Abstract
Glycans play a critical role in physiological and pathological processes through interaction with a variety of ligands. Altered expression and dysregulation of these molecules can cause aberrant cellular function such as malignancy. Glycomics provide information of the structure and function of glycans, glycolipids, and glycoproteins such as proteoglycans, and may help to predict cancer development and progression as biomarkers. In this report, we compared the expression of proteoglycans, the content and structure of glycosaminoglycans and glycolipids between patient-matched normal and cancer tissues obtained from colon cancer patients. Tumor-related proteoglycans, glypican-3, and syndecan-1 showed downregulation in cancer tissues compared to normal tissues. In cancer tissue, the total amount of chondroitin sulfate (CS)/dermatan sulfate and heparan sulfate were lower and, interestingly, the level of disaccharide units of both 4S6S (CS-E) and 6S (CS-C) were higher compared to normal tissue. Also, overall lipids including glycolipids, a major glycomics target, were analyzed by hydrophilic interaction liquid chromatography mass spectrometry. Increase of lyso-phosphatidylcholine (phospholipid), sphingomyelin (sphigolipid), and four types of glycolipids (glucosylceramide, lactosylceramide, monosialic acid ganglioside, and globoside 4) in cancer tissue showed the possibility as potential biomarkers in colon cancer. While requiring the need for careful interpretation, this type of broad investigation gives us a better understanding of pathophysiological roles on glycosaminoglycans and glycolipids and might be a powerful tool for colon cancer diagnosis.
Introduction
Colon and rectal cancers (also known as colorectal cancer and hereafter referred to as colon cancer) are the fourth leading cancer diagnosis in the United States, and the second leading cause of cancer death. In 2012, there were an estimated 143,000 colon cancer diagnosis and 51,000 deaths (Siegel et al., 2012). While the majority of patients (80%) present with a malignancy treatable by surgery, the remaining 20% of patients develop a metastatic form of the disease, often leading to liver and lung cancer (Cartwright, 2012).
Proteoglycans (PGs) consist of specific core proteins that are covalently linked to glycosaminoglycan (GAG) chains, including chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS), heparin (HP), keratan sulfate (KS), and hyaluronic acid (HA). PGs can be classified into subgroups based on their core specific protein. The biological functions of PGs include embryonic development, cell adhesion and recognition, tissue morphogenesis, and the regulation of signaling molecules as co-receptors (Linhardt and Toida, 2004). Evidence is emerging that abnormal PG expression and accumulation can alter cell signaling and growth patterns, leading to tumorigenesis including proliferation, invasion, angiogenesis, and metastasis (Fuster and Esko, 2005; Theocharis et al., 2010).
GAGs are linear and negatively charged polysaccharides linked to PG core proteins present on the cell surfaces and in the extracellular matrix. Unique among the GAGs is hyaluronic acid (HA), which is nonsulfated and not attached to a core protein. GAG chains are composed of disaccharide repeating units such as uronic acid (D-glucuronic acid (GlcA) or L-iduronic acid (IdoA)) or galactose (D-Gal) and amino sugar (D-galactosamine (GalN) or D-glucosamine (GlcN)). This structural diversity results in their differential binding to proteins (Capila and Linhardt, 2002) and allows GAGs to play selective roles in physiological and pathophysiological events, including cancer metastasis and invasion. For example, CS-E (sulfo groups at the C-4 and C-6 of GalNAc) is known to bind specifically to growth factors such as vascular endothelial growth factor, heparin-binding EGF-like growth factors, midkine, and pleiotrophin (Deepa et al., 2002). These interactions accelerate vessel formation and co-localization, followed by angiogenesis in ovarian cancer.
Lipids, including phospholipids and sphingolipids, the major structural components of cellular membranes, are also implicated in the genesis and development of cancerous tissue (Santos and Schulze, 2012). Lipids are known to play important roles as signaling molecules, acting in cellular proliferation, adhesion, migration, survival, angiogenesis, and differentiation (Furuya et al., 2011; Ishii et al., 2004). Aberrant glycosylation and accumulation of certain glycosphingolipids in specific types of cancer might correlate to altered cell–cell communication or cell–substratum interaction. Aberrant glycosylation of lipids may also reflect aberrant transmembrane signaling and cell motility causing cancer metastasis.
Investigating the molecular differences between normal and cancer tissue types can serve two basic purposes. First, molecular differences may be causal, activating some signaling or metabolic pathway that drives the development of the cancer. Alternatively, molecular differences might be diagnostic, serving as a marker or byproduct of underlying biochemical changes identifying cancerous tissue. Thus, molecules that are discovered to be different between normal and cancerous tissue may be useful to identify potential therapeutic drug targets or for screening tissues to confirm a diagnosis.
