Abstract
Genomic DNA in a bacterial cell is folded into a compact structure called a nucleoid, and nucleoid-associated proteins are responsible for proper assembly of active higher-order genome structures. The human gastric pathogen Helicobacter pylori express a nucleoid-associated protein encoded by the hup gene, which is the homolog to the Escherichia coli histone-like protein HU. An H. pylori hup mutant strain (X47 hup:cat) showed a defect in stationary phase survival. The X47 hup:cat mutant was more sensitive to the DNA damaging agent mitomycin C, and displayed a decreased frequency of DNA recombination, indicating Hup plays a significant role in facilitating DNA recombinational repair. The X47 hup:cat mutant was also sensitive to both oxidative and acid stress, conditions that H. pylori commonly encounters in the host. The hup mutant cells survived significantly (7-fold) less upon exposure to macrophages than the wild type strain. In a mouse infection model, the hup mutant strain displayed a greatly reduced ability to colonize host stomachs. The geometric means of colonization number for the wild type and hup mutant were 6 × 105 and 1.5 × 104 CFU/g stomachs, respectively. Complementation of the hup strain by chromosomal insertion of a functional hup gene restored oxidative stress resistance, DNA transformation frequency, and mouse colonization ability to the wild type level. We directly demonstrated that the purified His-tagged H. pylori Hup protein can protect (in vitro) an H. pylori-derived DNA fragment from oxidative damage.
Keywords: Helicobacter pylori, Histone-like protein, Oxidative stress, DNA protection, recombinational repair, Macrophage killing, Mouse colonization
1. Introduction
Helicobacter pylori infects the stomachs of approximately 50% of humans and results in a series of human gastric diseases including gastritis, peptic ulcers, and gastric cancer [1-4]. During the process of colonizing the host, H. pylori induces a strong inflammatory response mediated by a variety of host immune cells, leading to generation of a number of reactive oxygen species (ROS) [5-8]. H. pylori has excellent capacity and redundant mechanisms to directly detoxify ROS and to repair damaged macromolecules so that it can survive and colonize persistently in the harsh conditions within the gastric mucosa [9]. The most severe human gastric diseases are due to the persistent nature of the bacterium.
Studies in recent years have indicated that DNA recombinational repair plays a significant role in H. pylori's persistent colonization of the host [10-14]. In an attempt to identify additional components of the recombinational repair system in H. pylori, we screened for mitomycin C-sensitive clones from a random transposon mutagenesis library. In one of the mitomycin C-sensitive strains the transposon was shown to be inserted at the hup locus, suggesting that the hup gene is involved in DNA protection, recombination and/or repair processes. The hup gene encodes a protein that is homologous to the histone-like protein HU of Escherichia coli.
Genomic DNA in a bacterial cell is folded into a compact structure called a nucleoid, and the proper assembly of active higher-order genome structures requires accessory proteins, termed nucleoid-associated proteins (NAPs). In E. coli, several nucleoid-associated proteins, such as HU, IHF, H-NS, Fis, and Lrp, have been discovered and shown to play roles in DNA organization and protection [15, 16]. These proteins are sometimes referred to as histone-like because they have roles in nucleoid compaction comparable to eukaryotic histones. These proteins are not only involved in DNA supercoiling and compaction, but also modulate DNA functions such as replication, recombination, repair, and transcription [17]. Dps (DNA protection during starvation) is sometimes also referred to as a nucleoid-associated protein. Each bacterial species harbors a specific set of NAPs, with only HU-like proteins being ubiquitous among bacteria. The functions and DNA binding properties of HU-like proteins may vary depending on the specific NAP content and the host conditions [18].
Among the nucleoid-associated proteins, H. pylori harbors a HU-like protein Hup and a Dps homolog. The basic properties and substrate DNA binding specificity of H. pylori Hup protein were studied previously [19]. In this study, we first examined whether Hup is involved in DNA recombinational repair. Then we investigated the physiological roles of H. pylori Hup in DNA protection, particularly focusing on the role in protecting DNA from oxidative damage. Furthermore, the contribution of Hup for bacterial survival within the host stomach was examined herein with a mouse infection model.
2. Materials and Methods
2.1. H. pylori culture conditions
H. pylori was cultured on Brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 5% fetal bovine serum (called BA plates). Chloramphenicol (50 μg/ml) or kanamycin (40 μg/ml) was added to the medium for culturing mutants. Cultures of H. pylori were grown microaerobically at 37°C in an incubator under continuously controlled levels of oxygen (4% partial pressure O2, 5% CO2, and the balance was N2).
2.2. Construction of H. pylori hup mutant
A 1.04 kb fragment containing the H. pylori hup gene and the flanking regions was amplified by polymerase chain reaction (PCR) from genomic DNA of strain 26695 using the primer pair hupF1 (5’-AAAGTGTATTACGCCACGC-3’) and hupR1 (5’-AAAGGGATTTTAATGGCTTG-3’). The PCR product was cloned into pGEM-T vector to generate pGEM-hup. Subsequently, a chloramphenicol acetyl transferase (cat) cassette was inserted at the unique BbsI site within the hup sequence of pGEM-hup. The disrupted hup gene was then introduced into H. pylori wild type strains by natural transformation via allelic exchange and chloramphenicol-resistant colonies were isolated (hup::cat). The disruption of the gene in the genome of the mutant strain was confirmed by PCR showing an increase in the expected size of the PCR product and by direct sequencing of the PCR fragment.
2.3. Construction of H. pylori hup complementation strain
The complemented hup+ strain was constructed by inserting a wild-type copy of the hup gene in the rdxA locus of the hup::cat chromosome. Disruption of rdxA results in metronidazole resistance that will be used for selection in DNA transformation. PCR products corresponding to the 3’ end of the rdxA gene (266 bp), 523 bp of the full-length hup gene and the upstream sequence containing its promoter, and the 5’ end of the rdxA gene (256 bp) were amplified in three separate PCRs and then stitched together in subsequent PCR. The final PCR product (1019 bp) was used to transform the hup::cat strain by selecting for metronidazole (16 μg/ml) resistant colonies. Through homologous DNA recombination, an intact hup gene was inserted at the rdxA locus of the hup::cat strain.
