Abstract
In Escherichia coli, cytosine DNA methylation is catalyzed by the Dcm (DNA cytosine methyltransferase) protein and occurs at the second cytosine in the sequence 5′CCWGG3′. Although the presence of cytosine DNA methylation was reported over 35 years ago, the biological role of 5-methylcytosine in E. coli remains unclear. In order to gain insight into the role of cytosine DNA methylation in E. coli, we: (a) screened the 72 strains of the ECOR collection and 90 recently isolated environmental samples for the presence of the full-length dcm gene using the polymerase chain reaction; (b) examined the same strains for the presence of 5-methylcytosine at 5′CCWGG3′ sites using a restriction enzyme isoschizomer digestion assay; and (c) quantified the levels of 5-methyl-2′-deoxycytidine in selected strains using liquid chromatography tandem mass spectrometry. Dcm-mediated cytosine DNA methylation is conserved in all 162 strains examined, and the level of 5-methylcytosine ranges from 0.86% to 1.30% of the cytosines. We also demonstrate that Dcm reduces expression of ribosomal protein genes during stationary phase, and this may explain the highly conserved nature of this DNA modification pathway.
Keywords: Escherichia coli, 5-methylcytosine, dcm, LC MS/MS
Introduction
DNA bases are modified by post-replicative methylation by enzymes termed DNA methyltransferases. In prokaryotes, the most common modified DNA bases are 6-methyladenine and 5-methylcytosine (5mC). The most recognized role of modified DNA bases is in restriction-modification (R-M) systems (Ishikawa, et al., 2010). In each R-M system, there is a restriction endonuclease that cleaves foreign DNA and a site-specific DNA methyltransferase that prevents cleavage of host DNA, and in some cases controls expression of the R-M system (O’Driscoll, et al., 2005). However, some DNA methyltransferases are not found in conjunction with a cognate restriction enzyme, and are termed solitary DNA methyltransferases. In addition to DNA adenine methyltransferase (Dam), E. coli possesses another solitary DNA methyltransferase termed Dcm for DNA cytosine methyltransferase (Marinus & Lobner-Olesen, 2009).
The presence of Dcm was discovered in 1973 by Marinus and Morris (Marinus & Morris, 1973). The dcm gene of E. coli K-12 contains 1419 base pairs and the predicted protein is 472 amino acids (Bhagwat, et al., 1986, Hanck, et al., 1989). The protein contains the ten conserved motifs and a catalytic cysteine residue that is found in all cytosine-5 DNA methyltransferases (Posfai, et al., 1989). The Dcm protein methylates the internal C in the sequence 5′CCWGG3′ where W=A/T (Palmer & Marinus, 1994). 5mC is occasionally spontaneously deaminated in an existing C:G base pair and a T:G mismatch is formed. The dcm gene is in an operon with the very short patch repair (vsr) gene and is controlled by the same promoter. The vsr gene lies downstream of the dcm gene and the genes overlap by 7 codons (Sohail, et al., 1990). The Vsr protein is an endonuclease that is necessary to remove the new thymine residue (Hennecke, et al., 1991) and thus compensates for the mutagenic potential of 5mC. There is evidence that Dcm itself is required for robust very short patch repair of mismatched bacteriophage heteroplexes (Jones, et al., 1987, Lieb, 1987, Zell & Fritz, 1987), but this relationship has not been observed in all reports (Sohail, et al., 1990). Nonetheless, the sequence 5′CCWGG3′ is still a mutational hot-spot sequence, since not all mismatches are repaired (Lieb & Bhagwat, 1996).
