Abstract
Previous studies demonstrated that individuals with subtype D HIV infection who had been infected for 2 or more years were frequently misclassified as assay positive using cross-sectional incidence assays. Samples from 510 subjects (212 subtype A, 298 subtype D) who were infected for 2.2 to 14.5 years (median 5.4 years) and were not virally suppressed were tested using an LAg-Avidity enzyme immunoassay (LAg-Avidity EIA), Bio-Rad Avidity assay, and BED capture enzyme immunoassay (BED-CEIA). The performance of these three assays was evaluated using various assay cutoff values [LAg-Avidity EIA: <1.0 OD-n and <2.0 OD-n; Bio-Rad Avidity assay: <40% avidity index (AI) and <80% AI; BED-CEIA: <0.8 OD-n]. The mean LAg-Avidity EIA result was higher for subtype A than D (4.54±0.95 vs. 3.86±1.26, p<0.001); the mean Bio-Rad Avidity assay result was higher for subtype A than D (88.9%±12.5% vs. 75.1±30.5, p<0.001); and the mean BED-CEIA result was similar for the two subtypes (2.2±1.2 OD-n for subtype A, 2.2±1.3 OD-n for subtype D, p<0.9). The frequency of misclassification was higher for individuals with subtype D infection compared to those with subtype A infection, using either the LAg-Avidity EIA with a cutoff of <2.0 OD-n or the Bio-Rad Avidity assay with cutoffs of <40% or <80% AI. No subtype-specific differences in assay performance were observed using the BED-CEIA. Sex and age were not significantly associated with misclassification by any assay. The LAg-Avidity EIA with a cutoff <1.0 OD-n had the lowest frequency of misclassification in this Ugandan population.
Introduction
At a population level, HIV incidence, or the rate of new infections, is the most important quantity to measure when assessing the current state of the HIV epidemic. Determining where HIV transmission occurs provides important information on particular population subgroups and population-specific demographics.1 Knowledge of HIV incidence is necessary to understand transmission patterns, to provide a rational basis for targeting prevention efforts, to evaluate interventions, to reduce transmission, and to predict or project burden of HIV infection in different demographic and risk populations.2,3 There are three main approaches to determine HIV incidence in a population: direct measurement in cohort studies, inference from prevalence measurements, or estimation using tests for recent infection in cross-sectional surveys; multiple tests may be used in a recent infection testing algorithm.1 Most cross-sectional incidence studies use modified serologic assays for detecting HIV proteins, nucleic acids, and antibodies. However, there have been some new proposed biomarkers for measuring incidence that include other biomarkers, such as cytokine profiles, intraindividual virus genetic diversity, and changes in population-level genetic diversity. To date, none of these approaches has been subjected to rigorous evaluation with regard to their application in incidence estimation.4
Investigators use assays to measure biomarkers (especially those based on the maturation of the antibody response) to determine HIV incidence. Most cross-sectional HIV incidence assays measure characteristics of anti-HIV antibodies.5,6 The BED capture enzyme immunoassay (BED-CEIA)7 is currently used in the United States8,9 and other countries10 to estimate HIV incidence and identify high-incidence populations. A modified version of the Bio-Rad 1/2+O ELISA (Bio-Rad Avidity assay)11 has also been used for cross-sectional incidence estimation. Cross-sectional surveys based on a single serologic assay often overestimate HIV incidence, since some individuals with long-term infection are identified as assay positive (those who are likely to be recently infected and to have an assay result below an assay cutoff). HIV incidence testing assays that include both serologic and nonserologic biomarkers (multiassay algorithms, MAAs) have been developed for HIV subtype B that reduce or eliminate this type of misclassification.12–14