2.4. Assessment for susceptibility to mitomycin C
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in phosphate buffered saline (PBS) at a concentration of ~108 cells/ml. The cell suspensions were treated with 0, 50, 100, or 200 ng/ml mitomycin C for 20 min. The samples were serially diluted and spread on BA plates. After 4 days incubation in a microaerobic atmosphere (4% partial pressure O2) at 37°C, colonies are enumerated.
2.5. DNA transformation assay to assess inter-genomic recombination frequency
The donor DNA used in this study included: (i) a 330 bp PCR fragment of H. pylori rpoB gene fragment containing a site-specific mutation (at the center of the fragment) conferring rifampicin resistance, (ii) a linear DNA fragment containing a kanamycin resistance cassette (1.4 kb) flanked by H. pylori acnB gene sequences (about 550 bp on each side of the Kan cassette).
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in PBS at a concentration of ~ 108/ml (recipient cells for transformation). A 30 μl cell suspension sample was mixed with 100 ng of donor DNA and spotted onto a BA plate. After incubation for 18 hours under microaerobic condition at 37°C, the transformation mixture was harvested and suspended in 1 ml PBS. 100 μl portions of the suspension (or appropriate dilution as needed) were plated onto either BA plates or BA plates containing selective antibiotic (20 μg/ml rifampicin or 40 μg/ml kanamycin, depending on the donor DNA used). The plates were incubated for 4 days under the microaerobic condition at 37°C, and the numbers of colonies were counted. The transformation frequency was determined by the number of resistant colonies divided by the total number of CFU. In a normalized DNA transformation assay, the frequency of transformation is expressed as the number of transformants per 108 recipient cells. As negative controls, H. pylori strains with no DNA added were tested under this assay condition; no antibiotic-resistant colonies were observed.
2.6. Oxygen sensitivity (air survival) assay
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in PBS at a concentration of ~108 cells/ml. The cell suspensions were incubated at 37°C under normal atmospheric conditions (21% O2) with moderate shaking. Samples were then removed at various time points (2, 4, 6, 8, and 10 hours), serially diluted, and spread onto BA plates. Colony counts are recorded after 4 days of incubation in a microaerobic atmosphere (4% partial pressure O2) at 37°C.
2.7. Assessment for sensitivity to low pH condition
H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in the buffer (20 mM Tris-HCl, 150 mM NaCl) with different pH levels (pH 7.0, pH 5.0, or pH 3.0) at a concentration of ~108 cells/ml. The cell suspensions were incubated under a microaerobic condition (4% O2) at 37°C for 1 hour. The samples were serially diluted and plated for CFU counts (after 4 days incubation under microaerobic growth condition). The percentage of cell survival in pH 5.0 or pH 3.0 relative to that in pH 7.0 was calculated.
2.8. Macrophage killing assay
The survival of H. pylori cells within macrophages was investigated following the methods published [14, 20, 21] with minor modifications. Briefly, the macrophage RAW264.7 cells were seeded in 24-well plates in the culture medium (0.5 ml) and incubated at 37°C, 5% CO2 for 4 days (cell density is about 105 cells per well). The medium was replaced by fresh medium to remove the non-adherent cells. H. pylori cells were added at a ratio of 20 CFU bacteria per macrophage. Phagocytosis was synchronized by centrifugation at 600 × g for 5 min and then allowed to proceed for 1 h. Extracellular bacteria were removed by washing and incubation in the medium supplemented with gentamicin (100 mg/ml) for 1 h at 37°C, 5% CO2. After three washes to remove the antibiotics the cells were further incubated in fresh medium for 2 h. After removing the medium, the macrophage cells were lysed with ice-cold PBS with 0.1% saponin for 5 min. Appropriate dilutions of the supernatant were plated on BA plates and incubated at 37°C, 5% CO2, 2% O2 for 4 days to count the number of surviving bacteria. The number of surviving bacteria (CFU/ml) is compared with the number of viable bacteria initially added.
2.9. Mouse colonization
Mouse colonization assays were performed essentially as described previously [10, 12]. Briefly, the wild type X47 or isogenic hup mutant cells were harvested after 48 h of growth on BA plates (37°C, 4% oxygen) and suspended in PBS to an OD600 of 1.7. Headspace in the tube was sparged with argon gas to minimize oxygen exposure, and the tube was tightly sealed. The bacterial suspensions were administered to C57BL/6NCr mice (3 × 108 H. pylori cells / mouse) twice, with each of the oral deliveries made 2 days apart. Three weeks after the first inoculation, the mice were sacrificed and the stomachs were removed, weighed, and homogenized in argon-sparged PBS [22] to avoid O2 exposure. Stomach homogenate dilutions (dilutions conducted in sealed tubes containing argon-sparged buffer) were plated on BA plates supplemented with bacitracin (100 μg/ml), vancomycin (10 μg/ml) and amphotericin B (10 μg/ml). The plates were rapidly transported into an incubator containing sustained 4% partial pressure O2. After incubation for 5 to 7 days H. pylori colonies were counted and the data expressed as CFU per gram of stomach.
2.10. Overexpression and purification of H. pylori Hup protein
A DNA fragment containing H. pylori hup gene was amplified by PCR and cloned into pET-21a vector to generate pET-Hup-6His. E. coli BL21 Origami cells harboring pET-Hup-6His were grown at 37°C to an OD at 600 nm of 0.5 in 500 ml of LB medium with ampicillin (100 μg/ml) and kanamycin (40 μg/ml). Expression of the Hup protein was induced by adding 0.5 mM IPTG to the medium followed by further incubation for 3 h; cells were then harvested by centrifugation (5,000×g, 15min, 4°C). All subsequent steps were performed at 4°C. Cells were washed with 200ml of 20mM Na2HPO4 (pH7.4), 500mM NaCl, 5mM imidazole (buffer A) and re-suspended in 5 ml of the same buffer. Cells were lysed by two passages through a cold French pressure cell at 18,000 lb/in2. Cell debris was removed by centrifugation at 20,000 × g. The supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen), and buffer A was used to wash the resin until the A280 reached the baseline. Proteins were washed with buffer B (buffer A with 30mM imidazole) until the A280 reached the baseline, and finally eluted with buffer C (buffer A with 250mM imidazole). Extracts of E. coli BL21 Origami containing the vector only did not result in retrievable proteins from this purification (Ni-affinity) procedure. Fractions were analyzed by gel electrophoresis, and the Hup-positive fractions were pooled. Protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.).