The biological role of the dcm gene and 5′CCWGG3′ cytosine DNA methylation in E. coli remains unclear. The dcm gene is not essential as mutant, deletion, and knockout strains are viable (Marinus & Morris, 1973, Baba, et al., 2008). Interestingly, the dcm gene and cytosine DNA methylation is absent from E. coli B (Doskocil & Sormova, 1965, Fujimoto, et al., 1965), a host strain used extensively to study bacteriophages T1-T7 (Daegelen, et al., 2009). Genome sequencing of E. coli B (REL606) shows that when compared to E. coli strain K-12 MG1665, it has an IS1 associated 41 kbp deletion from uvrY to hchA that comprises ~0.9% of the genome including the dcm gene and 21 flagellar genes (Studier, et al., 2009). The loss of the dcm operon in E. coli B may have been coupled to the loss of the nearby flagellar and chemotaxis genes, as strains that lack flagellar and chemotaxis genes have an advantage during laboratory evolution experiments (Asakura, et al., 2011). Nonetheless, several pieces of evidence suggest that Dcm has a role in modulating the activity of the EcoRII R-M system in K-12 strains, which also targets 5′CCWGG3′ sequences. Experiments by Takahashi et al. indicate that loss of a plasmid containing the EcoRII methyltransferase and restriction enzyme genes is higher in dcm+ cells compared to dcm mutant cells, indicating that Dcm protects the genome against attack by this R-M system (Takahashi, et al., 2002). Furthermore, Dcm protects the cell from postsegregational killing due to loss of the EcoRII R-M system (Ohno, et al., 2008). Also, dcm partially protects DNA from cleavage during entry into a new host containing the EcoRII restriction enzyme (Hattman, et al., 1973). However, it is unclear if there are roles for Dcm beyond the role in the EcoRII R-M system. Therefore, we determined whether the dcm gene and 5mC were present in E. coli clinical strains and strains isolated from water and animal feces (environmental strains). We also tested the hypothesis that dcm influences the process of transcription, as cytosine-5 DNA methyltransferases often have this property.
Methods and Materials
Bacterial Strains
The Escherichia coli Reference Collection (ECOR), a set of E. coli strains isolated from a variety of hosts and geographical locations (Ochman & Selander, 1984), was obtained from the “Shiga-toxin producing E. coli” Center (STEC) at Michigan State University. Environmental strains of E. coli were isolated from seven different watersheds of Conesus Lake in New York over a five year period (Makarewicz, 2009, Simon & Makarewicz, 2009). Fifty-three strains were collected in the streams draining the watersheds, as well as at the mouth of the stream during all seasons of the year. Twenty-three independent strains were also collected from the Conesus Lake near-shore, focusing on those associated with the green alga Cladophora (Whitman, et al., 2003, Byappanahalli, et al., 2007). E. coli was isolated on m-ColiBlue24 plates (Millipore®)(Grant, 1997) and standard microbial testing was used to confirm the identification. All environmental isolates were positive for growth on lactose with gas formation, glucuronidase activity and the production of indole, while they were negative for growth on citrate and urea (APHA, 1999). Additional bacterial strains used in this study are listed in Table 1.
Table 1.
Additional bacterial strains and plasmids used in this study.
| Strains | Characteristic | Source/Reference |
|---|---|---|
| JM109 | wild-type dcm allele | New England BioLabs |
| ER2925 | dcm-6 | New England BioLabs |
| GM204 | Δ(supD-dcm-flaA) | Martin Marinus |
| BW25113 | wild-type dcm allele | (Baba, et al., 2008) |
| JW1944-2 | Δdcm-735::kan | (Baba, et al., 2008) |
| CP9 | ExPECa | (Russo, et al., 1993) |
| E234E69 | EPECb | (Levine, et al., 1978) |
| Popeye-1 | EHECc, O157:H7 | (Crane, et al., 2011) |
| Plasmidsd | ||
| pDcm-9 | dcm− vsr− | (Sohail, et al., 1990) |
| pDcm-21 | dcm+ vsr− | (Sohail, et al., 1990) |
Extraintestinal pathogenic E. coli
Enteropathogenic E. coli
Enterohemmoragic E. coli
both plasmids were originally constructed by enzyme mediated deletion of pDcm-4 which contains the entire dcm-vsr operon
Isolation and Genotype Analysis of Genomic DNA
Bacteria were propagated in Luria-Bertani broth overnight at 37°C with shaking at 250 rpm. Genomic DNA was isolated from 2 ml cultures of stationary phase cells using a DNeasy Blood and Tissue Kit (Qiagen) and RNase A was added at 200 μg/ml during lysis. Typical DNA preparations had A260/A280 readings of 1.8–2.1 and were 80–120 ng DNA/μl. A triplex PCR-based method for chuA, yjaA, and TSPE4.C2 was used to assign environmental isolates of E. coli to phylogenetic groups A, B1, B2, and D (Table 2) (Clermont, et al., 2000). Templates were either isolated genomic DNA or bacteria extracted in boiling TE buffer. Increasing Mg2+ to 3 mM in the PCR generated stronger products compared to 1.5 mM Mg2+.
Table 2.