Subtype designations have been powerful molecular epidemiologic tools to track the course of the HIV-1 pandemic. Group M is the predominant circulating HIV-1 group. It has been divided into the current nine subtypes: A–D, F–H, J, and K.15,16 HIV-2 is another strain of HIV that is less common. HIV-2 is predominantly found in Western Africa.17 Worldwide, it has been shown that 48% of HIV infections are caused subtype C, 12% by subtype A, 11% by subtype B, 5% by subtype G, 2% by subtype D, and 22% recombinants.18 Misclassification of HIV subtype D infection has marginal significance on a global level, but is extremely significant in Uganda where subtypes A and D predominate and in Sudan where subtype D is the most common.17,19 The performance of serologic HIV incidence assays has been shown to vary by HIV subtype.20–23 For example, among individuals infected for 2 or more years, being misclassified as assay positive is more likely in subtype D infection than subtype A infection.18,24 In addition, among those with long-standing subtype D infection, women are significantly more likely to be misclassified as assay positive than men. A limiting antigen avidity assay, LAg-Avidity EIA, was designed to reduce the frequency of misclassification; this assay includes a multisubtype recombinant HIV-1 target antigen (rIDR-M) present in limiting concentration.25 When compared to the BED-CEIA, the LAg-Avidity EIA had a lower frequency of misclassification among individuals with long-term infection for both subtypes A and D.25
In this report, we compared the performance of the LAg-Avidity EIA to the BED-CEIA and Bio-Rad Avidity assay by analyzing samples from individuals in Rakai, Uganda who were infected for more than 2 years.
Materials and Methods
Study population
This study included 510 individuals (212 HIV subtype A and 298 HIV subtype D) enrolled in the 2008–2009 Rakai Community Cohort Study (RCCS) who were known to have been infected for at least 2 years (2.2 to 14.5 years, median 5.4 years). These individuals had not previously received highly active antiretroviral therapy (HAART naive) and were not naturally virally suppressed (there was sufficient HIV RNA in the samples to permit amplification for sequencing).
Ethics statement
All study participants provided written informed consent. This study was approved by Institutional Review Boards in Uganda and the United States.
Laboratory Methods
HIV subtypes of the samples were determined previously.26,27 Subtype was limited to gp41 and only those with subtype A and D were included in this analysis. Individuals with intersubtype recombinant infection based on sequence analysis were excluded from this analysis. Samples were analyzed using three serologic assays: the LAg-Avidity EIA, the Bio-Rad Avidity assay, and the BED-CEIA. The LAg-Avidity EIA measures the quantity of high-avidity antibodies directed against an immunodominant domain of HIV. This assay was performed according to the methods of Duong et al.25; additional analysis was performed using a higher cutoff value of <2.0 OD-n. The Bio-Rad Avidity assay was performed using the Genetic Systems HIV-1/HIV-2 PLUS O EIA (Bio-Rad Laboratories, Redmond, WA), with the following minor modifications: a 30-minute initial incubation was added and the diethylamine reagent was diluted in water.11 The percent avidity (avidity index, AI) was calculated for each sample, as described.11 The BED-CEIA was performed according to the manufacturer's directions (Calypte Biomedical Corporation, Lake Oswego, OR), with one exception: samples were run in duplicate and results were reported as an average of normalized optical density units (OD-n).23,24 Samples were considered to be misclassified if they had the following test results: LAg-Avidity avidity <1.0 OD-n or <0.2 OD-n, Bio-Rad Avidity <40% or <80% AI, and BED-CEIA <0.8 OD-n.
Statistical analysis
Factors associated with assay misclassification (age, duration of infection, CD4 cell count, sex, subtype, and assay) were analyzed for each assay using the Fisher's exact test or Chi square test. Logistic regression was performed to determine the odds of misclassification for all factors analyzed at the following assay cutoffs: LAg-Avidity EIA <2.0 OD-n, Bio-Rad Avidity <80% AI, and BED-CEIA <0.80 OD-n. All factors associated with misclassification in the univariate analysis with p<0.10 were included in the multivariate logistic regression analysis. All analyses were performed using STATA v.11 (StataCorp, College Station, TX).