Here, we introduce some findings through PG expression in normal and cancer tissues and the contents of GAG disaccharides and glycolipids in the same tissues. A novel analytical method is also presented for the characterization of the lipid content of normal and cancerous colon.
Materials and Methods
Materials
Normal mucosa tissues and colon cancer tissues, collected from each of 10 different patients, were obtained at Konyang University Hospital Daejeon, South Korea (Table 1). The colon cancer tissues were obtained surgically from the tumor edge and the specimens of normal mucosa were taken at 5 cm or more from the tumor margin. All patients provided written informed consent before the collection tissues samples. Samples were stored in liquid nitrogen before analyses. The clinicopathologic data were collected by reviewing medical charts and pathology records. The collection and use of the samples were approved by the Institutional Review Board of the Konyang University Daejeon Hospital.
Table 1.
Patient Information for Colon Tissue Samples
| # Patient | # Sample | Tissue | Age | Sex | Diagnosis |
|---|---|---|---|---|---|
| 1 | 1 | Normal | 61 | female | Sigmoid colon cancer |
| 2 | Cancer | ||||
| 2 | 3 | Normal | 73 | male | Sigmoid colon cancer |
| 4 | Cancer | ||||
| 3 | 5 | Normal | 74 | female | Rectosigmoid colon cancer |
| 6 | Cancer | ||||
| 4 | 7 | Normal | 80 | male | Ascending colon cancer |
| 8 | Cancer | ||||
| 5 | 9 | Normal | 81 | female | Sigmoid colon cancer |
| 10 | Cancer | ||||
| 6 | 11 | Normal | 73 | female | Sigmoid colon cancer |
| 12 | Cancer | ||||
| 7 | 13 | Normal | 69 | male | Cecal cancer |
| 14 | Cancer | ||||
| 8 | 15 | Normal | 70 | female | Rectal cancer with lung and liver metastasis |
| 16 | Cancer | ||||
| 9 | 17 | Normal | 56 | female | Rectal cancer |
| 18 | Cancer | ||||
| 10 | 19 | Normal | 73 | male | Rectal cancer with liver metastasis |
| 20 | Cancer |
Sample preparation and isolation of lipids and GAGs
The isolated colon tissues were first lyophilized, and each dried tissue was crudely homogenized by rough segmentation into 10–20 pieces, depending on tumor size. A portion of each sample was retained for protein analysis, and the tissue remaining was used for lipid and GAG analysis.
For lipid and GAG analysis, tissues were first extracted with a series of organic solvents, and further digested to allow for GAG extraction. Tissues were extracted with a mixture of chloroform and methanol over a 6-day period. Tissues were placed in 10 mL glass vials and extracted with three separate solvent solutions (solution A, 2:1 chloroform:methanol; solution B, 1:1 chloroform:methanol; solution C, 1:2 chloroform:methanol) of increasing polarity. Tissues were extracted with 5 mL of each solution twice; tissues were incubated with each solution for 24 h for a single extraction. After incubation, the extraction solutions were transferred to a second glass vial where they were allowed to evaporate (under a filter to prevent contamination) at room temperature in a hood to a uniform final volume of 2 mL. Before further processing, all samples were allowed to dry completely for 48 h. For GAG analysis, dried tissue samples were weighed and individually proteolyzed using Actinase E (55°C, 800 μL of 2 mg/mL aqueous solution) for 2 days. GAGs were next isolated using Vivapure Mini Q-H spin columns. Each spin column was first equilibrated with a solution containing urea and 3-[(3-chloramidopropyl) dimethylammonio]-1-propane sulfate (CHAPS 2 wt% in 8 M urea). CHAPS urea solution was added to each tissue, undigested particulates were removed from these solutions by centrifugation (5000 g, 30 min), and the resulting tissue solutions were then loaded on the Vivapure spin columns (500 g). Spin columns were washed with three column volumes of 200 mM NaCl to remove impurities. GAGs were released from the spin columns using two washes of 16% NaCl (0.5 column volumes). Methanol (80% total volume) was added to the released GAG solutions, precipitating the GAGs overnight at 4°C. Precipitated GAGs were recovered by centrifugation (5000 g, 30 min) and the isolated GAG precipitate was then dissolved in doubly distilled water and analyzed.