Characteristics of environmental and ECOR strains used in this study.
| Source |
E. coli Phylogenomic Group
|
Total | ||||
|---|---|---|---|---|---|---|
| B1 | B2 | D | E | |||
| Algae | 9 | 14 | 23 | |||
| Animal | 25 | 21 | 23 | 13 | 4 | 86 |
| Water | 9 | 17 | 8 | 19 | 53 | |
|
| ||||||
| Total | 34 | 47 | 31 | 46 | 4 | 162 |
Polymerase Chain Reaction for the Dcm gene
PCR was carried out in 30 μl reactions containing 100 ng of genomic DNA or DNA from bacteria boiled in TE buffer, 0.3 μm of forward primer, 0.15 μM of reverse primer I, 0.15 μM of reverse primer II, 0.2 mM dNTPs, 1.5 mM MgCl2, and 0.75 units of TAQ DNA polymerase (Promega). Primer sequences are listed in Figure S1. The reaction conditions were 1 cycle of 95°C for 2 minutes, 32 cycles of 95°C for 1 minute, 55°C for 1 minute, 72°C for 1.5 minutes, and a final cycle of 72°C for 10 minutes. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Restriction Enzyme Isoschizomer Assay
The restriction enzymes BstNI and PspGI were purchased from New England BioLabs. Reactions were 20 μl and contained 1 μg of genomic DNA, and 0.3–0.5 units of enzyme. The DNAs were digested at 60°C for 2 hours and the products were analyzed by gel electrophoresis on 1% agarose gels and ethidium bromide staining. PspG1 was used at 60°C even though the optimal working temperature for the enzyme is 75°C (New England Biolabs) because the DNA degraded at 75°C (data not shown). Every experiment included DNA isolated from a dcm+ strain as a positive control (JM109 or BW25113) and DNA isolated from a dcm− strain as a negative control (ER2925, JW1944-2, or unmethylated phage lambda DNA).
Liquid Chromatography Tandem Mass Spectrometry
The levels of 5-methyl-2′-deoxcytidine (5mdC) in DNA samples were determined by high pressure liquid chromatography and tandem mass spectrometry (LC MS/MS) at the Roswell Park Pharmacodynamics/Pharmacokinetics Resource (Buffalo, NY) as previously described (Song, et al., 2005, Militello, et al., 2008). Briefly, isolated E. coli DNA (1 μg) from overnight cultures was digested to nucleosides using sequential treatment with S1 nuclease, snake venom phosphodiesterase, and alkaline phosphatase before separation on a dC18 column. Tandem mass spectrometry was used to detect the molecular ion (242.1 atomic mass units) and product ion (126.3 atomic mass units) for 5mdC. Simultaneously, the molecular ion and product ion for 2′-deoxyguanosine was detected. The ratios of 5mdC to 2′-deoxyguanosine in the experimental samples were compared to a standard curve of the same two nucleosides, to generate percent 5mdC. At least three distinct biological samples (separate cultures) were used for each strain, except for the commercial E. coli B preparation (four technical replicates).
Quantitative PCR
Overnight E. coli cultures were diluted 1:100 into fresh LB medium and grown at 37°C until early logarithmic phase (OD600 of ~0.4) and early stationary phase (OD600 of ~3.0). Total RNA was isolated using the MasterPure RNA Isolation kit (Epicentre). cDNA was made from 2–3 μg of RNA in presence of random primers. qPCR was performed on a Stratagene Mx3000P machine with Stratagene Brilliant Sybr Green qPCR master mix. Primer sequences are found in Figure S1. Reactions were performed in triplicate, and included at least two different RNA samples (biological replicates).
Results and Discussion
PCR Analysis for the Presence of dcm Loci
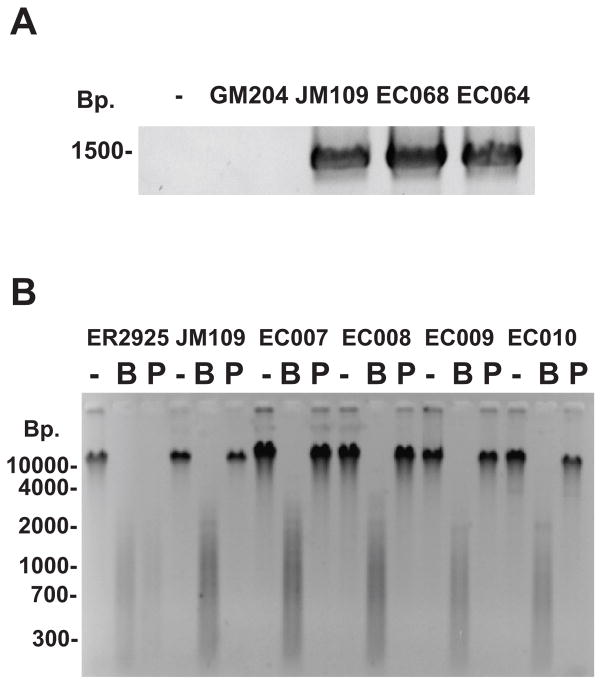
A PCR assay was developed to detect the presence of the dcm gene in E. coli. Forty one Escherichia coli and Shigella full length dcm DNA sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The sequences were aligned using ClustalX 2.0.10 (http://www.clustal.org/) and used to construct a N-J tree (Figure S2). In order to develop a set of PCR primers for the full length gene (1419 basepairs), the sequences at the beginning and the end of the alignment were examined. The first 88 nucleotides of all gene sequences were identical, and one forward primer was chosen. While there are three possible reverse primers, reverse primer III is present in only one sequence, and we therefore used a mixture of reverse primers I and II for all experiments. Initial PCRs were optimized using E. coli JM109 DNA (dcm+) as a positive control and the reactions routinely generated a product of the expected size of 1419 basepairs (Figure 1A). The assay was specific, as the dcm PCR product was not observed in reactions without DNA template or with DNA from E. coli GM204 DNA, a strain with a deletion of the dcm operon. To confirm that the PCR product truly represented the dcm gene, the PCR DNA from E. coli JM109 was purified and analyzed by DNA sequencing (data not shown).