Results
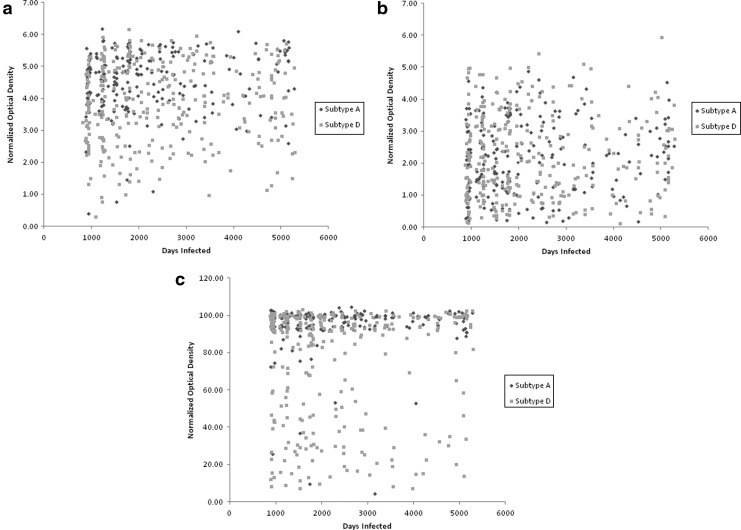
Characteristics of the study population are shown in Table 1. The mean LAg-Avidity EIA OD-n value was higher for subtype A than for subtype D (4.54±0.95 vs. 3.86±1.26, p<0.001, Fig. 1a). The mean Bio-Rad Avidity AI value was also higher for subtype A than subtype D (88.9%±12.5% vs. 75.1%±30.5%, p<0.001, Fig. 1b). The mean BED-CEIA OD-n value was similar for subtype A and subtype D (2.2±1.2 and 2.2±1.3, respectively, p<0.9, Fig. 1c). The frequency of misclassification was higher for subtype D than subtype A using both the LAg-Avidity EIA and the Bio-Rad Avidity assay; in contrast, there was no difference in the frequency of misclassification for these subtypes with the BED-CEIA (Table 1).
Table 1.
Factors Associated with Assay Performance in Specimens with Subtype A and D Infection
| Subtype A 41.6% (212/510) | Subtype D 58.4% (298/510) | |
|---|---|---|
| Age at infection, mean (years) | ||
| 18–28 | 23.6% (50/212) | 23.8% (71/298) |
| 29–34 | 34.0% (72/212) | 32.6% (97/298) |
| 35–40 | 19.7% (42/212) | 21.1% (63/298) |
| 41–51 | 22.5% (48/212) | 22.5% (67/298) |
| Duration of infection (years) | ||
| 2–5 | 52.1% (110/212) | 44.6% (133/298) |
| 6–9 | 27.7% (59/212) | 27.9% (83/298) |
| 10–15 | 20.2% (43/212) | 27.5% (82/298) |
| CD4 cell count (cells/μl) | ||
| >500 | 24.9% (53/212) | 30.5% (91/298) |
| 200–499 | 17.9% (38/212) | 23.8% (71/298) |
| 0–199 | 4.7% (10/212) | 4.7% (14/298) |
| No data | 52.1% (111/212) | 40.9% (122/298) |
| Sex | ||
| Male | 38.2% (81/212) | 35.2% (105/298) |
| Female | 61.8% (130/212) | 64.8% (193/298) |
| LAg-Avidity EIA (OD-n) | ||
| OD-n <1.0 | 0.9% (2/212) | 1.7% (5/298) |
| OD-n <2.0 | 1.9% (4/212) | 7.7% (23/298) |
| Bio-Rad Avidity assay (AI) | ||
| AI <40% | 1.9% (4/212) | 20.8% (62/298) |
| AI <80% | 4.7% (10/212) | 35.6% (106/298) |
| BED-CEIA (OD-n) | ||
| OD-n <0.80 | 11.8% (25/212) | 15.1% (45/298) |
Samples were from HIV-infected individuals from the Rakai Community Cohort Study (RCCS) who were infected 2+ years, were not on HAART, and had detectable virus.