Liquid chromatography-mass spectrometry (LC-MS) for disaccharide compositional analysis
Isolated GAGs were subject to enzymatic depolymerization. GAGs (20 μg) were digested with a mixture of chondroitinase ABC (10 mU) and chondroitinase ACII (5 mU) at 37°C for 10 h and the resulting dissacharide products were isolated in the flow-through by centrifugal filtration (10 kDa molecular weight cut-off (MWCO)), freeze-dried, and were ready for LC-MS analysis. The remaining undigested (retained) GAGs were then incubated with a mixture of heparinases I, II, and III at 35°C for 10 h. The resulting disaccharide products were similarly isolated in the flow-through by centrifugal filtration (10 kDa MWCO filter), freeze-dried, and subjected to LC-MS analysis (Yang et al., 2012). Fluorescent labeling of disaccharide was next undertaken to ensure highly sensitive detection and to eliminate potential complications associated with UV absorbing impurities coming from the biopsy tissues being analyzed. The freeze-dried sample containing the GAG-derived disaccharides (∼2 μg) was added 10 μL of a 0.1 M 2-aminoacridone (AMAC) solution in acetic acid dimethyl sulfoxide (3:17, v/v) and mixed by vortexing for 5 min. Next, 10 μL of 1 M NaBH3CN was added in the reaction mixture and incubated at 45°C for 4 h. LC-MS analyses were performed on an Agilent 1200 LC/MSD instrument (Agilent Technologies, Inc. Wilmington, DE) equipped with a 6300 ion-trap and a binary pump followed by a UV detector equipped with a high-pressure cell. The column used was a poroshell120 C18 column (2.1×150 mm, 2.6 μm, Agilent) at 45°C. Solution A was 80 mM ammonium acetate solution and Solution B was methanol. Solution A and 15% solution B was flowed (150 μL/min) through the column for 5 min, followed by a linear gradient of 15%–30% solution B from 5 to 30 min. The column effluent entered the electrospray ionization (ESI)-MS source for continuous detection by MS. The electrospray interface was set in negative ionization mode with a skimmer potential of −40.0 V, a capillary exit of −40.0 V, and a source temperature of 350°C, to obtain the maximum abundance of the ions in a full-scan spectrum (150–1200 Da). Nitrogen (8 L/min, 40 psi) was used as a drying and nebulizing gas.
Mass spectroscopic analysis for lipids
A UPLC HILIC column (2.0×150 mm2, Waters Corp., Milford, MA) was used to separate the lipids. Mobile phase A was 5 mM ammonium acetate prepared with HPLC grade acetonitrile and 2% of HPLC grade acetic acid. Mobile phase B was 5 mM ammonium acetate prepared in HPLC grade methanol with 2% of acetic acid. Lipid samples were diluted 10-fold in a 1:1 mixture of mobile phase A and B. After injection of 5 μL lipid extract through an Agilent 1200 autosampler, HPLC binary pump was used to deliver the gradient from 98% A to 40% A over 25 min at a flow rate of 150 μL/min. The LC column was directly connected online to the standard ESI source of LTQ-Orbitrap XL FT MS (Thermo Fisher Scientific, San Jose, CA). The optimized parameters used to generate good ionization, including a spray voltage of 3.5 kV, a capillary voltage of 34 V, a tube lens voltage of 100 V, a capillary temperature of 300°C, a sheath flow rate of 30, and an auxiliary gas flow rate of 6. External calibration of mass spectra routinely produced a mass accuracy of better than 3 ppm. All FT mass spectra were acquired at a resolution 60,000 with 300-2000 Da mass range. The error in the analysis of the lipid content for each sample was 8%.
Protein isolation and immunoblotting
Dried colon tissue samples were cut into small pieces and homogenized using a mechanical homogenizer in the presence of Nonidet-P40 lysis buffer (Boston Bioproducts, Ashland, MA) containing a cocktail of protease and phosphatase inhibitors (Pierce, Rockford, IL). Samples were then lysed on ice for 30 min. The protein concentration was determined using the BCA assay (Pierce, Rockford, IL). Approximately 20 μg of total protein was loaded (under reducing conditions) and separated on 4%–15% gradient SDS-polyacrylamide gel. After transferring to nitrocellulose membrane, proteins of interest were detected using relevant primary and HRP-conjugated secondary antibodies, followed by chemiluminescent (Pierce) exposure on high performance chemiluminescence film (GE Healthcare, Little Chalfont, UK). Primary antibodies used were anti-carcinoembryonic antigen, anti-epidermal growth factor receptor (Abcam, Cambridge, MA), anti-γ-tubulin (Sigma Aldrich), anti-decorin, anti-syndecan 1 and 2, anti-glypican 1and 3, and anti-β-actin (Santa Cruz Biotechnology, Dallas, TX).
Statistical analysis
The measured lipid abundance (ion abundance) was normalized to tissue dry weight and the cancerous and normal tissue distributions were compared using a Paired-Samples T-Test (IBM SPSS Statistics 20). Significance was taken to be p<0.05 for all tests, unless noted otherwise. Lipid structures that showed significant differences were then further analyzed to separate general metabolic shifts from potential as biomarkers. Good biomarkers showed consistent differences: 90% to 100% of patient data followed the overall tissue differences between normal and cancerous tissues. Lipid differences representing metabolic shifts showed clear differences in a majority of, but not all, sample sets.
