesses (Fig. 1D). Each nucleus was found to extend in a viscoelastic manner within just ~10 s (Fig. 7, A to C). As a function of lamin-A:B stoichiometry, the effective viscosity increased more rapidly than the effective elasticity (Fig. 7, D and E). Lamin-B thus acted like the elastic walls of a balloon, driving the nucleus to return to its original shape, whereas lamin-A contributed more as a highly viscous fluid within the balloon to impede deformation. Consistent with this physical distinction between lamin isoforms, fluorescence correlation spectroscopy has shown lamin-A to be mobile and lamin-B to be immobile (24).
Fig. 7. Lamin-A confers a viscous stiffness to nuclei that impedes nuclear remodeling by stress.
(A) Micropipette aspiration of an A549 cell nucleus expressing GFP-lamin-A shows extension of the lamina with time. (B) Schematic showing how nuclear compliance is calculated from image analysis as a function of time and aspiration pressure. (C) Modeling compliance over the first 12 s of deformation, with contributions from elasticity (G) and viscosity (η) in nuclei with different lamina compositions. (D and E) Relationship between the characteristic lamin-A:B ratio and (D) the elastic modulus or (E) the viscosity. The outlier points, A549 OE and MSC, indicated by open symbols, were omitted from the linear fits. (F) The response of the lamina can be considered as a combination of elastic and viscous components, with an elongation response time, τ (see Box 1). τ was calculated for nuclei extended to ~5 μm by micropipette aspiration over seconds-to-minutes time scales in cells with different lamin-A:B ratios (G) and in A549 cells overexpressing GFP-lamin-A (H). Lamin ratios were calculated from a combination of immuno-blotting and MS methods. The scaling of τ with changes in the lamin-A:B ratio, β, was found to be the same in both experiments. (I) A potential biological consequence of nuclear distension is the remodeling of chromosome territories and chromatin-envelope interactions. All points are significantly different where indicated (n ≥ 3 nuclei).
In vivo cell migration has been seen to dynamically distend a nucleus by twofold or more reversibly over tens of minutes (20). On such long time scales, stress might extend a nucleus locally to a length similar to a typical chromosome (e.g., 5 μm), but a stiffness-limited extension rate is likely important because elongation of chromatin within the extended nucleus (20) implies rearrangements of chromatin-lamina interactions (66, 67). We hypothesized therefore that across distinct cell types with very different epigenetic features, the time needed for the nucleus to rearrange or relax, τ, would depend primarily on lamin-A level (Fig. 7F). Indeed, for a broad range in lamin-A:B ratio across various glial, epithelial, and mesenchymal cell types with or without knockdown or over-expression of lamin-A, τ varied ~10,000-fold. This level of variation is similar to time-scale differences that would be obtained in aspirating water versus honey. As functions of lamin-A:B stoichiometry, the steeply positive power laws [τ ~ (Lamin-A: B)2.5] (fig. 7, G and H) were also consistent with predictions from polymer physics (Box 1). The scaling revealed a dominating contribution of lamin-A to nuclear viscosity relative to lamin-B’s contribution to nuclear elasticity. While low laminA proved insufficient to protect against extreme stresses that completely disrupted chromatin packing (Fig. 2D), the 30-fold higher lamin-A in stiff tissue relative to soft tissue (Fig. 1D) would tend to impede rapid nuclear distension. Thus, high stresses and/or stress fluctuations typical in a stiff tissue such as muscle, heart, or bone (high E in Fig. 1D) will not “shock” the nucleus and disrupt chromosome territories or chromatin-lamina interactions (Fig. 7I) that contribute to epigenetic regulation (66, 67) or, in the extreme, cause DNA breaks.
Box 1. Polymer physics of the nuclear lamina as a shock absorber.
Lamin-A’s nonlinear contribution to viscosity in nuclear mechanics relative to lamin-B’s contribution to solidlike elasticity can be understood from polymer physics theory. Such theories focus on averages (e.g., mean concentration) while neglecting heterogeneities in molecular states such as polydispersity (e.g., lamin-A splice-forms). For a viscoelastic object such as the model of Fig. 7F, the response time in elongation (92) is
This is the time required for the energy stored upon rapid stretching of the elastic component to be dissipated by the viscous component. For large polymers, τ is typically seconds to minutes as opposed to nanoseconds for small solutes (92). Moreover, as a function of concentration c of polymers ranging from filamentous proteins and DNA to synthetic polymers that are concentrated enough to interact (not dilute), theory and experiment show that Viscosity ~ ca with scaling exponent a = 3 ± 1 (93, 94). Simple polymer physics therefore predicts that Viscosity ~ [Lamin-A]3. The elasticity of a polymer network such as a lamin-B network, is expected to be proportional to chain density, and so Elasticity ~ [Lamin-B]1. Therefore
Experiment gave τ ~ [Lamin-A:B]2.5 (Fig. 7, G and H). Additional interactions of lamin-A with lamin-B might also affect network elasticity as Elasticity ~ [Lamin-A:B]b, and our measurements indicate b ≈ 1 for t → 0 (Fig. 7, C to E) and b ≈ 0.5 on minute time scales. Regardless, the nonlinear scaling result is similar, τtheory ~ c3 – b ≈ c2 to 2.5. Lamin-A thus contributes to nuclear mechanics primarily as a concentrated polymer that slowly flows when stressed.