CD4, cluster of differentiation 4; OD-n, normalized optical density; AI, avidity index; EIA, enzyme immunoassay; CEIA, capture enzyme immunoassay.
FIG. 1.
(a) LAg-Avidity enzyme immunoassay (EIA) results by duration of infection. (b) BED capture enzyme immunoassay (BED-CEIA) results by duration of infection. (c) Bio-Rad Avidity assay results by duration of infection.
Overall (for both subtypes combined), the misclassification frequencies for the assays were as follows: LAg-Avidity EIA: 1.4% (7/510) using a cutoff of <1.0 OD-n, 5.3% (27/510) using a cutoff of <2.0 OD-n; Bio-Rad Avidity assay: 12.9% (66/510) using a cutoff of <40% AI and 22.8% (116/510) using a cutoff of <80% AI; BED-CEIA: 13.7% (70/510) using a cutoff of <0.80 OD-n. All of the subtype A samples that were misclassified by the LAg-Avidity EIA using a cutoff of <1.0 OD-n (0.9%, 2/212) were also misclassified by the Bio-Rad Avidity assay using a cutoff of <40% AI and by the BED-CEIA. In a univariate analysis, misclassification by the LAg-Avidity EIA using a cutoff of <2.0 OD-n was associated with subtype D infection (p<0.01), a Bio-Rad Avidity assay result <80% AI (p<0.01), and a BED-CEIA result <0.80 OD-n (p<0.01). When adjusting for other factors, a Bio-Rad Avidity result of <80% AI and a BED-CEIA result of <0.80 OD-n were the only factors significantly associated with misclassification. In a univariate analysis, misclassification using the Bio-Rad Avidity assay was associated with CD4 cell counts between 200 and 499 cells/μl (p<0.05), HIV subtype D (p<0.01), an LAg-Avidity EIA result <2.0 OD-n (p<0.01), and a BED-CEIA result <0.80 OD-n (p<0.01).
When adjusting for other factors, CD4 cell counts between 200 and 499 cells/μl, subtype D infection, and an LAg-Avidity EIA result <2.0 OD-n were the only factors significantly associated with misclassification (Table 2). In a univariate analysis, BED-CEIA misclassification was associated with a duration of infection of 10–15 years (p<0.05), an LAg-Avidity EIA result <2.0 OD-n (p<0.01), and a Bio-Rad Avidity assay result <80% AI (p<0.01). When adjusting for other factors, a duration of infection 10–15 years and an LAg-Avidity EIA result <2.0 OD-n were the only factors significantly associated with misclassification. Sex and age were not associated with misclassification by any of the three assays.
Table 2.
Factors Associated with Misclassification of the BED-Capture Enzyme Immunoassay, the LAg-Avidity Enzyme Immunoassay, and the Bio-Rad Avidity Assay
| LAg-Avidity (OD-n <2.0) | LAg-Avidity OR (95% CI) | LAg-Avidity aOR (95% CI) | Bio-Rad Avidity (AI <80%) | Bio-Rad Avidity OR (95% CI) | Bio-Rad Avidity aOR (95% CI) | BED-CEIA (OD-n <0.8) | BED-CEIA OR (95% CI) | BED-CEIA aOR (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|
