Abstract
We report here a novel HIV-1 circulating recombinant form (CRF62_BC) that was isolated from three epidemiologically unlinked individuals [one from an injecting drug user (IDU); two from heterosexuals] in Dehong prefecture of western Yunnan province. CRF62_BC harbored two subtype B segments in the pol and vpu-env regions in a subtype C backbone. Subregion tree analysis demonstrated that subtype B regions originated from a Thai-B (subtype B′) lineage and the subtype C region was from an India C lineage. CRF62_BC is the fourth CRF composed of subtypes B′ and C known to date after CRF07_BC, CRF08_BC, and CRF57_BC, which were originally found among IDUs in China. The emergence of CRF62_BC may indicate the continual generation of new recombinant strains in various high-risk populations in western Yunnan. This may complicate the development of effective vaccines to limit the HIV-1 epidemic and increase the difficulty of AIDS prevention and control in China.
Recombination between HIV-1 subtypes is a significant mechanism that contributes to the genetic complexity of HIV-1, and new strains are frequently found in regions and populations where multiple subtypes are circulating. At least 20% of HIV-1 isolates sequenced worldwide are intersubtype recombinants.1 To date, more than 60 circulating recombinant forms (CRFs) and 100 unique recombinant forms (URFs) have been reported worldwide (www.hiv.lanl.gov).
Yunnan province of southwestern China, which shares a border with the heroin-producing areas (“Golden Triangle”), was thought to be the epicenter of the HIV-1 epidemic in China.2–4 The first HIV-1 epidemic in Yunnan was initiated with both HIV-1 subtype B and subtype B′ (Thai-B) strains,5,6 followed by two CRFs, CRF07_BC and CRF08_BC, that arose and began to circulate widely among injecting drug users (IDUs) in China.4 Multiple HIV-1 genotypes, including B, C, CRF01_AE, CRF07_BC, and CRF08_BC, are circulating in Yunnan7 and mosaic strains were arising frequently.8,9 In the present study, we identified a novel CRF designated CRF62_BC in high-risk populations in western Yunnan and characterized their structural characteristics.
Plasma was collected from three HIV-positive patients (one IDU, 10CN.YNFL13; two heterosexuals, 10CN.YNFL15 and 10CN.YNFL18) with different ethnicity in Dehong prefecture in western Yunnan (Table 1). They were diagnosed as HIV-1 positive in 2010 and were recruited from the National HIV-1 drug resistance cordon molecular epidemiology survey in Yunnan. The study was approved by the institutional review boards of the National Center for AIDS/STD Control and Prevention. The near full-length genome (NFLG) amplification and sequencing were performed as previously reported.10 Briefly, RNA was extracted using the QIAamp Viral Mini Kit (QIAGEN, Germany), then transcribed into cDNA using the Superscript III First-Strand Synthesis System (Invitrogen, USA).
Table 1.
Demographic Information of Study Subjects Who Harbored CRF62_BC
| Strain name | Sampling year | Geographic origin | Sex | Age | Ethnicity | Risk factor | Accession number |
|---|---|---|---|---|---|---|---|
| 10CN.YNFL13 | 2010 | Dehong, Yunnan | Male | 52 | Dai | IDU | KC870034 |
| 10CN.YNFL15 | 2010 | Dehong, Yunnan | Male | 25 | Han | Heterosexual | KC870035 |
| 10CN.YNFL18 | 2010 | Dehong, Yunnan | Male | 33 | Jingpo | Heterosexual | KC870037 |
IDU, injecting drug user.
With the near-endpoint diluted cDNA template, the NFLGs were amplified with TaKaRa LA Taq (TaKaRa, Dalian, China) using the same nested polymerase chain reaction (PCR) amplification conditions. The positive PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germany) and sequenced by ABI 3730XL sequencer using BigDye terminators (Applied Biosystems, Foster City, CA). The chromatogram data were cleaned and assembled using Sequencher v4.9 (Gene Codes, Ann Arbor, MI). Nucleotide sequences were first aligned with the HXB2 standard reference strain using Clustal W, merged into the reference file, and adjusted manually using BioEdit.11 Phylogenetic trees were constructed with MEGA 5.0 by using the neighbor-joining method with 1,000 bootstrap replications.12 The standard subtype reference alignment file including all HIV-1 group M was downloaded from the Los Alamos National Laboratory HIV Database (www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html#ref). Bootstrap values above 0.7 were considered a phylogenetic cluster. Recombination breakpoints were determined using the recombinant identification program (RIP) and jumping profile hidden Markov model (jpHMM) (www.hiv.lanl.gov) and SimPlot programs.10 Subregion tree analysis was used to confirm the genotype and to estimate the origin of each segment. The nucleotide position of the estimated recombination breakpoints relative to the HXB2 genome was determined by HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html).