Figure 1.
A) Detection of the dcm gene via PCR. E. coli genomic DNA from different strains was used as a template for PCR using one forward dcm primer and two reverse dcm primers as described in the Materials and Methods. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide straining. The negative sign (−) represents a no DNA control. E. coli GM204 DNA (GM204) contains deletion of the dcm operon. E. coli JM109 DNA (JM109) contains a wild-type dcm allele. EC068 and EC064 are DNAs from E. coli environmental isolates. B) Detection of 5-methylcytosine in 5′CCWGG3′ sequences using restriction enzyme isoschizomer pairs. DNA from E. coli ER2925 (dcm-6), JM109 (dcm+), and four environmental strains (EC007-EC010) were left undigested (−), digested with BstNI (B), or digested with PspGI (P). Reactions were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Subsequently, we used the PCR assay to screen the E. coli strains from multiple sources. The ECOR collection was initially screened, as it represents a diverse set of strains with respect to strain characteristics, phylogenetic groups, serotypes, and pathogenicity. Furthermore, new E. coli environmental samples were isolated as described in the materials and methods from a relatively small geographical region (Western New York, USA). These strains included representatives of the four main phylogenetic groups of A, B1, B2, and D (Clermont, et al., 2000). All 162 DNAs tested generated an appropriate size PCR product, indicating the presence of the dcm gene or a highly related dcm homolog. The presence of the dcm gene was independent of the source, pathogenicity, or phylogenetic group of the strain (Table S1).
Detection of Methylated 5′CCWGG3′ Sequences
While all strains tested contained a full length dcm gene, the PCR assay alone does not prove that each strain contains a functional cytosine DNA methylation and 5mC. Our PCR assay could not rule out dcm mutations that inactivate the enzyme, mutations in regulatory regions that inhibit transcription and translation, or the absence of other molecules required for cytosine DNA methylation. Therefore, a restriction enzyme isoschizomer assay was used to test for methylation of 5′CCWGG3′ sequences. Genomic DNAs were separately digested with the restriction enzymes BstNI and PspGI. Both enzymes cleave the sequence 5′CCWGG3′, but PspGI is blocked by Dcm-mediated cytosine methylation of the second cytosine. The assay was originally optimized with JM109 DNA (dcm+) and ER2925 DNA (dcm−). JM109 DNA was resistant to digestion with PspGI, which is consistent with DNA methylation of 5′CCWGG3′ sequences (Figure 1B). When ER2925 DNA was cut with PspGI, fragments that were heterogeneous in size were observed via gel electrophoresis, indicating ER2925 DNA is sensitive to this enzyme and lacks methylation at 5′CCWGG3′ sites. Titration of mixtures of methylated and unmethylated DNA indicated that the isoschizomer assay could detect partial cytosine DNA methylation down to 10%, but the assay is largely qualitative. DNA samples from all 162 ECOR and environmental strains were resistant to digestion by PspGI. This demonstrates that every strain of E. coli examined in this study has a dcm gene and 5mC in the sequence 5′CCWGG3′. Our data are in contrast to data on the solitary cytosine DNA methyltransferase M.Vch from Vibro cholera, as it was absent in 2 of 25 strains tested (Banerjee & Chowdhury, 2006). Our experiments cannot determine if all 5′CCWGG3′ sites are methylated, however there are reports suggesting the presence of rare, unmethylated 5′CCWGG3′ sites (Ringquist & Smith, 1992., Bormann Chung, et al., 2010). Nonetheless, each strain analyzed in our study has a functional cytosine DNA methylation pathway.