Results
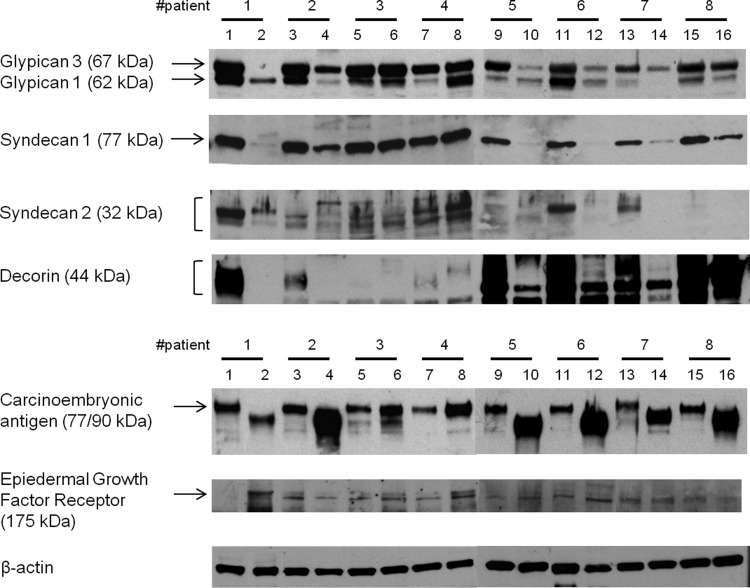
Western blotting was first used to check for the levels of carcinoembryonic antigen (CEA) and epidermal growth factor receptor (EGFR), two markers commonly associated with cancers (Kuriyama et al., 2009; Moetel et al., 1986). As expected, all 8 cancer samples showed higher CEA levels compared to those of normal tissues. EGFR levels increased in 5 of the 8 cancer tissues. Next, the presence of common PG core proteins was tested in 16 of 20 tissue samples, where sufficient tissue was available (Fig. 1). Six of 8 normal tissue samples showed a much higher expression of decorin than did cancer tissues. Decorin, a member of the small leucine-rich CS/DS PGs (Zhao et al., 2013), has been reported to be involved in cell growth and survival (Zhu et al., 2005) and to inhibit cancer cell proliferation by blocking tyrosine kinase activity such EGFR family and Met receptor (Goldoni et al., 2009). Lower expression of decorin and higher expression of EGFR are supportive of the reported interactions between decorin and EGFR in colon cancer. The major HS PGs are transmembrane-anchored syndecans and glycosylphophatidylinositol-anchored glypicans. Among them, syndecans-1, 2 and glypican-1, 3 were evaluated in both normal and cancer tissues. While glypican-3, known to be downregulated in several cancers, was observed in all normal tissues, little or no glypican-3 could be detected in 4 of 8 in cancer samples. Glypican-1 showed no clear difference in expression levels in normal and cancer tissues.
FIG. 1.
Expression of representative proteoglycans and protein markers associated with colon cancer in colon tissue samples. 16 out of 20 samples are shown. An increase in expression of both CEA and EGFR were shown in cancer tissue samples. Among proteoglycans, lower expression of decorin, glypican-3, and syndecan-1 was observed in cancer tissue samples compared to normal tissue samples. β-Actin was used as loading control. The biopsy tissue was limited in several cases, resulting in unclear Western blots.
Expression of syndecan-1 and syndecan-2 HSPG, known as tumor modulators, was next investigated. Syndecan-1 was observed in all (8 of 8) normal tissue samples and all cancer tissues, with the exception of one (sample 8) that showed enhanced levels of syndecan-1. Interestingly, the expression pattern of syndecan-2, which is normally highly expressed in colon carcinoma (Beauvais and Rapraeger, 2004; Han et al., 2004; Vicente et al., 2013), did not show enhanced levels in the cancer tissues studied. Our data seem to correlate better to a report indicating that colon cancer cell lines HT29 and Caco-2 that show reduced syndecan-2 expression, and the report that decreased levels of syndecan-2 do not influence proliferation and invasion by these cell lines (Han et al., 2004).
We next examined GAG amount and disaccharide composition in both normal and cancer tissues. The total GAG amounts, for both CS/DS and HS, were higher in normal tissues than in cancer tissues (Table 2). The total amount of CS/DS was 5.28±0.55 mg/g tissue in normal tissue samples, and 2.92±0.43 mg/g tissue in cancer samples. But, in the case of HS, a very slight elevation was found in normal (1.14±0.13) compared to cancer tissues (0.97±0.12). The total GAG content can be rationalized on the basis that all PGs tested in immunoblotting showed much higher expression in normal tissues than in cancer, thus, the total GAG content was higher in normal tissue. The composition of 4S disaccharide (CS-A) in both normal (89.23%) and cancer (74.85%) tissues was higher than any other disaccharides. Cancer tissues showed significant increases in the levels of 4S6S disaccharide (CS-E) and also 6S disaccharide (CS-C). Normal tissues showed higher levels of 2S4S disaccharide. The HS of normal tissues was elevated in TriS and 2S disaccharides. All numbers of disaccharide contents obtained from every patient (%, relative peak area) are shown in Supplementary Table S1 (for CS/DS) and Supplementary Table S2 (for HS) (Supplementary Material is available online at www.liebertpub.com). Despite some observed differences between normal and cancer tissue groups, it is unclear whether these differences are sufficient to be used as diagnostic or therapeutic as biomarkers.