| Percent (#/total tested) | 5.3% (27/510) | 22.8% (116/510) | 13.7% (70/510) | ||||||
| Age at infection | |||||||||
| 18–28 years | 3.3% (4/121) | 1 | — | 24.0% (29/121) | 1 | — | 17.4% (21/121) | 1 | — |
| 29–34 years | 7.1% (12/169) | 2.2 (0.70–7.1) | — | 19.5% (33/169) | 0.77 (0.44–1.4) | — | 14.8% (25/169) | 0.83 (0.44–1.6) | — |
| 35–40 years | 6.7% (7/105) | 2.1 (0.59–7.4) | — | 27.6% (29/105) | 1.2 (0.67–2.2) | — | 16.2% (17/105) | 0.92 (0.46–1.9) | — |
| 41–51 years | 3.5% (4/115) | 1.1 (0.26–4.3) | — | 21.7% (25/115) | 0.88 (0.48–1.6) | — | 6.1% (7/115) | 0.31 (0.13–0.76) | — |
| Duration of infection | |||||||||
| 2–5 years | 6.6% (16/243) | 1 | — | 24.3% (59/243) | 1 | — | 16.9% (41/243) | 1 | 1 |
| 6–9 years | 3.5% (5/142) | 0.52 (0.19–1.5) | — | 19.0% (27/142) | 0.73 (0.44–1.2) | — | 13.4% (19/142) | 0.76 (0.42–1.4) | 0.84 (0.46–1.5) |
| 10–15 years | 4.8% (6/125) | 0.72 (0.27–1.9) | — | 24.0% (20/125) | 0.98 (0.60–1.6) | — | 8.0% (10/125) | 0.43 (0.21–0.89)† | 0.43 (0.20–0.90)† |
| CD4 (cells/μl) | |||||||||
| ≥500 | 5.6% (8/144) | 1 | — | 29.2% (42/144) | 1 | 1 | 16.0% (23/144) | 1 | — |
| 200–499 | 4.6% (5/109) | 0.82 (0.26–2.6) | — | 17.4% (19/109) | 0.51 (0.28–0.95)† | 0.42 (0.21–0.86)† | 12.0% (13/109) | 0.71 (0.34–1.5) | — |
| <200 | 12.5% (3/24) | 2.4 (0.60–9.9) | — | 20.8% (5/24) | 0.64 (0.22–1.8) | 0.31 (0.07–1.4) | 25.0% (6/24) | 1.8 (0.63–4.9) | — |
| No data | 4.7% (11/233) | 0.84 (0.33–2.15) | — | 21.5% (50/233) | 0.66 (0.41–1.1) | 0.78 (0.44–1.4) | 12.0% (28/233) | 0.72 (0.40–1.3) | — |
| Sex | |||||||||
| Male | 5.4% (10/186) | 1 | — | 20.4% (38/186) | 1 | — | 11.8% (22/186) | 1 | — |
| Female | 5.3% (17/324) | 0.97 (0.44–2.2) | — | 24.1% (78/324) | 1.2 (0.80–1.9) | — | 14.8% (48/324) | 1.3 (0.76–2.2) | — |
| Subtype | |||||||||
| A | 1.9% (4/212) | 1 | 1 | 4.7% (10/212) | 1 | 1 | 11.8% (25/212) | 1 | — |
| D | 7.7% (23/298) | 4.4 (1.5–12.8)‡ | 0.78 (0.19–3.1) | 35.6% (106/298) | 11.2 (5.7–22.0)‡ | 13.9 (6.2–31.5)‡ | 15.1% (45/298) | 1.3 (0.79–2.2) | — |
| Misclassified by other assay | |||||||||
| LAg-Avidity ≥2.0 OD-n | — | — | — | 96.3% (26/27) | 1 | 1 | 48.2% (13/27) | 1 | 1 |
| LAg-Avidity <2.0 OD-n | — | — | — | 18.6% (90/483) | 114 (15–848)‡ | 109 (13–887)‡ | 11.8% (57/483) | 6.9 (3.1–15.5)‡ | 4.5 (1.8–11.5)‡ |
| AI ≥80% | 0.24% (1/394) | 1 | 1 | — | — | — | 24.1% (28/116) | 1 | 1 |
| AI <80% | 22.4% (26/116) | 113.5 (15–847)‡ | 108.2 (13–897)‡ | — | — | — | 10.7% (42/394) | 2.7 (1.6–4.5)‡ | 1.8 (0.94–3.3)* |
| BED-CEIA ≥0.8 OD-n | 3.2% (14/440) | 1 | 1 | 40.0% (28/70) | 1 | 1 | — | — | — |
| BED-CEIA <0.8 OD-n | 18.6% (13/70) | 6.9 (3.1–15.5)‡ | 4.5 (1.8–11.4)‡ | 20.0% (88/440) | 2.7 (1.6–4.5)‡ | 1.8 (0.9–3.6)* | — | — | — |
LAg-Avidity measured in normalized optical density units (OD-n); Bio-Rad Avidity measured as Avidity Index (AI); BED-CEIA measured in OD-n.