Detection of 5mdC in E. coli DNA using HLPC MS/MS
We were interested in determining the actual levels of 5-methylcytosine in different strains and used HPLC MS/MS to detect 5′-methyl-2′deoxycytidine (5mdC) levels in complete DNA digests. The dcm+ laboratory K-12 strains have ~1% 5mdC; JM109 has 0.92% (+/− 0.02) 5mdC and BW25113 has 1.02% (+/− 0.09) 5mdC. The data are consistent with previous results using different methodologies that indicate ~1% of the cytosines of E. coli K-12 strains are methylated (Vanyushin, et al., 1968). The level of 5mdC was not above the limit of detection (0.01%) in the dcm knockout strain JW1944-2, indicating that Dcm is the major or only enzyme that produces 5mC in laboratory E. coli strains. We also tested a commercial preparation of E. coli B DNA (Sigma) and did not detect 5mdC. We next tested nine ECOR and environmental isolates in this assay, representing pathogenic and non-pathogenic strains. In each case, 5mdC was detected, indicating that these strains do indeed contain 5mC. The levels of 5mdC ranged from 0.86% to 1.30% of the total cytosine in the DNA (Table 3). ANOVA analysis of all strains with 5mdC suggested that there is a statistically significant difference (p<0.05) between the amounts of 5mdC in all strains tested (p = 0.013). The small differences in levels of 5mdC could be due to small differences in GC content between strains, the lack of methylation of some 5′CCWGG3′ sites in some strains, the presence of 5mC at non-5′CCWGG3′ sites, and/or the presence of another DNA methyltransferase in some strains (e.g. R-M systems).
Table 3.
5-methyl-2′deoxycytidine levels in E. coli strains as determined by LC MS/MS.
| Strain | Description | % 5mdC | St Dev |
|---|---|---|---|
| BW25113 | K-12 laboratory strain | 1.02 | 0.09 |
| JM109 | K-12 laboratory strain | 0.92 | 0.02 |
| JW1944-2 | dcm knockout strain | under LOD1 | |
| E. coli B | B laboratory strain | under LOD1 | |
| EC6001 | environmental isolate | 1.10 | 0.05 |
| EC6002 | environmental isolate | 1.03 | 0.05 |
| EC6006 | environmental isolate | 1.08 | 0.15 |
| EC6026 | environmental isolate | 1.03 | 0.12 |
| ECOR11 | Human urinary tract infection | 0.92 | 0.18 |
| ECOR27 | Giraffe isolate | 0.86 | 0.03 |
| ECOR47 | Sheep isolate | 0.92 | 0.11 |
| ECOR52 | Orangutan isolate | 1.30 | 0.02 |
| Popeye-1 | Human O157:H7 strain | 1.13 | 0.27 |
The limit of detection (LOD) is 0.01% 5mdC
Potential Roles for Dcm and Cytosine DNA Methylation
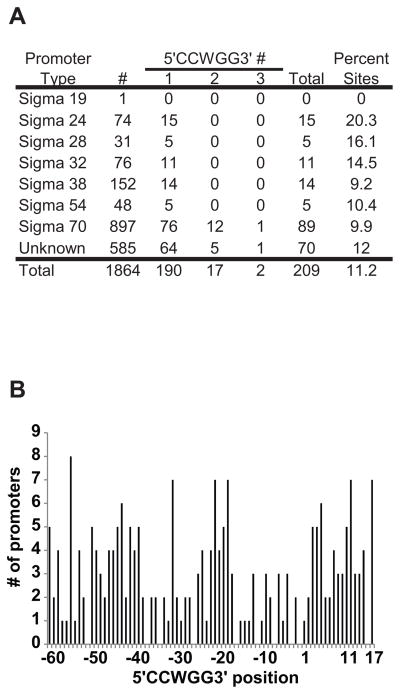
Our data indicate that the dcm gene and cytosine DNA methylation at 5′CCWGG3′ sequences are highly conserved in E. coli, which suggests that cytosine DNA methylation has an important role in E. coli biology. There are reports implicating 5mC in a role in phage lambda recombination, Tn10 insertion, and R-M system maintenance (Korba & Hays, 1982, Lee, et al., 1987, Takahashi, et al., 2002). Yet, there is no consensus model for dcm function and there is little known regarding the relationship between dcm and E. coli biological processes beyond protection from the EcoRII restriction enzyme. Methylated DNA bases are associated with transcriptional silencing in eukaryotes (Feng, et al., 2010). There are reports that some E. coli genes contain Dcm recognition sites within their promoters (Gomez-Eichelmann & Ramirez-Santos, 1993, Palmer & Marinus, 1994). We have extended this observation by analyzing a large number of the promoter sites (1864) in the complete genome of E. coli K-12 MG1655 (Gama-Castro, et al., 2011). Promoter sites associated with Sigma 24, 28, 32, 38, 54, and 70 all have examples of 5′CCWGG3′ sequences (Figure 2A), suggesting that DNA methylation could influence transcription initiation. 190 promoters have one 5′CCWGG3′ site, 17 promoters have two 5′CCWGG3′ sites, and two promoters have three 5′CCWGG3 sites. The distribution of all 5′CCWGG3′ sites in the promoter region relative to the transcription start site is given in Figure 2B. Based on the analysis of the variance to mean ratio (1.53) the distribution of 5′CCWGG3′ locations in promoters is clumped (neither random nor uniform) (p=0.0018). As expected there are fewer 5′CCWGG3′ sites associated with the −35 and −10 regions as these regions contain the conserved sequences for sigma factor binding. An analysis of clusters of orthologous genes (COGs) indicates that 5′CCWGG3′ sites are abundant in numerous COG categories and the highest percentages are in transcription, amino acid transport and metabolism, and translation (Table S2).