Many progeroid syndromes—beyond progeria due to defective lamin-A—involve mutations that impair proteins important to DNA repair (68). The finding by IP-MS (Fig. 6D) that one DNA repair factor, XRCC6 (Ku70) (69), pulled down with lamin-A from two cell types using high-affinity antibody and that XRCC6 was slightly but consistently decreased upon LMNA knockdown (fig. S13C) suggests a mechanochemical link of DNA repair to lamin-A. Further study is motivated by a previous MS finding that both XRCC6 and ELYS pull down with biotinylated lamin-A bound with low affinity (μM) to a streptavidin analog (70). Regardless of a possible molecular link to DNA repair factors, lamin-A clearly protects the nucleus against stress.
Systems Mechanobiology: Core Gene Circuit Yields Steady-State Scaling for Lamin-A
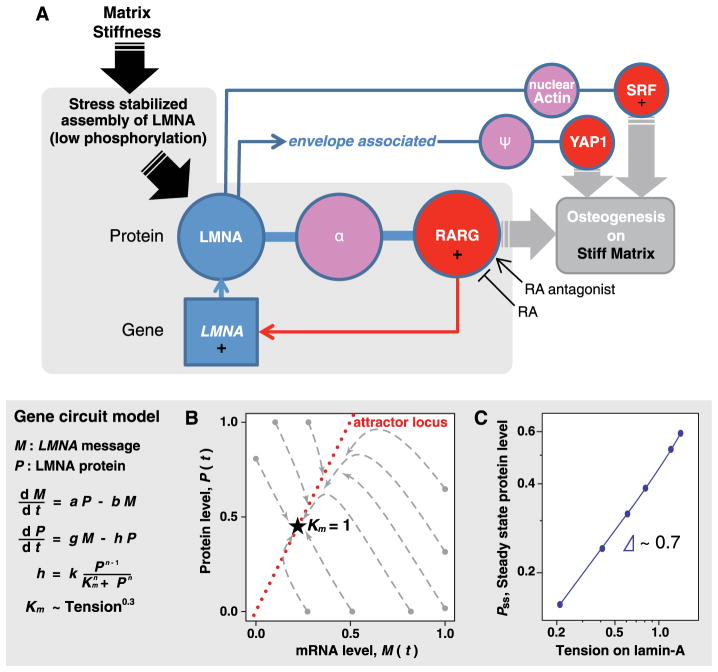
High matrix stiffness is associated with an increased stress or tension on the nucleus and promotes lamin-A expression and a physically stiffer nucleus. If we thus assume that tension in the rope-like supercoil assemblies of lamin-A filaments suppresses the affinity of an enzyme that initiates phosphorylation/solubilization/degradation of lamin-A, then a parsimonious gene circuit (Fig. 8A) could be modeled mathematically (Fig. 8, B and C), with lamin-A protein effectively feeding back on its own message. Mechanics was explicitly included only in the stress-dependent protein turnover term; synthesis of message and protein as well as degradation of message were all assumed to be linear, with all rate constants chosen to be of order unity. As a test of whether such a model could capture key experimental trends, computational results showed that steady-state lamin-A levels scaled with (Tension)0.7, which parallels the scaling of lamin-A with tissue microelasticity E (Fig. 1D), noting that Tension ~ E. Kinetic measurements of laminA changes with cell mechanical perturbations are clearly needed to further develop such a model.
Fig. 8. A feedback-based gene circuit for lamin-A exhibits polymer physics scaling if cell tension suppresses protein turnover.
(A) Gene circuit connecting matrix stiffness to osteogenesis; not shown is an overlapping circuit for adipogenesis on soft matrix that includes the positive regulator SREBP1. LMNA protein level is regulated by a stress-sensitive phosphorylation mechanism and feeds back into LMNA transcript through interaction with RARG, possibly through an intermediary, α, and can be perturbed with antagonist (AGN) or agonist (RA). LMNA protein also influences location of YAP1 (through a possible intermediary, Ψ) to drive cell fate (11), and LMNA regulates SRF through interaction with nuclear actin (49). A simple model was generated based on this circuit: Time evolution of LMNA mRNA (M) level is dependent on the LMNA protein level (P), whereas the protein level itself is regulated by a tension-dependent degradation term, h. The model shows that tension-regulated protein turnover can produce steady-state (SS) protein levels that scale with cell tension. (B) Trajectories of lamin-A message and protein as the model converges from a range of initial conditions to a single steady-state solution appropriate to the tension. (C) Setting the kinase/protease binding coefficient, Km, to be proportional to (Tension)0.3 allows the model to generate steady-state lamin scaling with tension consistent with experiment (Fig. 1D).
Discussion
Matrix elasticity is upstream of lamin-A levels in the 3D xenograft model as well as in the 2D culture models. Lamin-A knockdown in MSCs indeed did not affect expression of the key collagens that scale with tissue stiffness but did suppress the SRF pathway that promotes expression of abundant actin-myosin cytoskeletal components. Soft matrix likewise appears to minimize cytoskeletal stress or tension on the wrinkled nucleus, which thus minimizes stress on lamin-A and thereby favors its phosphorylation and turnover. Low lamin-A protein limits its own transcription by altering nuclear localization of RA transcription factors: Soft matrix and low laminA produce the highest cytoplasmic levels of RARG, and the same conditions not only showed the lowest LMNA promoter activity but also showed no significant changes of lamin-A protein levels with added RA and AGN. With adipogenic stimuli that include a soft matrix, partial knockdown of lamin-A in MSCs (to lamin-A:B ~ 3) maximized in vitro adipogenesis, consistent with A:B scaling in tissue profiling.