Levels of significance: *p<0.10, †p<0.05, and ‡p<0.01.
OR, univariate odds ratio; aOR, multivariate adjusted odds ratio.
Discussion
This report examined the impact of HIV subtype on misclassification of long-standing infections as recent infection using three serologic incidence assays. Samples were obtained from adults in Uganda enrolled in a longitudinal cohort study who were known to be infected for at least 2 years with either subtype A or subtype D HIV. Because viral suppression is known to be associated with misclassification by serologic HIV incidence assays,28 samples were selected for this study from individuals who were HAART naive and were not virally suppressed (see Materials and Methods). Cutoff values of cross-sectional incidence assays can be varied. Lower cutoffs identify fewer true recently infected individuals but reduce misclassification, while higher cutoffs have the opposite effect. We presented both high and low values. In this study population, the LAg-Avidity EIA correctly classified 98.6% (503/510) of subtype A and D-infected individuals as assay negative using an assay cutoff of 1.0 OD-n. Using this cutoff, the LAg-Avidity EIA had the lowest frequency of misclassification among the three assays evaluated. When the LAg-Avidity EIA cutoff was increased to 2.0 OD-n, there was a significant association between infecting subtype and misclassification, with a 4-fold higher misclassification frequency for subtype D. The Bio-Rad Avidity assay correctly classified 87.1% (444/510) of samples using a cutoff of 40% AI; most cases of misclassification (94%=62/66 cases) were in subtype D samples. When the Bio-Rad Avidity assay cutoff was increased to 80% AI, there was a significantly higher frequency of misclassification in subtype D compared to subtype A. The BED-CEIA correctly classified only 86.3% (440/510) of individuals as assay negative using a cutoff of 0.8 OD-n.
In a previous study of individuals from the RCCS, samples from those infected for at least 2 years had different frequencies of misclassification for subtypes A and D in women, but not in men, using the Bio-Rad Avidity assay and the BED-CEIA.24 In this study, we also found a significant association between misclassification by the Bio-Rad Avidity assay and HIV subtype. While we did observe a high frequency of misclassification with the BED-CEIA, the misclassification frequencies were similar in the two subtypes. In this study, neither of these two assays displayed a significant association between misclassification and sex.
Our findings demonstrate the problems in using all three of these incidence assays as stand-alone assays in subtype D endemic areas. An association between low LAg-Avidity EIA results (OD-n <2.0) and low Bio-Rad avidity results (AI <80%) was observed in individuals with subtype D infection. Since these assays use different target antigens, this suggests individuals with subtype D infection are less likely to produce high avidity anti-HIV antibodies. Additionally, this suggests these assays should not be used together [e.g., in a multiassay algorithm (MAA)] in populations with a significant frequency of subtype D infections. The choice of assays to include in an MAA should take into consideration the factors associated with misclassification by each assay.
These three cross-sectional incidence assays detect antibodies against HIV, which should not impact this selected population of long-standing infected individuals but might be relevant when applying these assays in an open population where very recent (Ag-positive only) infections are observed. Even so, misclassification was observed in this selected population, which indicates that the results of these assays should be carefully monitored depending on the population under study. When single incidence assays with high specificity are used for cross-sectional surveys, the proportion of false-recent infections will be reduced; however, the sensitivity for identifying individuals with recent infection will also be reduced. Multiassay algorithms have been developed that include serological assays with relaxed cutoff values; these algorithms maximize the identification of recently infected individuals, while providing a high level of specificity.12 Further studies are needed to determine whether the lower LAg-Avidity EIA values seen in subtype D reflect a muted early antibody response to HIV infection (similar to what was observed in previous studies of the BED-CEIA and Bio-Rad Avidity assay29) or a waning of an initially robust antibody response. Additional studies are needed to determine the impact of viral suppression (natural or HAART induced) on the performance of the LAg-Avidity EIA.