2.11. DNA protection assay
A linear DNA fragment (800 bp long) was PCR amplified from H. pylori genomic DNA. 0.2 μM DNA was incubated with 2 μM purified H. pylori Hup protein, or BSA as a control. Then the samples were treated with 20 mM FeSO4 plus 5 mM H2O2 or control buffer for 5 minutes. The samples were analyzed on an ethidium bromide stained 1% agarose gel.
3. Results and discussion
3.1. Construction of an H. pylori hup mutant and its effect on cell growth
Using a random transposon mutagenesis library of H. pylori [23], we identified a dozen genes that confer resistance to mitomycin C (data not shown). In one of the MMC-sensitive strains, the transposon was found to be inserted within the HP0835 locus. In the published H. pylori genome sequence [24], HP0835 was annotated as a hup gene homolog. The gene product of H. pylori hup displays 34% and 36% amino acid identity to E. coli HupA and HupB, respectively. In some bacteria including E. coli, HU is a heterodimer composed of two highly homologous subunits (HupA and HupB), whereas in many other bacteria including H. pylori HU is present as a homodimer [19, 25].
To study the physiological role of H. pylori Hup, we constructed a hup mutant by inserting a chloramphenicol resistance cassette (CAT) at the restriction site BbsI within the hup gene. The hup mutant strains can be easily obtained if transformants are screened at a low oxygen (2% partial pressure) condition. The hup mutants were constructed in several different strains such as 26695, 43504, and X47. As X47 is a well-adapted strain for mouse colonization assay, most of the in vitro and all the in vivo assays reported herein were done with X47 and its isogenic hup mutant.
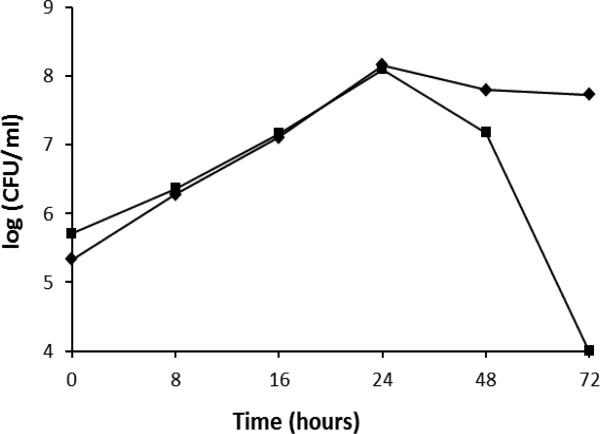
Under the normal in vitro growth condition (4% O2, 5% CO2), the H. pylori hup mutant grew similarly to the wild type strain. During the exponential growth phase (before reaching 24 hours), a similar growth rate was observed for the WT and mutant strain (Fig. 1). Upon entering stationary phase, however, the survival of the hup mutant decreased much faster than the wild type strain. As shown in Fig.1, the hup mutant lost viability completely after 3 days, while the majority of the wild type cells were still viable. Thus, H. pylori Hup protein appears to play a significant role in protecting its genomic DNA during stationary phase when toxic metabolites are accumulated. A similar role of a histone-like protein Hlp in Mycobacterium smegmatis was reported; Hlp is essential for cell viability while cells are at a dormant state (long-term stationary phase) [26]. A stationary phase survival defect was also observed for E. coli HU-like IHF mutant strain [27]. Upon entering stationary phase, a Salmonella enterica mutant lacking IHF displayed down-regulated expression of classic stationary phase genes; and this down-regulation was not caused by a reduction in the level of the RpoS sigma factor [28]. H. pylori Hup protein may play a similar role in up-regulating some genes responsible for stationary phase survival.
Fig. 1. Growth curve of H. pylori cells.
H. pylori X47 wild type (diamond) and its isogenic hup mutant (square) cells were grown microaerobically (4% O2, 5% CO2, and balanced with N2) at 37°C. At the indicated time points, the number of live cells was determined by serial dilution and plate counting for colony forming units (CFU). The data are from a representative experiment.
3.2. H. pylori Hup is involved in DNA recombinational repair
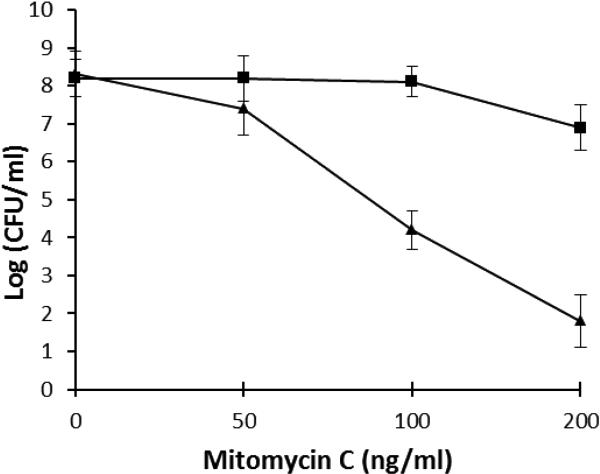
Random transposon insertion in the hup gene resulted in sensitivity to mitomycin C. In this study, we further examined the sensitivity of the X47 hup::cat mutant to MMC, compared to the wild type. The result shown in Fig.2 confirmed that the hup mutant is much more sensitive to MMC than the wild type strain. For example, after treatment with 200 ng/ml MMC for 20 min, the viability of the wild type cells is 5 orders of magnitude greater than that of the hup mutant.