At least for stiff tissue cells with abundant RARG, the response in the vitamin A pathway was also downstream of matrix stiffness. Stiff matrix and high lamin-A led to the highest nuclear levels of RARG, and the same conditions showed not only the highest LMNA promoter activity but also about a twofold variation in lamin-A levels upon addition of RA and AGN. AGN enhanced lamin-A, typical of a stiff tissue, and MSC osteogenesis also increased. Differentiation might also be sensitive to splice-forms because AGN increased the A splice-form of LMNA, as did forced overexpression, and tissue profiling showed bone has more A than C splice-form, all of which motivates further study. In human bone tissue, lamin-A is among the 20 most abundant proteins detected by MS, together with several SRF-regulated gene products (ACTB, ACTA2, and MYL9) (71), and whereas SRF positively regulates a key transcription factor (RUNX2) in osteogenesis (45), chromatin-IP has thus far identified the RUNX2 gene to be a direct target of RA transcription factors and not of SRF. Lamin-A overexpression in MSCs on stiff matrix not only enhanced osteogenesis but also produced a modest decrease in nuclear YAP1 consistent with the measured nonmonotonic responses of YAP1 across tissues. A complex interplay between YAP1 and/or lamin-A might reconcile past observations that YAP1 promotes osteogenesis (11) but also inhibits RUNX2 (72); switching between YAP1 and TAZ (WWTR1) activities is also possible, because TAZ can promote osteoblast differentiation of MSCs by enhancing RUNX2-dependent transcriptional activation (73), and the IP-MS data suggested a YAP1-TAZ interaction. Regardless, the proposed gene circuit for lamin-A seems to be one important core module that modulates various transcriptional pathways (RA, SREBP1, SRF, and YAP1) in enhancing matrix elasticity–directed differentiation.
Our multifaceted approach to a broad range of solid tissues using proteomics and transcriptomics plus knockdown in a highly plastic stem cell that expresses abundant lamin-A seems useful for generic pathway analyses. Indeed, polymerase I and transcript release factor (PTRF) was one of the few factors that not only correlated well with tissue E, and decreased with lamin-A knockdown, but also is a known target of RA and SRF transcription factors. The additional fact that mutations in human PTRF cause muscular dystrophies and lipodystrophies that also result from LMNA mutations (74) suggests an additional feedback into intersecting pathways. Understanding the physiological determinants of normal lamin levels could thus begin to clarify why some laminopathies have phenotypes that dominate in a particular subset of tissues.
Beyond a role in regulating transcription factors, the lamins interact directly with many other nuclear proteins and form lamina-associated domains (LADs) that are repressive regions rich in heterochromatin (66, 67). LADs have roles in some aspects of lineage-specific differentiation, and so the 30-fold variations in lamin-A with tissue stiffness imply a physical regulation of LADs. The relative abundance of Aversus B-type lamins could be critical, and despite the weak scaling observed for lamin-B1 and -B2 in solid tissues, nucleated blood cells show large variations in lamin-B (55). Mechanisms of regulation of lamin-Bs within soft marrow are thus far unclear, but human hematopoietic stem cells (HSCPs) do exhibit a soft nuclear phenotype (20) consistent with soft marrow. The MSCs studied herein were also human bone marrow–derived, and their potential to contribute to both rigid bone and softer marrow fat is relevant to marrow microenvironments and to a distinct influence of osteoblasts and adipocytes on HSCPs (75).
Lamins as Stress-Modulated Lineage Enhancers
Neither A- nor B-type lamins are essential for lineage induction or specification. LMNA knockout mice develop all tissues but die weeks after birth with growth retardation of connective tissues and also muscular dystrophy (59–61) that is very severe compared with the prototypical mouse model of muscular dystrophy (Dmdmdx), which lives for 2 years (76). LMNA expression is therefore essential for maturation and survival against the incessant stressing in adult tissues, and once the lamin level matches to the stress, then the optimal level can enhance a prespecified lineage. The weakly modulated B-type lamins are also dispensable in embryonic development; lamin-B knockout mice die at birth as neurons apoptose (34) in migration through the dense midcortex, which stretches normal nuclei by four- to fivefold (20). The lamin knockouts thus suggest important structure-stabilizing roles for the lamins, but the findings reveal lamin-A to be the most stress-inducible differentiation-enhancing factor and lamin-B2 to be the most refractory. Distinct regulation of stress-inducible isoforms versus constitutive isoforms is reminiscent of the heat shock protein 90 isoform family that constitutes roughly 2% of protein mass in mammalian cells (77). This seems no coincidence because mechanical work and heat are, after all, two key terms in inescapable thermodynamic laws that cells have evolved and adapted to.