Table 2.
LC-MS Analysis of CS/DS and HS Disaccharide Content1
| Percentage (%, relative peak area)* | Total amount (mg/g tissue) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CS/DS | Tri S | 2S4S | 2S6S | 4S6S | 2S | 4S | 6S | 0S | CS/DS | HA |
| Normal | – ** | 1.70±0.36 | 0.20±0.04 | 0.66±0.22 | – | 89.23±1.77 | 6.10±0.95 | 2.09±0.58 | 5.28±0.55 | 2.43±0.21 |
| Cancer | – | 0.49±0.13 | 0.32±0.05 | 1.52±0.21 | – | 74.85±2.53 | 19.84±2.37 | 2.98±0.53 | 2.92±0.43 | 1.25±0.26 |
| HS | Tri S | NS6S | NS2S | NS | 2S6S | 6S | 2S | 0S | HS |
|---|---|---|---|---|---|---|---|---|---|
| Normal | 13.38±1.82 | 3.81±0.47 | 14.01±1.44 | 21.79±0.80 | – | 4.19±0.63 | 3.44±1.73 | 39.38±1.82 | 1.14±0.13 |
| Cancer | 7.99±1.74 | 4.40±1.07 | 12.54±1.35 | 23.21±0.67 | – | 6.09±0.53 | 1.10±0.16 | 44.67±1.80 | 0.97±0.12 |
See Supplementary Tables S1 and S2 for complete data set.
average±standard error; **(–), not detected.
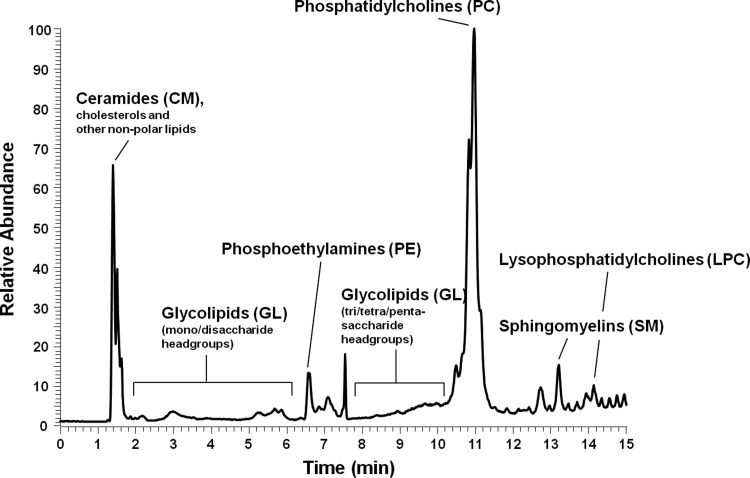
Lipids were next extracted from twenty colon tissue samples, injected onto and separated by a UPLC HILIC column, and followed by direct injection into a mass spectrometer, and the lipid content was determined by ion response. Method reproducibility was checked within 10% variation. A typical lipid separation profile, in which lipid content was measured from relative ion abundance, is shown in Figure 2. The lipids exhibit a wide variety structures due to the polydisperse lengths of the lipid side chains. In this analysis, the most common lipid side chain structures for each lipid type were analyzed. Phospholipids compose the majority of the cell membrane, and as such were the most abundant lipid measured. Glycolipids (Fig. 3), a subset of sphingolipids, showed some of the most interesting differences between cancer and normal tissue samples despite being only a minor membrane component. For each type of lipid, the content of several structures and the total content, or the sum of all structures in that type, were compared between the normal and cancer tissues. Unless otherwise specified, lipid content was found to increase in cancerous tissue compared to normal tissue (Table 3).
FIG. 2.
Profile of lipid analyzed from colon tissues by HILIC-FTMS. Lipids can be separated by polarity and determined by relative ion response, which the lipid type with higher polarity showed late retention time.