Fig. 2. Sensitivity of H. pylori X47 wild type (square) and its isogenic hup mutant (triangle) to mitomycin C.
H. pylori cell suspensions in PBS were treated with different concentrations (as indicated in the × axis) of mitomycin C for 20 min. The surviving CFU were determined after 4 days incubation under a microaerobic condition. The data are the means of three experiments with standard deviation as indicated.
Mitomycin C causes predominantly DNA intra-strand cross-links, leading ultimately to DNA double strand breaks. Repair of DNA double strand breaks usually requires homologous DNA recombination. Histone-like protein HU from other bacteria was shown to bind preferentially to DNA recombination intermediates [29, 30]. Based on the DNA binding specificity, Ghosh and Grove [30] proposed an in vivo role of the deinococcal HU in stabilizing homologous recombination intermediates rather than to function as an architectural element. Similarly, the most preferable binding substrate of H. pylori Hup protein was shown to be four-way junction DNA, the recombination intermediate [19]. Thus, H. pylori Hup protein is likely involved in DNA recombinational repair process. To test this hypothesis, we examined whether loss of Hup function had an adverse effect on DNA recombination by determining the frequency of DNA transformation.
As described previously [10-12], we used two different types of DNA for examining DNA transformation of H. pylori. A specific A-to-G mutation in the H. pylori rpoB gene (rpoB3 allele) confers rifampicin resistance [31]. A 330-bp PCR fragment containing this specific mutation at the center of the fragment was used to transform H. pylori strains by using rifampicin resistance as a selective marker. Another type of DNA used for transformation was the sequence of H. pylori acnB gene (a housekeeping gene, 1.1 kb) in which a kanamycin resistance cassette (Kan, 1.4 kb) was inserted at the center (acnB:Kan).
The results for transformation are shown in Table 1. Using rpoB3 as donor DNA, the X47 hup::cat mutant had a transformation frequency of 2.7 × 10−5, which was 15-fold lower than that for the wild type strain X47 (4.16 × 10−4). Using acnB:Kan as donor DNA, wild type H. pylori X47 had a transformation frequency of 1.09 × 10−5. In contrast, the transformation frequency for the X47 hup::cat mutant was 1.1 × 10−6, which is 10-fold lower than that of the WT strain. For both types of DNA donor (rpoB3 and acnB:Kan), the mutant strain data (transformation frequency) are significantly lower than those of the WT strain, according to Student t-test (P<0.01). These results provided evidence that Hup plays a significant role in the DNA recombination process of H. pylori. Further investigations are required to understand how Hup protein interacts with other components of the recombinational repair machinery.
Table 1.
Transformation frequency with different types of donor DNA
| Donor DNA |
||
|---|---|---|
| H. pylori strains | rpoB3 (330 bp) | acnB:Kan (2.5 kb) |
| X47 WT | 41630 ± 6190 | 1090 ± 166 |
| X47 hup::cat | 2710 ± 693 | 110 ± 13 |
| X47 hup::cat-hup+ | 48750 ± 8738 | ND |
The transformation frequencies are presented as the number of transformants (resistant colonies) per 108 recipient cells. Data are means ± standard errors from three independent determinations. ND, not determined.
3.3. H. pylori hup mutant is highly sensitive to oxidative and acid stress conditions
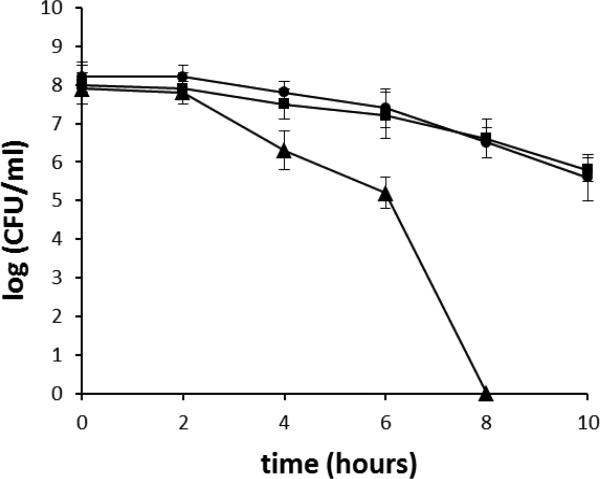
Oxidative stress is a major stress condition that H. pylori encounters in its physiological niche. H. pylori induces strong host inflammatory responses that involve recruitment of neutrophils, lymphocytes and macrophages; these immune cells release reactive oxygen species that damage DNA. H. pylori DNA was shown to be a target for host-generated oxidative stress based on studies of H. pylori nth strains that are unable to repair oxidized pyrimidines [32]. Further studies showed that mutant cells of ruvC [14], mutS [33], mutY [34], recN [11], addA (recB) [12, 13] or recRO [10] were more sensitive to oxidative stress, indicating important roles of DNA recombination and repair in H. pylori for the bacterial survival of oxidative damage. To examine the role of the Hup protein in protecting DNA from oxidative damage, we examined the sensitivity of the hup mutant to oxidative stress by an air survival assay. The cell suspensions (~ 5×108 cells/ml) were exposed to air, and the numbers of surviving cells were determined at various time points (Fig.3). The number of wild type cells decreased slowly; and at the 10 h time point, about 5×106 cells (~1% of that at the time zero) survived. In contrast, the hup mutant showed a greater sensitivity to air exposure. Two hours after exposing cells to air, the number of surviving mutant cells started to decrease at a rate much faster than that of the wild type cells. At the 8 h time point, the hup mutant cells were eliminated (i.e. no viable cells recovered). Previous studies demonstrated that oxidative stress causes damage to H. pylori genomic DNA [35, 36]. Thus, H. pylori Hup appears to play a significant role in protecting its DNA from oxidative stress-induced damage.
Fig. 3. Survival of H. pylori cells upon exposure to air.