Although softening or stiffening the nucleus through lamin-A knockdown or overexpression could physically modulate the cytoskeleton and cell spreading in a manner similar to matrix elasticity, nuclear effects were not as large as matrix effects. This could be because cytoskeleton-nuclear interactions are several times smaller in net area than cell-matrix interactions; from this purely physical point of view, matrix elasticity should be upstream of nuclear mechanics and lamin regulation. In addition, tissue elasticity does not appear greatly affected by nuclear mechanics: In a stiff tissue such as skeletal muscle (E ~ 12 kPa), nuclei make up just 0.15% of muscle volume, whereas cytoskeletal volume is almost 60% (78), and in a much softer tissue such as liver (E ~ 1.5 kPa), the nuclear-to-cytoplasmic percentage is much higher but still small at around 6 to 7% (79). Nonetheless, lamin-A:B levels did adjust by about twofold in primary adult cells (MSCs) as well as cell lines (U251 and A549) in response to tissue microenvironments in vivo, or else soft gels and stiff substrates (including rigid plastic), and/or soluble factors in the RA pathway. Although this typical range for the nuclear “mechanostat” is a fraction of the lamin-A:B range found across adult tissues, fibroblasts with abundant lamin-A not only lose nearly all of it when reprogrammed to induced pluripotent stem cells (iPSCs) (80) but also are more efficiently reprogrammed (>20-fold) by RA agonists (81) that our results suggest suppress lamin-A. Modifiers beyond the RA pathway could thus couple to the lamin mechanostat and further amplify its range up or down to better match microenvironment stiffness and stress.
Common Structures Under Tension: Supercoiled Helical Assemblies
The α-helical coiled coil and the collagen triple helix are the two known motifs for supercoiled multistranded proteins (82). Both structures tend to assemble into rope-like fibers that are often subjected to tension, but how tension affects structure or turnover is understudied. Collagen-I fibers are degraded more slowly by matrix protease when tension is applied (83), consistent with the well-known atrophy of disuse of connective tissue. The initial findings that lamin-A’s filamentous coiled-coil assembly is hyperphosphorylated in cells with low tension on soft matrix is consistent with higher turnover and requires in-depth study but seems to suggest a more general model of biostabilization by tension. Lamin-B1 and -B2 appear less susceptible to this mechanochemical regulation, perhaps because farnesylation of these proteins concentrates them at the nuclear envelope.
Additional structural modulators of supercoil assembly are also likely to contribute to pathophysiological responses. The Ig domain of lamin-A, which affects filament assembly (84) and interacts with a range of nuclear and nuclear-membrane proteins in addition to the SUNs (62) responds conformationally to stress. Phosphorylation at the nearby Ser390, a target of CDK1 (85), was found to differ between wild-type lamin-A and the less stable mutant. Force-driven structural changes in the cytoskeletal protein p130Cas (86) also regulate its phosphorylation by tyrosine kinases, although stress increases phosphorylation of p130Cas and decreases it in lamin-A. Density of lamin-A is ultimately regulated to limit the stress per molecule, thereby adjusting nuclear strength and stiffness consistent with polymer physics and with expectations for a mechanostat. The multifunctionality of a nuclear IF protein such as lamin-A thus results in mechanosensitive feedback on multiple pathways that contribute to differentiation.
Materials and Methods
Whole-Mouse Tissue Lysis
NSG mice (NOD/SCID/IL-2Rγ−/−) were 4 to 8 months in age (n = 5 mice). Protocols approved by Penn’s Institutional Animal Care and Use Committee (IACUC) were used throughout. Sections of fresh tissue (~50 mm3 heart, brain, liver, lung, kidney, and cartilage from ear) were finely chopped with a razor blade on ice. Additional experiments used flash-frozen tissue (~50 mm3 heart, brain, liver, lung, skeletal muscle, kidney, cartilage from ear, skull, and marrow-free thigh bone), which was finely ground in a pestle and mortar on dry ice. The resulting pastes were suspended in 300-μL ice-cold lysis buffer [1x RIPA buffer (radio immunoprecipitation assay buffer), 1x NuPAGE LDS running buffer (polyacrylamide gel electrophoresis, lithium dodecyl sulfate, from Invitrogen), 0.1% protease inhibitor cocktail (PIC)]. After 30 min incubation at 4°C, samples were subjected to 5 × 20 × 1 s pulses with a probe sonicator (intermediate setting, on ice). A reducing agent was added [β-mercaptoethanol (BME) to 1%] and the samples were heated to 80°C for 10 minutes before ultra-centrifugation [1 hour at 90,000 revolutions per minute (rpm) in a TLA120.1 rotor (Beckman) at 4°C]. The blue aqueous layer was then carefully extracted from between layers of fat and any insoluble pelleted material.
Nuclear Enrichment from Mouse Tissue [adapted from (87)]
Sections of flash-frozen tissue (~50 mm3 heart, brain, liver, lung, skeletal muscle, and fat) were thawed on ice, finely chopped with a razor blade and suspended in 10 mL buffer A [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT) with 0.1% (PIC, Sigma Aldrich)]. The tissue was then disrupted on ice with a Dounce homogenizer [10 strokes with a loose-fitting pestle (B grade), 10 strokes with a tight-fitting pestle (A grade)] and persistent detritus was removed by passage through a 40-μm cell filter (Fisher Scientific). Crude nuclear material was then pelleted by centrifugation (10 min at 5000 rpm at 4°C), resuspended in 1 mL buffer S1 (10 mM HEPES, 0.25 M sucrose, 10 mM MgCl2, and 0.1% PIC) and layered over 1 mL buffer S3 (10 mM HEPES, 1.2 M sucrose, 0.5 mM MgCl2, and 0.1% PIC). Nuclear material was pelleted by centrifugation [10 min at 5000 rpm in a TLS-55 swinging-bucket rotor (Beckman) at 4°C] and resuspended in 50 μL ice-cold lysis buffer [1x RIPA, 1x NuPAGE LDS running buffer (Invitrogen), 0.1% PIC]. After 30 min incubation at 4°C, samples were subjected to 15 × 1 s pulses with a probe sonicator (low setting, on ice). Before gel electrophoresis, a reducing agent was added (BME to 1%), heated to 80°C for 10 min, and centrifuged to pellet any insoluble material (10 min at 13,300 rpm).