Acknowledgments
We would like to thank Dr. Gary Murphy for his critical review. We thank Drs. Bharat S. Parekh and Yen Duong, for providing LAg kits and advice on use of the LAg-Avidity EIA, and thank Dr. S. Michele Owen for her advice on use of the Bio-Rad Avidity assay.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institutes of Health.
Sources of funding: (1) The HIV Prevention Trials Network (HPTN), funded by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Child Health and Human Development (NICH/HD), National Institute of Drug Abuse (NIDA), and the National Institute of Mental Health (NIMH), Office of AIDS Research, National Institutes of Health (NIH), Department of Health and Human Services (UM1-AI068613–Eshleman), and (2) R01-AI095068 (Eshleman/Brookmeyer). The studies that collected samples used for analysis were funded by the NIH, NIAID (R01-A134826, K22-AI092150-01, and R01-A134265), NICHD (R01-HD 050180), and the Bill and Melinda Gates Foundation (Grant 22006). Additional support was provided by the Division of Intramural Research, NIAID, NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Mastro TD, Kim AA, Hallett T, et al. : Estimating HIV incidence in populations using tests for recent infection: Issues, challenges and the way forward. J HIV AIDS Surveill Epidemiol 2010;2(1):1–14 [PMC free article] [PubMed] [Google Scholar]
- 2.Busch MP, Pilcher CD, Mastro TD, et al. : Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS 2010;24(18):2763–2771 [DOI] [PubMed] [Google Scholar]
- 3.Mastro TD: Determining HIV incidence in populations: Moving in the right direction. J Infect Dis 2013;207(2):204–206 [DOI] [PubMed] [Google Scholar]
- 4.Busch MP, Pilcher CD, Mastro TD, et al. : Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS 2010;24(18):2763–2771 [DOI] [PubMed] [Google Scholar]
- 5.Brookmeyer R, Quinn T, Shepherd M, Mehendale S, Rodrigues J, and Bollinger R: The AIDS epidemic in India: A new method for estimating current human immunodeficiency virus (HIV) incidence rates. Am J Epidemiol 1995;142(7):709–713 [DOI] [PubMed] [Google Scholar]
- 6.Thomas HI, Wilson S, O'Toole CM, et al. : Differential maturation of avidity of IgG antibodies to gp41, p24 and p17 following infection with HIV-1. Clin Exp Immunol 1996;103(2):185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbs T, Kennedy S, Pau CP, McDougal JS, and Parekh BS: Performance characteristics of the immunoglobulin G-capture BED-enzyme immunoassay, an assay to detect recent human immunodeficiency virus type 1 seroconversion. J Clin Microbiol 2004;42(6):2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall HI, Song R, Rhodes P, et al. : Estimation of HIV incidence in the United States. JAMA 2008;300(5):520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prejean J, Song R, Hernandez A, et al. : Estimated HIV incidence in the United States, 2006–2009. PLoS One 2011;6(8):e17502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mermin J, Musinguzi J, Opio A, et al. : Risk factors for recent HIV infection in Uganda. JAMA 2008;300(5):540–549 [DOI] [PubMed] [Google Scholar]
- 11.Masciotra S, Dobbs T, Candal D, et al. :. Antibody Avidity-Based Assay for Identifying Recent HIV-1 Infections Based on Genetic Systems TM 1/2 plus O EIA. 17th Conference on Retroviruses and Opportunistic Infections, San Francisco, CA, February16–19, 2010 Abstract 937 [Google Scholar]
- 12.Laeyendecker O, Brookmeyer R, Cousins MM, et al. : HIV incidence determination in the United States: A multiassay approach. J Infect Dis 2013;207(2):232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brookmeyer R, Konikoff J, Laeyendecker O, and Eshleman SH: Estimation of HIV incidence using multiple biomarkers. Am J Epidemiol 2013;177(3):264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brookmeyer R, Laeyendecker O, Donnell D, and Eshleman S: Cross-sectional HIV incidence estimation in HIV prevention research. J Acquir Immune Defic Syndr 2013;63(Suppl 2):S233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peeters M, Toure-Kane C, and Nkengasong JN: Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS 2003;17(18):2547–2560 [DOI] [PubMed] [Google Scholar]