H. pylori cell suspensions in PBS were incubated at 37°C under normal atmospheric conditions (21% partial pressure O2). Samples were removed at the times indicated in the x-axis and were used for plate count determinations upon incubation in a 5% oxygen environment. The data are the means of three experiments with standard deviation as indicated. Symbols: square, wild type; triangle, hup::cat; circle, hup complementation strain. Based on statistical analysis (Student t-test), the cell survival differences between the WT and the mutant strains are significant (P<0.01) for all the data points except for the 2 h time point.
H. pylori survives and colonizes an acidic niche on the gastric surface [37]. Therefore, low pH is another stress condition that H. pylori encounters in its physiological niche. Despite the existence of sophisticated pH homeostasis systems and acid tolerance mechanisms, bacteria may still suffer DNA damage from acid stress. Indeed, E. coli O157:H7 chromosomal DNA was shown to be significantly damaged by acid stress, and the Dps protein plays an important role in protecting DNA from acid-induced damage [38]. Previously, we showed that H. pylori DNA recombination proteins RecN and RecRO are involved in repair of acid-induced DNA damage [10, 11]. In this study, we characterized the hup mutant for its sensitivity to low pH conditions.
The wild type H. pylori or the hup mutant cells were treated for 1 h by suspending cells in the buffer at different pH (pH 7.0, pH 5.0, or pH 3.0), and the survival was subsequently determined. As shown in Table 2, treatment at pH 5.0 for 1 h did not have a significant effect on survival of the wild type cells, while the same treatment killed approximately 90% of the hup mutant cells. About 40% of the wild type H. pylori cells survived the pH 3.0 condition for 1 h, whereas the hup mutant cells were almost completely killed (>98% lethality) by the same treatment. Thus, the H. pylori hup mutant is more sensitive to acid stress compared to the wild type.
Table 2.
Acid sensitivity of H. pylori strains.
| % survival at a: |
|||
|---|---|---|---|
| Strains | pH 7.0 | pH 5.0 | pH 3.0 |
| X47 WT | 100 | 94 ± 7 | 39 ± 6 |
| X47 hup::cat | 100 | 8.0 ± 3.7 | 1.6 ± 1.2 |
| X47 hup::cat-hup+ | 100 | 95 ± 8 | 41 ± 4 |
The values for pH less than 7.0 are the percentages of cell survival after treatment for 1 h at pH 5.0 or pH 3.0 relative to the survival at pH 7.0. The data are the means ± standard errors from three independent determinations.
Sensitivity of the hup mutant to oxidative stress and acid stress could be interpreted as the Hup protein has an ability to physically protect DNA from stress damage (See section 3.7 below). Alternatively, Hup may regulate expression of other genes involved in stress resistance. In E. coli, HU influences the expression of genes involved in anaerobic respiration, the response to osmotic stress, the acid stress, and the response to DNA damage [39-41]. By modeling DNA topology, E. coli HU is known to play a role in modulation of transcription profiles which has important impacts on cellular physiology [39, 42]. S. enterica HU controls three regulons that coordinate virulence, response to stress and general physiology [43]. Thus, determination of the Hup regulon in H. pylori is of interest for future studies.
3.4. H. pylori hup mutant is more sensitive to macrophage killing
H. pylori infection induces a strong inflammatory response by the host, with the recruitment of lymphocytes, macrophages, and polymorphonuclear cells; however the bacterium is oftentimes able to resist this immune response and establish a persistent gastric infection. In particular, although H. pylori can be efficiently ingested by the different types of phagocytic cells, it is able to survive for prolonged periods within these cells and presumably is able to resist damage by free radicals derived from the phagocytic respiratory burst [8, 21]. The mechanisms known to contribute to survival within macrophages include enzymatic detoxification of reactive oxygen species and DNA repair. For example, absence of the H2O2-detoxification enzyme catalase [20] or DNA recombinational repair enzyme RuvC [14] in H. pylori resulted in reduction of survival ability within macrophages. Given the sensitivity of an H. pylori hup mutant to DNA damaging agents and oxidative stress, we investigated whether the Hup protein contribute to survival of H. pylori within macrophages.
A macrophage killing assay was performed using a murine macrophage cell line RAW264.7 (See Materials and Methods). Similar numbers (5 × 108 CFU/ml) of H. pylori WT X47 or X47 hup:cat mutant cells were inoculated to the macrophage (Fig. 4). After extracellular killing by gentamycin and 2 hours incubation within macrophages, the numbers of surviving cells of the WT and of the hup mutant strain were compared. As shown in Fig.4, a mean number of 3.0 × 106 CFU/ml wild type cells survived. In contrast, the same treatment resulted in recovery of a mean number of 4.4 × 105 CFU/ml of the hup mutant cells (7 fold less than the WT). Based on statistical analysis (Student t-test), the cell survival differences between the WT and the hup mutant strains are significant (P<0.01). These results indicate a role for Hup in survival of H. pylori within macrophages.
Fig. 4. Survival of H. pylori cells in macrophage RAW264.7 cells determined with the gentamycin killing assay.
Similar numbers of the WT X47 (gray bars) or X47 hup:cat mutant cells (black bars) were inoculated to the macrophages. After extracellular killing by gentamycin and 2 hours incubation within macrophages, the numbers of surviving cells of the WT and the mutant strain were determined. Data are means from three independent determinations with standard deviations.
The roles of histone-like proteins in macrophage survival have been recently observed in other bacteria. The histone-like protein Lsr2 knockout strain of M. smegmatis survived significantly less well than the wild type strain [44]. The hupAhupB double mutant of S. enterica was shown to be defective in invasion of epithelial cells and in its ability to survive in macrophages [43]. The observed effects on macrophage survival were ascribed either to direct protection of DNA from oxidative damage by histone-like protein [44] or to HU-mediated global gene regulation in response to stress [43]. Interestingly, expression of a mutant version of HU protein can convert the commensal E. coli K-12 to an invasive form with characteristics of host cell invasion, phagosomal disruption, and intracellular replication, suggesting the role of HU in modulating bacterial survival within host cells [45].