Mass Spectrometry
Gels from SDS–polyacrylamide gel electrophoresis (SDS-PAGE) (NuPAGE 4 to 12% bis-tris, Invitrogen) were loaded with 12 μL of lysate per lane and run at 10 min × 100 V, followed by 20 min × 160 V. Coomassie-stained polyacrylamide gel sections were washed [50% 0.2 M ammonium bicarbonate (AB) solution, 50% acetonitrile (ACN), 30 min at 37°C], dried by lyophilization, incubated with a reducing agent [20 mM tris(2-carboxyethyl)phosphine (TCEP) in 25 mM AB solution at pH 8.0, 15 min at 37°C], and alkylated [40 mM iodoacetamide (IAM) in 25 mM AB solution at pH 8.0, 30 min at 37°C]. The gel sections were dried by lyophilization before in-gel trypsinization [20 μg/mL sequencing grade modified trypsin in buffer as described in the manufacturer’s protocol (Promega), 18 hours at 37°C with gentle shaking]. The resulting solutions of tryptic peptides were acidified by the addition of 50% digest dilution buffer (60 mM AB solution with 3% methanoic acid).
Peptide separations (5 μL injection volume) were performed on 15-cm PicoFrit column (75 μm inner diameter, New Objective) packed with Magic 5 μm C18 reversed-phase resin (Michrom Bioresources) using a nanoflow high-pressure liquid chromatography system (Eksigent Technologies), which was coupled online to a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ion source. Chromatography was performed with solvent A (Milli-Q water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Peptides were eluted at 200 nL/min for 3 to 28% B over 42 min, 28 to 50% B over 26 min, 50 to 80% B over 5 min, and 80% B for 4.5 min before returning to 3% B over 0.5 min. To minimize sample carryover, a fast blank gradient was run between each sample. The LTQ-Orbitrap XL was operated in the data-dependent mode to automatically switch between full-scan MS [in terms of mass m and charge z: m/z = 350 to 2000 in the Orbitrap analyzer (with resolution of 60,000 at m/z 400)] and the fragmentation of the six most intense ions by collision-induced dissociation in the ion-trap mass analyzer.
Raw mass spectroscopy data was processed using Elucidator (version 3.3, Rosetta Biosoftware). The software was set up to align peaks in data from samples derived from corresponding molecular weight regions of the 1D gels. Peptide and protein annotations were made using SEQUEST (version 28, Thermo Fisher Scientific) with full tryptic digestion and up to two missed cleavage sites. Peptide masses were selected between 800 and 4500 amu with peptide mass tolerance of 1.1 amu and fragment ion mass tolerance of 1.0 amu. Peptides were searched against a database compiled from UniRef100 (November 2010) mouse and human, plus contaminants and a reverse decoy database. A deltaCn of 0.01 and mass error limit of 20 parts per million (ppm) was used, resulting in a false positive rate of ~10%. In these experiments, only proteins detected with three or more peptides were considered. [Therefore, for positive identification of a protein, P < (0.1)3.] The peptide database was modified to search for alkylated cysteine residues (monoisotopic mass change, Δ = +57.021 Da) and oxidized methionine (Δ = +15.995 Da). In proteomic profiling experiments, we also considered the acetylation of lysine (Δ = +42.011 Da), methylation of lysine and arginine (Δ = +14.016 Da), hydroxylation of lysine, proline, aspartate, and asparagine (Δ = +15.995 Da), and phosphorylation of serine, tyrosine, threonine, histidine, and aspartate (Δ = +79.966 Da). In experiments in which cysteine residues were labeled with monobromobimane (mBBr), the modification was searched for in three possible oxidation states (Δ = +133.053 Da, Δ = +150.056 Da, and Δ = +151.064 Da). Peptides derived from the autolysis of trypsin were considered to be contaminants and were not used in subsequent calculations. When evaluating total ion current, only signals from annotated peptides were summed. The Peptide Ratio Fingerprint (PRF) algorithm was coded for Mathematica (version 8, Wolfram Research) and was used for all MS protein quantitation (88). Synthetic, HPLC-purified versions of key peptides were purchased from GenScript (figs. S4 and S8, E and F) and were used to confirm MS detection and linearity of response.