Figure 2. The number and location of 5′CCWGG3′ sites in Escherichia coli K-12 MG1655 promoters.
A) 5′CCWGG3′ abundance in E. coli promoters. Promoter sequences available in the Regulon database (http://regulondb.ccg.unam.mx/) were downloaded and queried for the number of 5′CCWGG3′ sites in different E. coli promoters. # represents the number of promoters in each category and total is the total number of promoters with 5′CCWGG3′ sequences. B) Histogram of the frequency of 5′CCWGG3′ sites with respect to the transcription start site. The numbers represent the distance to the transcription start site where the number 1 is the transcription start site. The position refers to the first C in the sequence 5′CCWGG3′.
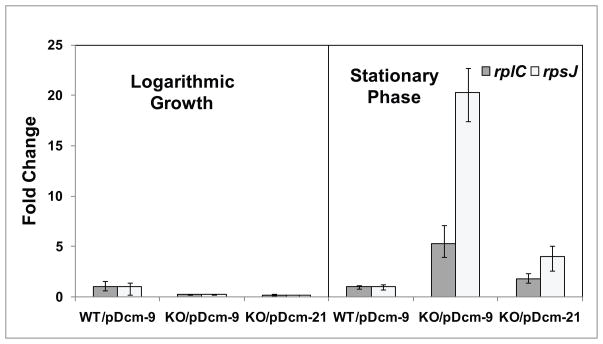
To determine if Dcm plays a role in transcription, RNA levels in wild-type bacteria with a plasmid with a inactive dcm truncation (BW25113/pDcm-9), dcm knockout bacteria with a plasmid with an inactive dcm truncation (JW1944-2/pDcm-9), and dcm knockout bacteria with a plasmid containing a functional dcm gene (JW1944-2/pDcm-21) were compared using qPCR. We focused on ribosomal protein gene expression, as previous reports indicate that ribosomal protein S16 gene contains a large number of 5′CCWGG3′ sites (Gomez-Eichelmann & Ramirez-Santos, 1993), and many genes in the translation COG have 5′CCWGG3′ sites in their promoters (Table S2). We started with the rplC and rpsJ genes; these genes code for large and small ribosomal subunits and are part of an operon controlled by the rpsJp promoter. Indeed, there are three 5′CCWGG3′ sites within the rplC gene, one site within the rpsJ gene, and one site 364 basepairs upstream of the rpsJ start codon. At early logarithmic phase, there was relatively no change in rplC and rpsJ RNA levels when comparing the three strains (Figure 3). However, at early stationary phase, there was a marked increase in both rplC and rpsJ RNA levels in JW1944-2/pDcm-9 cells, and the RNA levels were reduced in the complemented JW1944-2/pDcm-21 cells. These data indicate that Dcm is necessary for repression of these genes, and thus potentially influence stationary phase fitness or viability. Expression of the rplC and rpsJ genes is increased in the presence of 5-azacytidine, an inhibitor of cytosine DNA methylation (ML VanHorne and KT Militello, unpublished data). Thus, we hypothesize that depression of ribosomal protein gene expression is due directly to the loss of DNA methylation. These data are important as they indicate that Dcm has a role in the cell beyond protection from restriction enzymes that cleave the same sequence.