3.5. H. pylori hup mutant displays an attenuated ability to colonize mouse stomachs
As the H. pylori hup mutant showed sensitivity to conditions that H. pylori encounters in its physiological niche, namely (oxidative and acid) stress and to macrophage killing, we sought to determine the effect of the Hup on H. pylori colonization in the host. We performed an assay using a mouse infection model as described previously [11, 12, 33]. The wild type X47 or the isogenic hup mutant strain were inoculated into C57BL/6J mice, and the colonization of H. pylori cells in the mouse stomachs was examined 3 weeks after inoculation (Fig.5). H. pylori was recovered from all 12 mice that had been inoculated with the wild type strain, with a geometric mean number of 6.0 ×105 CFU/g stomach. In contrast, 9 of 12 mice that were inoculated with the hup strain were found to harbor H. pylori. The geometric mean of the colonization number for the hup strain in the 12 mice was 1.5 ×104 CFU/g stomach. According to Wilcoxin rank test analysis, the range of colonization values of the hup strain is significantly lower than that of the wild type at the 99% confidence level (P<0.01). These results indicate that H. pylori Hup plays a significant role in bacterial survival/colonization in the host.
Fig. 5. Mouse colonization results of H. pylori strains.
The mice were inoculated with H. pylori two times (two days apart) with a dose of 1.5 × 108 viable cells administered per animal each time. Colonization of H. pylori in mouse stomachs was examined 3 weeks after the first inoculation. Data are presented as a scatter plot (log scale) of colony forming units per gram of stomach as determined by plate counts. Each point represents the CFU count from one mouse stomach, and the solid horizontal lines represent the geometric means of the colonization numbers for each group. The detection limit of the assay is 500 CFU/g stomach, corresponding to log10 (CFU/g) =2.7.
3.6. Complementation of the hup strain
It is unlikely that disruption of the hup gene has a polar effect on the downstream genes, as the hup gene alone forms a transcriptional unit within the H. pylori genome [24, 46]. To ensure that the phenotypes (both in vitro and in vivo) observed for the hup strain were completely attributed to inactivation of hup, we introduced a functional copy of the hup gene back into the hup strain. The strain X47 hup::cat-hup+ contains a mutated hup gene at the original locus and an intact hup gene at the rdxA locus (See Materials and Methods). The growth characteristics of the complemented strain were similar to the wild type (data not shown).
The sensitivity of the complemented strain to stress conditions was shown to be similar to that of the wild type. For example, both the wild type and the complemented strain can survive exposure to air for 10 h, while the hup mutant strain was completely killed by this treatment (Fig. 3). The complementation of hup function restored the acid sensitivity to the wild type level (Table 2). The complementation of hup function also restored the transformation frequency to the wild type level, as determined by using rpoB3 fragment as DNA donor (Table 1).
The complemented strain was also examined for the mouse colonization ability. H. pylori bacteria were recovered from all 6 mice that had been inoculated with the hup complementation strain, with a geometric mean number of 5.2 ×105 CFU/g stomach (Fig.5). According to Wilcoxin rank test analysis, the range of colonization values of the hup complementation strain is not significantly different from that of the wild type, but significantly (P<0.01) higher than that of the hup mutant strain. This indicates that the complementation of the hup strain restored its ability to colonize mouse stomachs.
3.7. H. pylori Hup protein protects DNA against hydroxyl radical damage
All the phenotypes above observed for the H. pylori hup mutant strain, such as stationary phase survival defect, sensitivity to oxidative stress, and lower survival within macrophages, pointed to a role for the Hup in protecting bacterial DNA from oxidative damage. To further support this notion, we sought to demonstrate its biochemical activity using the purified H. pylori Hup protein. H. pylori Hup is a small protein with a molecular weight of 10 kDa. Chen et al. [19] showed that H. pylori Hup protein exists as a dimer in solution and exhibits greater thermal stability compared with other bacterial HU homologues. Substrate specificity for H. pylori Hup protein binding was mainly investigated in that study [19]. Here, we focus on the function of the Hup in protecting bacterial DNA from oxidative damages.
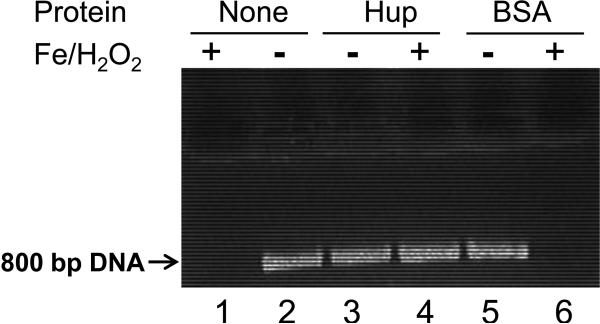
A His-tagged H. pylori Hup protein was overexpressed in E. coli and purified using Ni-NTA affinity column (not shown). The ability of the Hup protein to protect DNA against damage by hydroxyl radicals was studied. A linear 800-bp DNA fragment was PCR amplified by using H. pylori genomic DNA as template. The DNA was incubated in vitro with H2O2 and Fe(II) in the presence or absence of the purified H. pylori Hup protein or a BSA control protein. As shown in Fig.6, the DNA was completely destroyed by treatment with 20 mM FeSO4 plus 5 mM H2O2 for 5 min (lane 1 vs. lane 2). Notably, DNA was substantially protected from degradation by pretreatment with the Hup protein (lane 4). As a control, pretreatment with an equivalent concentration of BSA did not result in any protection (lane 6).
Fig. 6. DNA protection activity of H. pylori Hup protein.
Linear DNA (~0.2 μM, 800 bp PCR fragment of H. pylori genomic DNA) was used alone (lane 1, 2) or after incubation with either 2 μM of purified Hup (lane 3, 4) or BSA (lane 5, 6). Both proteins were suspended in identical imidazole buffers. Samples were treated for 5 min with either 20 mM FeSO4 plus 5 mM H2O2 (lane 1, 4, 6) or left untreated (lane 2, 3, 5). The samples were analyzed on an ethidium bromide stained 1% agarose gel
Eukaryotic histones are known to protect DNA against hydroxyl radical-induced DNA strand breaks by binding DNA and organizing it into higher order chromatin structures [47-49]. A prokaryotic histone-like protein, Mycobacterium tuberculosis Lsr2 was also shown to be able to protect DNA against reactive oxygen intermediates (ROI) [44]. Electron microscopy and DNA binding studies suggested that Lsr2 shields DNA from ROI by binding bacterial DNA and physically protecting it [44]. Here we showed that H. pylori Hup protein has the capability to physically protect DNA against hydroxyl radical damage, in addition to its potential role in DNA recombinational repair and gene regulation. This is in agreement with the oxygen sensitivity of the hup mutant observed in vitro. This also helps explain the decreased survival of the hup mutant within macrophages and its attenuated ability in colonizing host stomachs.