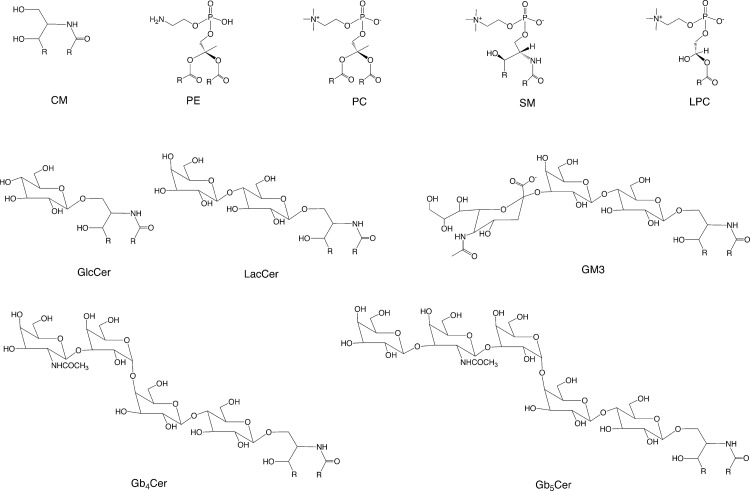
FIG. 3.
Generalized structures of phospholipids, sphigolipids, and glycolipid analyzed. Lipid polarity increases with the numbers of sugar groups. R is the lipid side chain.
Table 3.
Overview of Lipid Structures and its Responses Analyzed from Colon Tissue Samples
| Lipid family | Lipid structure | Structure analyzed* | Average normal tissue response | Average cancerous tissue response | Significant difference |
|---|---|---|---|---|---|
| Phospholipids | Phosphoethylamines | Sum of all structures | 1,200,000 | 1,900,000 | No significance found |
| Phosphatidylcholines | PC 32 | 3,400,000 | 6,600,000 | p<0.01 | |
| PC 32:1 | 1,000,000 | 4,400,000 | p<0.01 | ||
| Lyso-phosphatidylcholines | Sum of all structures | 2,700,000 | 4,800,000 | p<0.05 | |
| Sphingolipids | Ceramides | Sum of all structures | 1,800,000 | 2,800,000 | No significance found |
| Glycolipids | Sum of GlcCer structures | 71,000 | 160,000 | p<0.01 | |
| Sum of LacCer structures | 40,000 | 250,000 | p<0.01 | ||
| GM3 | GM3 16:0 | 3,500 | 6,700 | p<0.05 | |
| GM3 24:0 | 3,900 | 6,900 | p<0.05 | ||
| Sum of Gb4Cer structures | 9,100 | 22,000 | p<0.01 | ||
| Sum of Gb5Cer structures | 28,000 | 13,000 | p<0.05 | ||
| Sphingomyelins | Sum of all structures | 8,800,000 | 11,000,000 | p<0.05 |
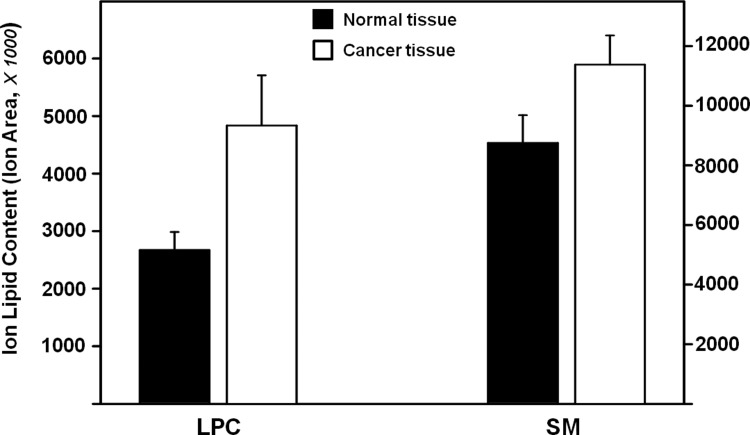
In the phospholipid family, few significant differences were seen. No significant differences were seen in the phosphoethylamine structures analyzed, and only two structures (PC 32 and PC 32:1) in the phosphatidylcholine group showed significant differences. In the lyso-phosphatidylcholine (LPC) group, four of the five structures analyzed showed significant differences. When the sums of the LPC contents were compared, lipid levels were found to increase in the cancerous tissues (Fig. 4). In the sphingolipid family, no differences were found in ceramide (CM) structures, but significant differences were found in all other lipid types. A significant increase was found in the lipid content of sphingomyelin (SM) lipids isolated from cancerous tissue (Fig.4), but much like the LPC lipids, differences between the normal and cancerous tissues seem to be driven by increased levels in relatively few patients with seven out of ten patients showing increased levels and three patients showing decreased levels.
FIG. 4.
Total ion lipid contents of lyso-phosphatidylcholines in the phospholipid family and sphingomyelins in the sphingolipid family. The sum of LPC and SM content, respectively, showed significant increase in cancer tissues compared to normal tissues. Error of analysis for each sample is 8% (not shown).