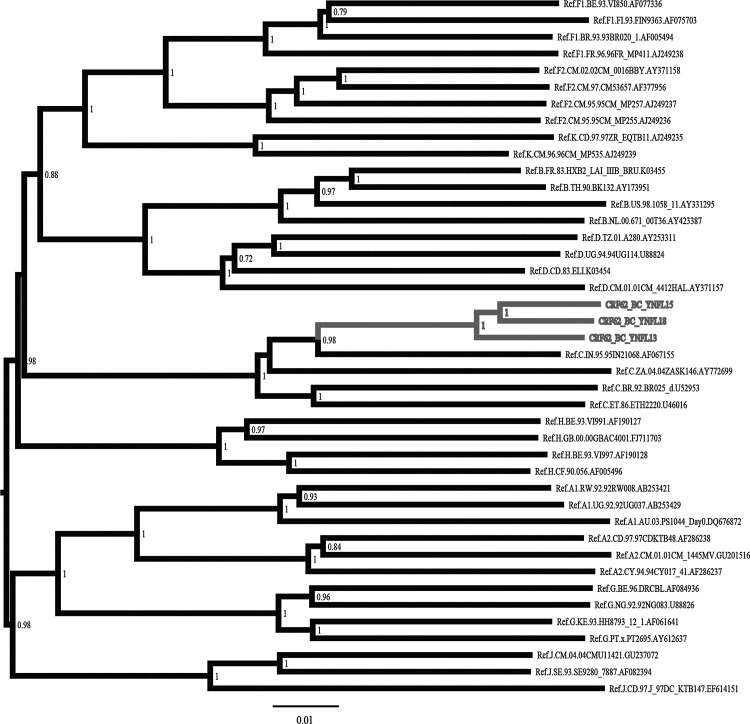
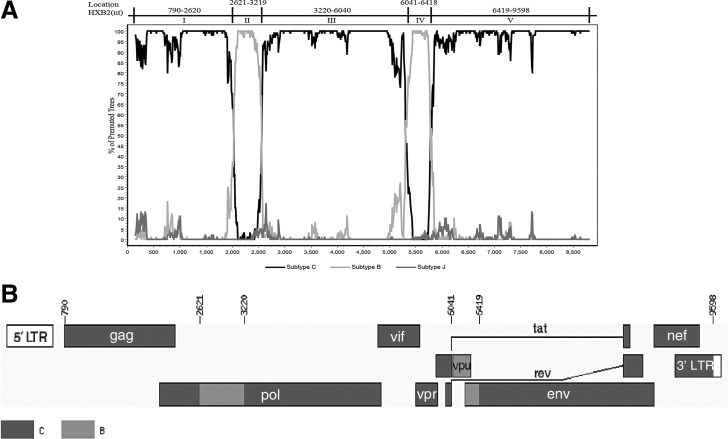
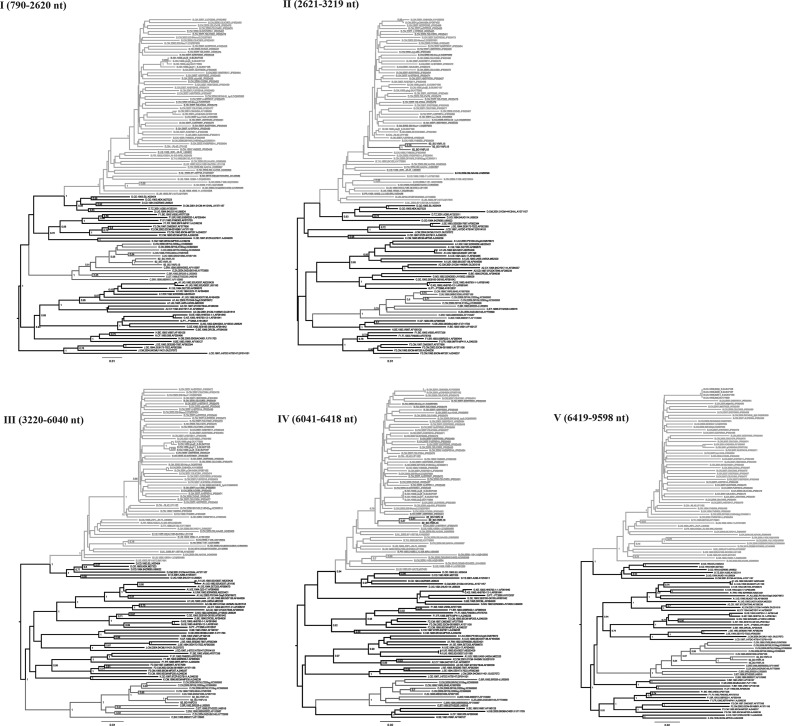
As shown in Fig. 1, these three strains formed a distinct monophyletic cluster, distantly related to all known HIV-1 subtypes/CRFs. Recombination analysis showed that the NFLG sequences of these three strains were composed of two subtype B regions (Regions II and IV) in a subtype C backbone (Regions I, III, and V) (Fig. 2). Subregion tree analyses further confirmed the parental origins of each segment of the recombinant genome as follows: Region I (nucleotide position relative to HXB2: 790–2,620 nt)=subtype C; Region II (2,621–3,219 nt)=subtype B; Region III (3,220–6,040 nt)=subtype C; Region IV (6,041–6,418 nt)=subtype B; Region IV (6,419–9,598 nt)=subtype C. Subregion segments also indicated that the parental origins of the subtype B and C regions are Thai B (subtype B′) and India C lineages, respectively (Fig. 3). The recombinant structure is distinct from any known CRFs to date and is now designated CRF62_BC.