Another group of bacterial proteins that can protect DNA from oxidative damage is Dps family proteins [50, 51]. Dps homologues are expressed in many bacterial species and accumulate to high levels under conditions of oxidative stress or nutritional stress. Dps appears to protect DNA through the dual mechanisms of iron sequestration (preventing Fenton mediated ROI generation) and in DNA binding (creating a protective physical barrier) [50, 52]. Histone-like proteins share a number of physical properties with Dps, including small size, high isoelectric point, and an ability to bind DNA with little specificity. In cintrast to Dps, histone-like proteins do not bind and sequester iron. H. pylori has a Dps homolog named NapA (neutrophil activating protein). H. pylori NapA has been well documented to play roles in human neutrophil recruitment and in stimulating host cell production of ROI [53-55]. On the other hand, as a nucleoid-associated protein (NAP), H. pylori NapA plays a separate role in protecting its DNA from oxidative stress damage [56, 57]. In a bacterium harboring multiple NAPs, these NAPs usually have overlapping and complementary functions. For example, deletion of HU from the E. coli genome is not lethal unless IHF and H-NS are deleted as well [58]. In contrast, the disruption of HU is lethal for organisms in which it is the only NAP available [59-61]. For H. pylori, we can easily obtain hup single mutant strains which show a normal growth at the exponential growth phase in vitro, but we failed in attempts to knock out both hup and napA at the same time (i.e. to create a double mutant, our preliminary result), suggesting partially complementary functions of the two proteins.
There is keen interest in host DNA damage and the etiology of carcinogenesis due to chronic inflammation caused by persistent pathogens [62]. Indeed, increased DNA damage of human host cells was shown to be related to H. pylori infection saturating host cell repair capacity [63]. At the same time, H. pylori seems capable of protecting its own DNA from damage. We propose that Hup and NapA play a critical role in this protection. Interestingly, a recent proteomic analysis identified Hup and NapA as two of only three identified H. pylori proteins that showed a greater expression level among strains associated with gastric cancer, suggesting their involvement in gastric carcinogenesis [64].
In conclusion, the results from this study provided evidence that the histone-like protein Hup in H. pylori facilitates DNA recombinational repair, and has the capability to physically protect DNA against hydroxyl radical damage. It plays an important role in protecting H. pylori DNA from damage elicited by oxidative stress and acid stress, conditions that the bacterium commonly encounters in the host. Furthermore, this protein contributes to H. pylori survival in the host stomach, which could be the combined effect due to its roles in DNA protection (physically), recombinational repair, and modulation of gene expression in response to stress. Further investigations are required to determine the roles of Hup in global gene regulation, particularly in possible regulation of virulence genes.
Highlights.
A nucleoid associated histone-like protein is identified in the gastric pathogen Helicobacter pylori
It protects DNA from acid and oxidative stress
It permits the pathogen to be more virulent in a mouse model.
Acknowledgements
This work was supported by NIH grants R21AI076569 and R21AI078096. We thank Sue Maier for her expertise and assistance on mouse colonization assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720–41. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 4.Uemura N, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 5.Bagchi D, Bhattacharya G, Stohs SJ. Production of reactive oxygen species by gastric cells in association with Helicobacter pylori. Free Radic Res. 1996;24(6):439–50. doi: 10.3109/10715769609088043. [DOI] [PubMed] [Google Scholar]
- 6.Baik SC, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996;56(6):1279–82. [PubMed] [Google Scholar]
- 7.Farinati F, et al. Oxidative DNA damage accumulation in gastric carcinogenesis. Gut. 1998;42(3):351–6. doi: 10.1136/gut.42.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramarao N, Gray-Owen SD, Meyer TF. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity. Mol Microbiol. 2000;38(1):103–13. doi: 10.1046/j.1365-2958.2000.02114.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61(4):847–60. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Lo LF, Maier RJ. The RecRO pathway of DNA recombinational repair in Helicobacter pylori and its role in bacterial survival in the host. DNA repair. 2011;10(4):373–9. doi: 10.1016/j.dnarep.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun. 2008;76(1):153–60. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Maier RJ. A RecB-like helicase in Helicobacter pylori is important for DNA repair and host colonization. Infect Immun. 2009;77(1):286–91. doi: 10.1128/IAI.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amundsen SK, et al. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69(4):994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loughlin MF, et al. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect Immun. 2003;71(4):2022–31. doi: 10.1128/IAI.71.4.2022-2031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol. 2005;56(4):858–70. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 16.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. The Journal of biological chemistry. 1999;274(46):33105–13. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 17.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nature reviews. Microbiology. 2010;8(3):185–95. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 18.Grove A. Functional Evolution of Bacterial Histone-Like HU Proteins. Current issues in molecular biology. 2010;13(1):1–12. [PubMed] [Google Scholar]
- 19.Chen C, Ghosh S, Grove A. Substrate specificity of Helicobacter pylori histone-like HU protein is determined by insufficient stabilization of DNA flexure points. The Biochemical journal. 2004;383(Pt 2):343–51. doi: 10.1042/BJ20040938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu M, Czinn SJ, Blanchard TG. Absence of catalase reduces long-term survival of Helicobacter pylori in macrophage phagosomes. Helicobacter. 2004;9(3):211–6. doi: 10.1111/j.1083-4389.2004.00226.x. [DOI] [PubMed] [Google Scholar]
- 21.Odenbreit S, et al. Interaction of Helicobacter pylori with professional phagocytes: role of the cag pathogenicity island and translocation, phosphorylation and processing of CagA. Cell Microbiol. 2001;3(1):21–31. doi: 10.1046/j.1462-5822.2001.00088.x. [DOI] [PubMed] [Google Scholar]
- 22.Seyler RW, Jr., Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69(6):4034–40. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186(23):7926–35. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 25.Oberto J, Rouviere-Yaniv J. Serratia marcescens contains a heterodimeric HU protein like Escherichia coli and Salmonella typhimurium. Journal of bacteriology. 1996;178(1):293–7. doi: 10.1128/jb.178.1.293-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anuchin AM, et al. The role of histone-like protein, Hlp, in Mycobacterium smegmatis dormancy. FEMS microbiology letters. 2010;308(2):101–7. doi: 10.1111/j.1574-6968.2010.01988.x. [DOI] [PubMed] [Google Scholar]