Determination of Absolute Lamin Ratio (A + C)/(B1 + B2)
When comparing the lamin composition in samples from two conditions—for example, a control and a knockdown—MS analysis allowed the relative (fold) change in lamin level to be established in the lamin-A,C overlap region, lamin-B1, lamin-B2, and an overlap peptide that is common to all lamins, LLEGEEER (fig. S4). These ratios were combined to calculate the absolute ratio between A-type and B-type lamin
where f represents the fold change in protein level in going from condition 1 (e.g., the control) to condition 2 (e.g., the knockdown). Because this measurement was dependent on detection of a single peptide, it was repeated a number of times to obtain a confident measure of the A:B ratio in condition 1. Subsequent derivation of the ratio in condition 2 had no additional dependence on detection of LLEGEEER.
The parameter fLamin B1,B2 represents the overlap between the two B-type lamins. Although we show that lamin-B1 and lamin-B2 were detected in similar quantities and that neither change to a great extent compared with lamin-A,C (Fig. 1F and fig. S4D), we derived this value from median ion current values.
The tissue experiments shown in Fig. 1D were measured relative to a “condition 1” average of brain and heart denoted as <brain, heart>, a reference point chosen in the middle of the elasticity scale for which we had many tissue duplicates (n = 4 mice for heart and brain). A lysate of A549 cells (n = 3 culture replicates) was used as a reference point for all cell measurements in Fig. 1D. MS measurements of isolated nuclear material were made in technical triplicates.
Fluid Shear on Isolated Nuclei
Isolated A549 nuclei suspended in nuclei wash buffer were diluted to about 250 nuclei per μL. Nuclei samples were labeled with 150 μM mBBr and immediately loaded into a cone and plate rheometer (Bohlin Gemini) with the stage heated to 37°C. To control for baseline labeling at this temperature an identical sample was placed in a 37°C water bath. Samples were spun in the rheometer for 10 to 40 min with a 10-μm gap. Shear force varied from 0.5 to 5 Pa in different samples. After the run, labeling in both sample and control was quenched with 2 mM glutathione. For each condition, a sample was taken to be imaged. Samples were prepared for MS as described above. To facilitate identification of the labeled lamin Ig domain peptide by MS, recombinantly expressed, mBBr labeled, and trypsinized lamin Ig domain was spiked into control A549 lysate.
Labeling Cysteines with mBBr in Adherent Cells
Low-passage primary human MSCs were seeded on soft (0.3 kPa), intermediate (10 kPa), and stiff (40 kPa) polyacrylamide gels and cultured for 2 days in Dulbecco’s minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum. A549 cells cultured on plastic were transfected with wild-type and R453W-mutant GFP-lamin constructs using Lipofectamine 2000 (LF2k, Invitrogen) per the manufacturer’s protocol. Cells were washed with phosphate-buffered saline (PBS) before labeling with freshly prepared 400 μM mBBr in PBS at 37°C. After 10 min, the cells were harvested by trypsinization, suspended in ice-cold media, pelleted, and frozen at −20°C. Samples were enriched before MS analysis by immunoprecipitation of lamin-A or GFP, as described below. In addition, a portion of cells were washed thoroughly after mBBr labeling, fixed with formaldehyde, and immunostained for lamin-A (mouse monoclonal sc-7292, Santa Cruz), α-smooth muscle actin (mouse monoclonal A5228, Sigma Aldrich), and nonmuscle myosin-IIA (rabbit polyclonal M8064, Sigma Aldrich) for imaging at high resolution.
Model of Lamin Transcript and Protein Levels
The rate equations for lamin-A protein (P) and mRNA (M) concentrations, respectively, include synthesis and degradation
where a is first-order protein-induced mRNA production rate constant, b is first-order mRNA degradation rate constant, g is first-order mRNA translational rate constant, h is force-dependent protein degradation, modeled as
where k is maximal protein degradation rate, n is cooperativity coefficient ≥2 typical of dimer-based interactions, and Km isaffinity of kinase/protease for the lamin meshwork, which increases with stress or tension sustained by the meshwork. Like pulling on a coiled rope, the key idea is that tension on this meshwork of lamin-A coiled-coil protein squeezes out free volume and sequesters the enzyme’s binding site on lamin-A. At steady-state
thus yielding nonzero steady-state values for P and M
Based on the steady-state analysis above, a solution only exists if
Time evolution of P and M was modeled in Mathematica (Wolfram) with example trajectories in Fig. 8B as a phase plot of P(t) versus M(t) converging to {PSS, MSS}. Although steady-state values depend on the various rate constants, we assumed all to be important and of order ~1 as we focus on Km: At high stresses where lamin-A assembly is favored, Km increases so that lamin-A phosphorylation/degradation decreases. Plotting PSS against different values for Km fit a power-law . If Km = (tension)0.3, then PSS ~ (tension)0.7, as found experimentally for lamin-A (Fig. 8C).
Supplementary Material
Acknowledgments
We greatly appreciate support from the U.S. National Institutes of Health (NIH R01HL062352, P01DK032094, R01EB007049, and NCATS-8UL1TR000003), the U.S. National Science Foundation (grant 1200834, the University of Pennsylvania’s Materials Research Science and Engineering Center, and Nano Science and Engineering Center, Nano/Bio Interface Center), the Human Frontier Science Program (to I.L.I. and D.E.D.), and the Stem Cell Xenograft Core (to A. Secreto, J. Glover, and G. Danet-Desnoyers) and Microarray Cores (P30-DK090969). We thank the Wistar Institute Proteomics Core for assistance with MS and standard data analyses. This work was also supported in part by U.S. NIH grants HL038794 (to D.W.S.) and CA010815 (National Cancer Institute core grant to the Wistar Institute). We thank D. Gilbert (Florida State University, Department of Biological Science), H. Herrmann (Heidelberg, DKFZ), and H. Worman (Columbia University, Department of Medicine), respectively, for GFP-lamin-A, recombinant lamins, and SUN2 plasmids.