- 16.Robertson DL, Anderson JP, Bradac JA, et al. : HIV-1 nomenclature proposal. Science 2000;288(5463):55–56 [DOI] [PubMed] [Google Scholar]
- 17.Lihana RW, Ssemwanga D, Abimiku A, and Ndembi N: Update on HIV-1 diversity in Africa: A decade in review. AIDS Rev 2012;14(2):83–100 [PubMed] [Google Scholar]
- 18.Hemelaar J, Gouws E, Ghys PD, and Osmanov S: Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011;25(5):679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hierholzer M, Graham RR, El Khidir I, et al. : HIV type 1 strains from East and West Africa are intermixed in Sudan. AIDS Res Hum Retroviruses 2002;18(15):1163–1166 [DOI] [PubMed] [Google Scholar]
- 20.Young CL, Hu DJ, Byers R, et al. : Evaluation of a sensitive/less sensitive testing algorithm using the bioMerieux Vironostika-LS assay for detecting recent HIV-1 subtype B' or E infection in Thailand. AIDS Res Hum Retroviruses 2003;19(6):481–486 [DOI] [PubMed] [Google Scholar]
- 21.Parekh BS, Hanson DL, Hargrove J, et al. : Determination of mean recency period for estimation of HIV Type 1 incidence with the BED-capture EIA in persons infected with diverse subtypes. AIDS Res Hum Retroviruses 2011;27:265–273 [DOI] [PubMed] [Google Scholar]
- 22.Parekh BS, Hu DJ, Vanichseni S, et al. : Evaluation of a sensitive/less-sensitive testing algorithm using the 3A11-LS assay for detecting recent HIV seroconversion among individuals with HIV-1 subtype B or E infection in Thailand. AIDS Res Hum Retroviruses 2001;17(5):453–458 [DOI] [PubMed] [Google Scholar]
- 23.Laeyendecker O, Brookmeyer R, Mullis C, et al. : Specificity of four laboratory approaches for cross-sectional HIV incidence determination: Analysis of samples from adults with known non-recent HIV infection from five African countries. AIDS Res Hum Retroviruses 2012;28(10):1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullis CE, Munshaw S, Grabowski MK, et al. :. Differential Misclassification of HIV-1 Cross-Sectional Incidence Assays by Subtype in Rakai, Uganda: 19th Conference on Retroviruses and Opportunistic Infections, Seattle, Washington, 2012. Abstract 541 [Google Scholar]
- 25.Duong YT, Qiu M, De AK, et al. : Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: Potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012;7(3):e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabowski M, Redd A, Mueller A, et al. :. The Natural Selection of HIV Subtypes Due to Differential Infectivity and Pathogenicity Is Accelerated by ART: Rakai, Uganda: 19th Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 2012. Abstract 521 [Google Scholar]
- 27.Collinson-Streng AN, Redd AD, Sewankambo NK, et al. : Geographic HIV type 1 subtype distribution in Rakai district, Uganda. AIDS Res Hum Retroviruses 2009;25(10):1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendel SK, Mullis CE, Eshleman SH, et al. : Effect of natural and ARV-induced viral suppression and viral breakthrough on anti-HIV antibody proportion and avidity in patients with HIV-1 subtype B infection. PLoS One 2013;8(2):e55525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longosz AF, et al. :. Characterization of Immune Responses in Ugandan Women Infected with Subtype A and D HIV Using the BED Capture Immunoassay and an Antibody Avidity Assay. 20th Conference on Retroviruses and Opportunistic Infections, Atlanta, Georgia, 2013. Abstract 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]