- 27.Nystrom T. Glucose starvation stimulon of Escherichia coli: role of integration host factor in starvation survival and growth phase-dependent protein synthesis. Journal of bacteriology. 1995;177(19):5707–10. doi: 10.1128/jb.177.19.5707-5710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangan MW, et al. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Molecular microbiology. 2006;59(6):1831–47. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- 29.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. The EMBO journal. 2000;19(23):6527–35. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh S, Grove A. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. Journal of molecular biology. 2004;337(3):561–71. doi: 10.1016/j.jmb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, et al. Spontaneous mutations that confer antibiotic resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2001;45(3):727–33. doi: 10.1128/AAC.45.3.727-733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Rourke EJ, et al. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc Natl Acad Sci U S A. 2003;100(5):2789–94. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, et al. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol. 2005;58(1):166–76. doi: 10.1111/j.1365-2958.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 34.Eutsey R, Wang G, Maier RJ. Role of a MutY DNA glycosylase in combating oxidative DNA damage in Helicobacter pylori. DNA Repair (Amst) 2007;6(1):19–26. doi: 10.1016/j.dnarep.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park AM, et al. Oxygen tension regulates reactive oxygen generation and mutation of Helicobacter pylori. Free Radic Biol Med. 2004;36(9):1126–33. doi: 10.1016/j.freeradbiomed.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, et al. Oxidative stress defense mechanisms to counter iron-promoted DNA damage in Helicobacter pylori. Free Radic Res. 2005;39(11):1183–91. doi: 10.1080/10715760500194018. [DOI] [PubMed] [Google Scholar]
- 37.Scott DR, et al. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci U S A. 2007;104(17):7235–40. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong KC, et al. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 2008;8:181. doi: 10.1186/1471-2180-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(45):16397–402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberto J, et al. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One. 2009;4(2):e4367. doi: 10.1371/journal.pone.0004367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi H, et al. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Current microbiology. 2009;58(5):443–8. doi: 10.1007/s00284-008-9340-4. [DOI] [PubMed] [Google Scholar]
- 42.Kar S, et al. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. The Journal of biological chemistry. 2006;281(52):40144–53. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 43.Mangan MW, et al. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology. 2011;157(Pt 4):1075–87. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- 44.Colangeli R, et al. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4414–8. doi: 10.1073/pnas.0810126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koli P, et al. Conversion of commensal Escherichia coli K-12 to an invasive form via expression of a mutant histone-like protein. mBio. 2011;2(5) doi: 10.1128/mBio.00182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 47.Kornberg RD, Lorch Y. Chromatin structure and transcription. Annual review of cell biology. 1992:563–87. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 48.Furukawa A, et al. Guanine-specific DNA damage induced by gamma-irradiated histone. The Biochemical journal. 2005;388(Pt 3):813–8. doi: 10.1042/BJ20050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enright HU, Miller WJ, Hebbel RP. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic acids research. 1992;20(13):3341–6. doi: 10.1093/nar/20.13.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao G, et al. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem. 2002;277(31):27689–96. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 51.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. Journal of bacteriology. 1997;179(16):5188–94. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta S, Chatterji D. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. The Journal of biological chemistry. 2003;278(7):5235–41. doi: 10.1074/jbc.M208825200. [DOI] [PubMed] [Google Scholar]
- 53.Brisslert M, et al. Helicobacter pylori induce neutrophil transendothelial migration: role of the bacterial HP-NAP. FEMS Microbiol Lett. 2005;249(1):95–103. doi: 10.1016/j.femsle.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Evans DJ, Jr., et al. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun. 1995;63(6):2213–20. doi: 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satin B, et al. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med. 2000;191(9):1467–76. doi: 10.1084/jem.191.9.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooksley C, et al. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J Med Microbiol. 2003;52(Pt 6):461–9. doi: 10.1099/jmm.0.05070-0. [DOI] [PubMed] [Google Scholar]
- 57.Wang G, et al. Dual Roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect Immun. 2006;74(12):6839–46. doi: 10.1128/IAI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuzawa K, et al. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992;122(1):9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 59.Micka B, Marahiel MA. The DNA-binding protein HBsu is essential for normal growth and development in Bacillus subtilis. Biochimie. 1992;74(7-8):641–50. doi: 10.1016/0300-9084(92)90136-3. [DOI] [PubMed] [Google Scholar]
- 60.Liu D, et al. The essentiality and involvement of Streptococcus intermedius histone-like DNA-binding protein in bacterial viability and normal growth. Molecular microbiology. 2008;68(5):1268–82. doi: 10.1111/j.1365-2958.2008.06232.x. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen HH, et al. The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Molecular microbiology. 2009;73(2):240–52. doi: 10.1111/j.1365-2958.2009.06766.x. [DOI] [PubMed] [Google Scholar]
- 62.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Current opinion in immunology. 2011;23(4):473–80. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toller IM, et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(36):14944–9. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khoder G, et al. Proteomic Helicobacter pylori biomarkers discriminating between duodenal ulcer and gastric cancer. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2009;877(11-12):1193–9. doi: 10.1016/j.jchromb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]