Footnotes
References and Notes
- 1.Frost HM. Bone “mass” and the “mechanostat”: A proposal. Anat Rec. 1987;219:1–9. doi: 10.1002/ar.1092190104. [DOI] [PubMed] [Google Scholar]
- 2.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 3.Chen BPC, et al. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics. 2001;7:55–63. doi: 10.1152/physiolgenomics.2001.7.1.55. [DOI] [PubMed] [Google Scholar]
- 4.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 5.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 6.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 8.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;90:3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 12.Engler AJ, et al. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. 10.1083/jcb. 200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler AJ, et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: Scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci USA. 2005;102:4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connelly JT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 20.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammerding J, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 22.De Vos WH, et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet. 2011;20:4175–4186. doi: 10.1093/hmg/ddr344. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann H, Strelkov SV, Burkhard P, Aebi U. Intermediate filaments: Primary determinants of cell architecture and plasticity. J Clin Invest. 2009;119:1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimi T, et al. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 26.Shivashankar GV. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys. 2011;40:361–378. doi: 10.1146/annurev-biophys-042910-155319. [DOI] [PubMed] [Google Scholar]
- 27.Broers JLV, et al. A- and B-type lamins are differentially expressed in normal human tissues. Histochem Cell Biol. 1997;107:505–517. doi: 10.1007/s004180050138. [DOI] [PubMed] [Google Scholar]
- 28.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 29.Gardel ML, et al. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–1305. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 30.Kim T, Hwang W, Lee H, Kamm RD. Computational analysis of viscoelastic properties of crosslinked actin networks. PLOS Comput Biol. 2009;5:e1000439. doi: 10.1371/journal.pcbi.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strobl G. The Physics of Polymers: Concepts for Understanding their Structure and Behavior. Springer; New York: 2007. [Google Scholar]
- 32.Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunnah D, Sewry CA, Vaux D, Schirmer EC, Morris GE. The apparent absence of lamin B1 and emerin in many tissue nuclei is due to epitope masking. J Mol Histol. 2005;36:337–344. doi: 10.1007/s10735-005-9004-7. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y, et al. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang YL, Leone LM, Kaufman LJ. Elastic moduli of collagen gels can be predicted from two-dimensional confocal microscopy. Biophys J. 2009;97:2051–2060. doi: 10.1016/j.bpj.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krimm I, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. 10.1016/S0969-2126 0200777-3. [DOI] [PubMed] [Google Scholar]
- 38.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: Building blocks of nuclear architecture. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 39.Bertacchini J, et al. The protein kinase Akt/PKB regulates both prelamin A degradation and Lmna gene expression. FASEB J. 2013;27:2145–2155. doi: 10.1096/fj.12-218214. [DOI] [PubMed] [Google Scholar]
- 40.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 41.Akter R, Rivas D, Geneau G, Drissi H, Duque G. Effect of lamin A/C knockdown on osteoblast differentiation and function. J Bone Miner Res. 2009;24:283–293. doi: 10.1359/jbmr.081010. [DOI] [PubMed] [Google Scholar]
- 42.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raab M, et al. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu PPY, Ramanan N. Serum response factor is required for cortical axon growth but is dispensable for neurogenesis and neocortical lamination. J Neurosci. 2011;31:16651–16664. doi: 10.1523/JNEUROSCI.3015-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, et al. Serum response factor regulates bone formation via IGF-1 and Runx2 signals. J Bone Miner Res. 2012;27:1659–1668. doi: 10.1002/jbmr.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 47.Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus. 2010;1:264–272. doi: 10.4161/nucl.1.3.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 49.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: Implications for partial lipodystrophy and other laminopathies. Hum Mol Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- 51.Ayala-Sumuano JT, et al. Srebf1a is a key regulator of transcriptional control for adipogenesis. Sci Rep. 2011;1:178. doi: 10.1038/srep00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, et al. SiRNA against Fabp5 induces 3T3-L1 cells apoptosis during adipocytic induction. Mol Biol Rep. 2010;37:4003–4011. doi: 10.1007/s11033-010-0059-5. [DOI] [PubMed] [Google Scholar]
- 53.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okumura K, Hosoe Y, Nakajima N. c-Jun and Sp1 family are critical for retinoic acid induction of the lamin A/C retinoic acid-responsive element. Biochem Biophys Res Commun. 2004;320:487–492. doi: 10.1016/j.bbrc.2004.05.191. [DOI] [PubMed] [Google Scholar]
- 55.Olins AL, et al. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 56.Agarwal C, Chandraratna RAS, Johnson AT, Rorke EA, Eckert RL. AGN193109 is a highly effective antagonist of retinoid action in human ectocervical epithelial cells. J Biol Chem. 1996;271:12209–12212. doi: 10.1074/jbc.271.21.12209. [DOI] [PubMed] [Google Scholar]