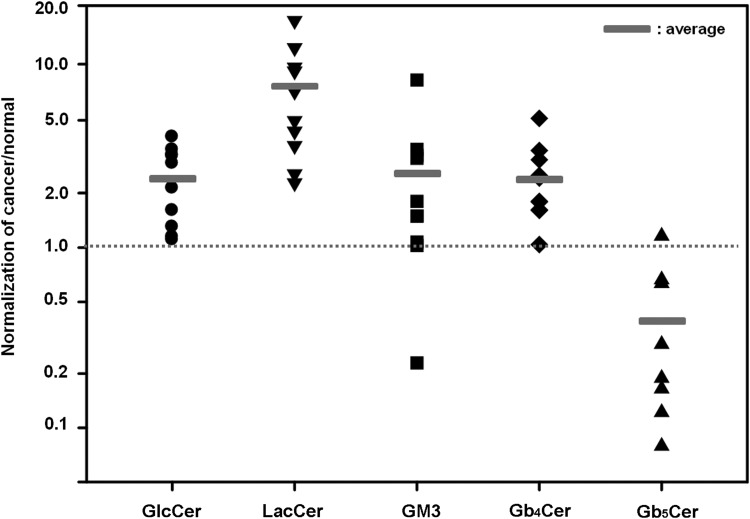
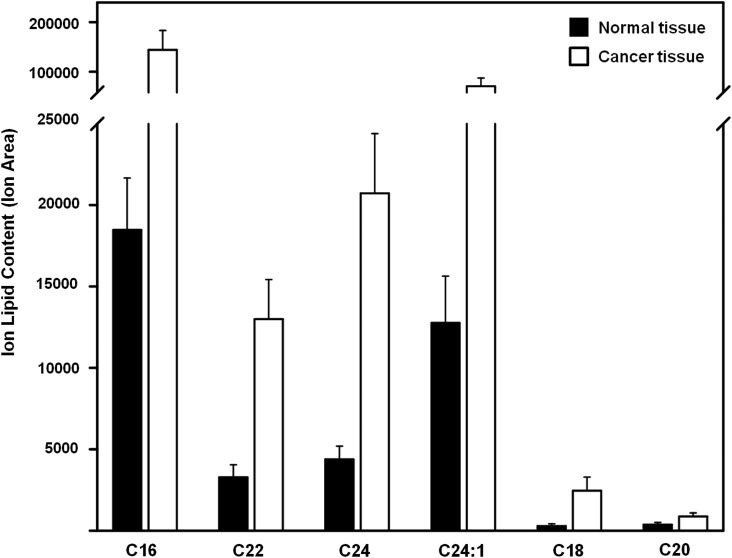
Next, glycolipids in the sphingolipid family were analyzed. The structures of glucosylceramide (GlcCer), lactosylceramide (LacCer), monosialic ganglioside 3 (GM3), globoside 4 (Gb4Cer), and globoside 5 (Gb5Cer) are shown in Figure 3. All showed significant differences in normal and cancerous tissues. Four glycolipid types (GlcCer, LacCer, GM3, and Gb4Cer) showed the significant increases in cancerous tissue (Table 3). Only Gb5Cer showed a significant decrease in the cancerous tissues compared to normal tissues. The amount of each glycolipid (normalized by tissue weight) from individual patient cancer tissue sample was divided by the amount of each glycolipid found in the matching normal tissue sample (Fig. 5). Each glycolipid exhibited a statistically significant difference in cancerous tissue in nine out of ten patients studied. Most pronounced was the average ratio of 7.27 for LacCer (Table 4). Because the LacCer content increased more in cancer tissue than the content of other lipids and also showed a lipid increase in every single patient tested, we decided to examine the sub-structures (resulting from different lipid chain length) present in LacCer. The lipid response levels for the LacCer substructures are shown in Figure 6. All of these substructures showed a significant increase in cancer tissues with massive differences seen in LacCer -C16 and LacCer -C24, suggesting that the LacCer structures might represent the best potential biomarker for colon cancer uncovered in the current study.
FIG. 5.
Distribution of patients according to the average ratio of glycolipid contents normalized by cancer over normal tissue. The number more than one (dotted line) means that the lipid expression level in cancer tissue is higher than in normal tissue. Small rectangle represents the average of each glycolipid normalized.
Table 4.