FIG. 1.
Phylogenetic tree analysis of the near full-length genome (NFLG) sequences of CRF62_BC. All HIV-1 group M reference sequences were used to construct the neighbor-joining phylogenetic tree. The sequences of CRF62_BC are marked in gray. The stability of the nodes was assessed by bootstrap analysis with 1,000 replications, and only bootstrap values above 0.7 are shown at the corresponding node. The scale bar represents 1% genetic distance.
FIG. 2.
Recombination breakpoint analyses of CRF62_BC. (A) Bootscanning plots of CRF62_BC (10CN.YNFL13) with subtype B (U71182), subtype C (AF067155), and subtype J (GU237072) as reference genotypes with a window size of 300 nt and step size of 10 nt. The nucleotide positions relative to HXB2 are indicated, (B) Recombinant structure of CRF62_BC. The mosaic map was generated using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_mapper.html).
FIG. 3.
Subregion tree analyses of the CRF62_BC genome. The phylogenetic trees of the five mosaic segments defined by bootscanning plot analysis were constructed with MEGA 5.0 using the neighbor-joining method. The subtype references are derived from the Los Alamos HIV Sequence Database. The reliability of tree topology was assessed by bootstrap analysis with 1,000 replications, and only bootstrap values above 0.7 are shown at the corresponding nodes. The sequences of CRF62_BC are marked in gray.
Multiple lineages of HIV-1 strains are circulating in Dehong prefecture of western Yunnan7 where the earliest HIV-1 epidemic in China began among IDUs. Previous studies suggested that two closely related CRFs, CRF07_BC and CRF08_BC, may have originated from western Yunnan.8,13 CRF07_BC and CRF08_BC subsequently spread across China mainly through drug trafficking routes. The coexistence of multiple strains along with the heavy epidemic increase the probability of recombination. However, different from the national widespread epidemic of CRF07_BC and CRF08_BC7, CRF62_BC appears to circulate on only a small scale in the region. The reasons may be that the mobility of the local population is relatively small, drug trafficking is currently strictly controlled, and HIV-1 spread through other means (heterosexual transmission) may not be efficient.
The emergence of CRF62_BC may suggest the continued generation of new recombinant strains in western Yunnan. This may further complicate the development of effective vaccines to limit the HIV-1 epidemic in China. And whether recombination may confer selective advantages over parental viruses remains to be investigated.
Sequence Data
The nucleotide sequences of the three isolates 10CN.YNFL13, 10CN.YNFL15, and 10CN.YNFL18 have been submitted to GenBank with the accession numbers KC870034, KC870035, and KC870037, respectively.
Acknowledgments
This work was supported by national major projects for Infectious Diseases Control and Prevention (grants 2012ZX10001-008 and 2012ZX10001-002), National Natural Science Foundation of China (grants 81020108030 and 81261120393), Chinese State Key Laboratory for Infectious Disease Development Grant (grant 2012SKLID103), the International Development Research Center of Canada (grant 104519-010), and partly by the Japan-China Medical Association. We would like to thank Brian Foley from the Los Alamos National Laboratory for advice on CRF nomenclature.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Zhang M, Foley B, Schultz AK, et al. : The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 2010;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Y: HIV infection and AIDS in China. Arch AIDS Res 1992;6(1–2):1–5 [PubMed] [Google Scholar]
- 3.Sun X, Nan J, and Guo Q: AIDS and HIV infection in China. AIDS 1994;8(Suppl 2):S55–S59 [PubMed] [Google Scholar]
- 4.Su L, Graf M, Zhang Y, et al. : Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B′) recombinant strain in China. J Virol 2000;74(23):11367–11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graf M, Shao Y, Zhao Q, et al. : Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses 1998;14(3):285–288 [DOI] [PubMed] [Google Scholar]
- 6.Li Z, He X, Wang Z, et al. : Tracing the origin and history of HIV-1 subtype B′ epidemic by near full-length genome analyses. AIDS 2012;26(7):877–884 [DOI] [PubMed] [Google Scholar]
- 7.He X, Xing H, Ruan Y, et al. : A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 2012;7(10):e47289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang R, Kusagawa S, Zhang C, Xia X, Ben K, and Takebe Y: Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J Virol 2003;77(1):685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Chen L, Yang S, et al. : Recombination form and epidemiology of HIV-1 unique recombinant strains identified in Yunnan, China. PLoS One 2012;7(10):e46777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei H, Su L, Feng Y, et al. : Near full-length genomic characterization of a novel HIV Type 1 CRF07_ BC/01_AE recombinant in MSM from Sichuan, China. AIDS Res Hum Retroviruses 2013;29(8):1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall TA: BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic Acids Symposium, Series 1999 [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, and Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24(8):1596–1599 [DOI] [PubMed] [Google Scholar]
- 13.Tee KK, Pybus OG, Li XJ, et al. : Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J Virol 2008;82(18):9206–9215 [DOI] [PMC free article] [PubMed] [Google Scholar]