- 57.Shimono K, et al. Potent inhibition of heterotopic ossification by nuclear retinoic acid receptor-γ agonists. Nat Med. 2011;17:454–460. doi: 10.1038/nm.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, et al. Implantation of adult bone marrow-derived mesenchymal stem cells transfected with the neurotrophin-3 gene and pretreated with retinoic acid in completely transected spinal cord. Brain Res. 2010;1359:256–271. doi: 10.1016/j.brainres.2010.08.072. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jahn D, et al. A truncated lamin A in the Lmna− − mouse line: Implications for the understanding of laminopathies. Nucleus. 2012;3:463–474. doi: 10.4161/nucl.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubben N, et al. Post-natal myogenic and adipogenic developmental: Defects and metabolic impairment upon loss of A-type lamins. Nucleus. 2011;2:195–207. doi: 10.4161/nucl.2.3.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haque F, et al. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci USA. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franz C, et al. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 67.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dechat T, et al. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kubben N, et al. Identification of differential protein interactors of lamin A and progerin. Nucleus. 2010;1:513–525. doi: 10.4161/nucl.1.6.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alves RDAM, et al. Unraveling the human bone microenvironment beyond the classical extracellular matrix proteins: A human bone protein library. J Proteome Res. 2011;10:4725–4733. doi: 10.1021/pr200522n. [DOI] [PubMed] [Google Scholar]
- 72.Zaidi SK, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 74.Hayashi YK, et al. Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J Clin Invest. 2009;119:2623–2633. doi: 10.1172/JCI38660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shin JW, et al. Mechanobiology of bone marrow stem cells: From myosin-II forces to compliance of matrix and nucleus in cell forms and fates. Differentiation. 2013;2013 doi: 10.1016/j.diff.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chamberlain JS, Metzger J, Reyes M, Townsend DW, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H, Burrows F. Targeting multiple signal transduction pathways through inhibition of Hsp90. J Mol Med. 2004;82:488–499. doi: 10.1007/s00109-004-0549-9. [DOI] [PubMed] [Google Scholar]
- 78.Eisenberg BR, Kuda AM. Stereological analysis of mammalian skeletal muscle. II. White vastus muscle of the adult guinea pig. J Ultrastruct Res. 1975;51:176–187. doi: 10.1016/s0022-5320(75)80146-8. 10.1016/S0022-5320 7580146-8. [DOI] [PubMed] [Google Scholar]
- 79.Hope J. Stereological analysis of the ultrastructure of liver parenchymal cells during pregnancy and lactation. J Ultrastruct Res. 1970;33:292–305. doi: 10.1016/s0022-5320(70)90023-7. 10.1016/S0022-5320 7090023-7. [DOI] [PubMed] [Google Scholar]
- 80.Mattout A, Biran A, Meshorer E. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Cell Biol. 2011;3:341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- 81.Wang W, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beck K, Brodsky B. Supercoiled protein motifs: The collagen triple-helix and the alpha-helical coiled coil. J Struct Biol. 1998;122:17–29. doi: 10.1006/jsbi.1998.3965. [DOI] [PubMed] [Google Scholar]
- 83.Flynn BP, et al. Mechanical strain stabilizes reconstituted collagen fibrils against enzymatic degradation by mammalian collagenase matrix metalloproteinase 8 (MMP-8) PLoS ONE. 2010;5:e12337. doi: 10.1371/journal.pone.0012337. 10.1371/journal. pone.0012337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shumaker DK, et al. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc Natl Acad Sci USA. 2005;102:15494–15499. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. 10.1016/0092-8674 9090470-Y. [DOI] [PubMed] [Google Scholar]
- 86.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lamond AI. Nucleolar isolation protocol. LamondLab. com; Dundee, Scotland: 2009. http://www.lamondlab.com/f7nucleolarprotocol.htm. [Google Scholar]
- 88.Shin JW, Swift J, Spinler KR, Discher DE. Myosin-II inhibition and soft 2D matrix maximize multinucleation and cellular projections typical of platelet-producing megakaryocytes. Proc Natl Acad Sci USA. 2011;108:11458–11463. doi: 10.1073/pnas.1017474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Su D, Gudas LJ. Gene expression profiling elucidates a specific role for RARgamma in the retinoic acid-induced differentiation of F9 teratocarcinoma stem cells. Biochem Pharmacol. 2008;75:1129–1160. doi: 10.1016/j.bcp.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Welch ID, Cowan MF, Beier F, Underhill TM. The retinoic acid binding protein CRABP2 is increased in murine models of degenerative joint disease. Arthritis Res Ther. 2009;11:R14. doi: 10.1186/ar2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu SL, Levi L, Siegel R, Noy N. Retinoic acid induces neurogenesis by activating both retinoic acid receptors (RARs) and peroxisome proliferator-activated receptor β/δ (PPARβ/δ) J Biol Chem. 2012;287:42195–42205. doi: 10.1074/jbc.M112.410381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barnes HA. Handbook of Elementary Rheology. University of Wales, Institute of Non-Newtonian Fluid Mechanics; Cardiff: 2000. [Google Scholar]
- 93.Rubinstein M, Colby R. Polymer Physics. Oxford University Press; Oxford: 2003. [Google Scholar]
- 94.Heo Y, Larson RG. The scaling of zero-shear viscosities of semidilute polymer solutions with concentration. J Rheol. 2005;49:1117. doi: 10.1122/1.1993595. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.