Figure 3. Expression of ribosomal protein genes in the absence and presence of dcm.
E. coli wild-type bacteria with a plasmid containing an inactive dcm truncation (BW25113/pDcm-9), dcm knockout bacteria containing an inactive dcm truncation (JW1944-2/pDcm-9), and dcm knockout bacteria containing a plasmid with a functional dcm gene (JW1944-2/pDcm-21) were grown to early logarithmic phase and early stationary phase. Total RNA was isolated and converted to cDNA. The levels of rplC and rpsJ were measured by qPCR, and normalized to the levels of malate dehydrogenase (mdh) RNA. The BW25113/pDcm-9 samples were set to a value of one. Error bars represent one standard deviation.
Bacterial ribosome number is correlated with growth rate. In addition to translational control of ribosomal protein gene expression during growth, there is new evidence for widespread transcriptional control of ribosomal protein genes (Lemke, et al., 2011). Dcm may participate in reducing or fine-tuning ribosome biosynthesis during stationary phase via methylation-dependent reduction in transcription of ribosomal protein genes during stationary phase. Methylated 5′CCWGG3′ sites in promoters or genes bodies could represent the binding sites for repressors of transcription initiation or elongation. Alternatively, activators may exist that are not able to bind to 5′CCWGG3′sites when they are methylated. In both models, absence of methylation at these sites will be correlated with increased gene expression. Identifying gene regulatory molecules that are affected by methylation of 5′CCWGG3′ sites will be a high priority. The link between DNA methylation and ribosome biosynthesis could be at the heart of the interaction between a host and a parasitic R-M system. As a large number of DNA methyltransferases found in REBASE modify 5′CCWGG3′sites, it is possible that R-M systems influence expression of ribosomal protein genes and/or other genes to promote their maintenance. The effect of Dcm and other DNA methyltransferases on the entire E. coli transcriptome is currently under investigation.
Supplementary Material
Acknowledgments
We thank Dr. John Crane (SUNY Buffalo) and Dr. Martin Marinus (University of Massachusetts Medical School) for providing E. coli strains. We thank Dr. Ashok Bhagwat (Wayne State University) for providing the pDcm-9 and pDcm-21 plasmids. We thank Ping Wang and Joshua Prey at the Roswell Park Cancer Institute for the LC MS/MS analysis. Support for this work was provided by the Geneseo Foundation and NIH grant R15AI074035-01 (K.T.M).
Footnotes
Authors’ Contributions
K.T.M. and R.D.S. contributed equally to the work.
References
- APHA. Standard Methods for the Examination of Water and Wastewater. American Public Health Association; New York: 1999. [Google Scholar]
- Asakura Y, Kojima H, Kobayashi I. Evolutionary genome engineering using a restriction-modification system. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr585. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Huan HC, Datsenko K, Wanner BL, Mori H. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods Mol Biol. 2008;416:183–194. doi: 10.1007/978-1-59745-321-9_12. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Chowdhury R. An orphan DNA (cytosine-5-)-methyltransferase in Vibrio cholerae. Microbiology. 2006;152:1055–1062. doi: 10.1099/mic.0.28624-0. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Sohail A, Roberts RJ. Cloning and characterization of the dcm locus of Escherichia coli K-12. J Bacteriol. 1986;166:751–755. doi: 10.1128/jb.166.3.751-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann Chung CA, Boyd VL, McKernan KJ, Fu Y, Monighetti C, Peckham HE, Barker M. Whole methylome analysis by ultra-deep sequencing using two-base encoding. PLoS One. 2010;5:e9320. doi: 10.1371/journal.pone.0009320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Ferguson J, Ishii S, Sadowsky MJ. Population structure of Cladophora-borne Escherichia coli in nearshore water of Lake Michigan. Water Research. 2007;41:3649–3654. doi: 10.1016/j.watres.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daegelen P, Studier FW, Lenski RE, Cure S, Kim JF. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3) J Mol Biol. 2009;394:634–643. doi: 10.1016/j.jmb.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Doskocil J, Sormova Z. The sequences of 5-methylcytosine in the DNA of Escherichia coli. Biochem Biophys Res Commun. 1965;20:334–339. doi: 10.1016/0006-291x(65)90369-4. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto D, Srinivasan PR, Borek E. On the nature of the deoxyribonucleic acid methylases. Biological evidence for the multiple nature of the enzymes. Biochemistry. 1965;4:2849–2855. doi: 10.1021/bi00888a041. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Salgado H, Peralta-Gil M, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Eichelmann MC, Ramirez-Santos J. Methylated cytosine at Dcm (CCATGG) sites in Escherichia coli: possible function and evolutionary implications. J Mol Evol. 1993;37:11–24. doi: 10.1007/BF00170457. [DOI] [PubMed] [Google Scholar]
- Grant MA. A new membrane filtration medium for simultaneous detection and enumeration of Escherichia coli and total coliforms. Appl Environ Microbiol. 1997;63:3526–3530. doi: 10.1128/aem.63.9.3526-3530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanck T, Gerwin N, Fritz HJ. Nucleotide sequence of the dcm locus of Escherichia coli K12. Nucleic Acids Res. 1989;17:5844. doi: 10.1093/nar/17.14.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S, Schlagman S, Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973;115:1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke F, Kolmar H, Brundl K, Fritz HJ. The vsr gene product of E. coli K-12 is a strand- and sequence-specific DNA mismatch endonuclease. Nature. 1991;353:776–778. doi: 10.1038/353776a0. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Fukuda E, Kobayashi I. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010;17:325–342. doi: 10.1093/dnares/dsq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Wagner R, Radman M. Mismatch repair of deaminated 5-methyl-cytosine. J Mol Biol. 1987;194:155–159. doi: 10.1016/0022-2836(87)90724-8. [DOI] [PubMed] [Google Scholar]
- Korba BE, Hays JB. Partially deficient methylation of cytosine in DNA at CCATGG sites stimulates genetic recombination of bacteriophage lambda. Cell. 1982;28:531–541. doi: 10.1016/0092-8674(82)90208-2. [DOI] [PubMed] [Google Scholar]
- Lee SY, Butler D, Kleckner N. Efficient Tn10 transposition into a DNA insertion hot spot in vivo requires the 5-methyl groups of symmetrically disposed thymines within the hot-spot consensus sequence. Proc Natl Acad Sci USA. 1987;84:7876–7880. doi: 10.1073/pnas.84.22.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Sanchez-Vazquez P, Burgos HL, Hedberg G, Ross W, Gourse RL. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci U S A. 2011;108:5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J Bacteriol. 1987;169:5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M, Bhagwat AS. Very short patch repair: reducing the cost of cytosine methylation. Mol Microbiol. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- Makarewicz J. Nonpoint source reduction to the nearshore zone via watershed management practices: nutrient fluxes, fate, transport and biotic responses — background and objectives. J Great Lakes Res. 2009;35:3–9. [Google Scholar]
- Marinus M, Lobner-Olesen A. DNA Methylation. In: Böck A, Curtiss R III, Kaper J, et al., editors. EcoSal - Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 2009. pp. 1–88. [Google Scholar]
- Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973;114:1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militello KT, Wang P, Jayakar SK, Pietrasik RL, Dupont CD, Dodd K, King AM, Valenti PR. African trypanosomes contain 5-methylcytosine in nuclear DNA. Eukaryot Cell. 2008;7:2012–2016. doi: 10.1128/EC.00198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll J, Fitzgerald GF, van Sinderen D. A dichotomous epigenetic mechanism governs expression of the LlaJI restriction/modification system. Mol Microbiol. 2005;57:1532–1544. doi: 10.1111/j.1365-2958.2005.04769.x. [DOI] [PubMed] [Google Scholar]
- Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Handa N, Watanabe-Matsui M, Takahashi N, Kobayashi I. Maintenance forced by a restriction-modification system can be modulated by a region in its modification enzyme not essential for methyltransferase activity. J Bacteriol. 2008;190:2039–2049. doi: 10.1128/JB.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli--a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- Posfai J, Bhagwat AS, Posfai G, Roberts RJ. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989;17:2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringquist S, Smith CL. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc Natl Acad Sci USA. 1992;89:4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon RD, Makarewicz JC. Impacts of manure management practices on stream microbial loading into Conesus Lake, NY. J Great Lakes Res. 2009;35:66–75. [Google Scholar]
- Sohail A, Lieb M, Dar M, Bhagwat AS. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990;172:4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, James SR, Kazim L, Karpf AR. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem. 2005;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
- Studier FW, Daegelen P, Lenski RE, Maslov S, Kim JF. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J Mol Biol. 2009;394:653–680. doi: 10.1016/j.jmb.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Naito Y, Handa N, Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin BF, Belozersky AN, Kokurina NA, Kadirova DX. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968;218:1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. Occurrence of Escherichia coli and Enterococci in Cladophora (Chlorophyta) in Nearshore Water and Beach Sand of Lake Michigan. Appl Environ Microbiol. 2003;69:4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R, Fritz HJ. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987;6:1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.