Occurrence of Glycolipids from Individual Patient
| # Patient | GlcCer | LacCer | GM3 | Gb4Cer | Gb5Cer |
|---|---|---|---|---|---|
| 1 | 2.91 | 4.97 | 1.50 | 1.61 | 0.66 |
| 2 | 4.06 | 2.27 | 3.48 | 1.79 | – |
| 3 | 1.15 | 9.63 | 1.80 | 3.42 | 0.63 |
| 4 | 2.13 | 12.17 | 1.50 | 1.62 | 1.16 |
| 5 | 3.20 | 7.15 | 1.03 | 2.42 | 0.08 |
| 6 | 1.30 | 9.17 | 1.07 | 1.80 | 0.19 |
| 7 | 3.24 | 2.55 | 3.22 | 3.05 | 0.29 |
| 8 | 1.61 | 16.80 | 3.10 | 5.13 | – |
| 9 | 1.10 | 3.64 | 0.23 | 1.04 | 0.16 |
| 10 | 3.46 | 4.36 | 8.24 | 2.50 | 0.12 |
| *Ave±S.E. | 2.42±0.34 | 7.27±1.48 | 2.52±0.72 | 2.44±0.38 | 0.41±0.12 |
FIG. 6.
Lipid content of LacCer substructures in normal and cancerous tissues. All types of LacCer substructures in cancer tissue samples showed significantly higher content than in normal tissue samples (p<0.01). Error of analysis for each sample is 8% (not shown).
Discussion
PGs and GAGs are involved in numerous biological and physiological processes. Each tissue produces specific repertoires of GAG chains covalently linked to PGs for the binding and regulation of distinct proteins, including chemokines, cytokines, growth factors, morphogens, enzymes, extracellular matrix proteins, or adhesion molecules (Capila and Linhardt, 2002). Dysregulation and changes in expression of PGs and content of GAGs can lead to abnormal circumstances such as malignancy, and the events may become diagnostic and prognostic factors in cancer.
In this study, we analyzed 20 clinically obtained tissue samples, 10 from normal, and 10 from cancer tissues (Table 1). The difference in CEA and EGFR protein in normal and cancer tissues were identified. As expected, an increase of expression levels of CEA, γ-tubulin (data not shown), and, to a lesser extent, EGFR were observed in cancer tissues. Known tumor-related PGs, including glypican-1,-3, syndecans-1,-2, and decorin, were investigated. The expression of glypican-3 and syndecan-1 was reduced in colon cancer tissues compared with normal tissues. Glypican-3, a well-studied PG in cancer research, showed very different patterns of expression during tumor progression, it has exhibited reduced or undetectable expression level in several types of cancers and has played a role as an inhibitor of cell growth and an inducer of apoptosis (Gonzalez et al., 1998; Lin et al, 1999; Murthy et al, 2000). The expression of syndecan-1 was reduced in cancer tissue as compared with normal tissue. Syndecan-1 is reportedly decreased or absent in 87% of adenocarcinomas (Hashimoto et al., 2008). Interestingly, the levels of syndecan-2 in both normal and cancer tissues showed no clear trend. There is still controversy over the expression of syndecans in cancer. The Karamanos group showed that syndecans-2 and -4 as well as glypican-1 were overexpressed in the breast cancer lines compared to normal mammary cells (Tsonis et al., 2013). In contrast, Vicente et al. reported that syndecan-2 was highly expressed and the mRNA expression level of syndecan-1 and -4 were significantly reduced in highly metastatic colon carcinoma cells (Vicente et al., 2013). Oh and co-workers (Han et al., 2004) showed that Caco-2 and HT29 cells endogenously expressed as low levels of syndecan-2, whereas HCT116 cells expressed high levels of syndecan-2, in the same type of human colon adenocarcinomas. These dual roles of syndecans in cancer may reflect tissue phenotype, stage-specificity, and their complex interaction with other extracellular components. Although our immunoblotting data do not fully support PGs as independent biomarkers in colon cancer, it is still important to examine further the correlation between PGs and colon tissues with different clinical observations in human cancer. Further investigation is still needed, along with large datasets and detailed histological evaluation.
We also investigated the total amount of GAG and GAG disaccharide composition in both normal and cancer tissues (Table 2 and Supplementary Table S1). Overall, the amount of CS/DS and HS in normal tissue samples were lower, as compared to cancer tissue. Interestingly, the relative amounts of 4S6S (CS-E) and 6S (CS-C) CS/DS GAGs were higher in cancer tissue. At the same time, TriS and 2S disaccharides in HS GAGs were reduced in cancer tissues compared to normal tissues. These data are consistent with other reported research on GAGs in cancer. Most adherent cancer cells have shown higher levels of HS-0S and significantly lower levels of HS sulfation (Yamada et al., 2012). This has been explained by increased levels of HS sulfatases in multiple types of tumors, resulting in inhibition of signaling, and the onset of invasion and metastasis in cancer (Castro et al., 2008; Kudo et al., 2006; Lai et al., 2008). Also, a recent publication of Sugahara group reported that the proportion of CS-E unit increased in some types of cancers such as ovarian and pancreatic cancers, resulting in the promotion of cancer cell proliferation, motility, and angiogenesis through the function of CS-E binding to various growth factors (Mizumoto and Sugahara, 2013). Also, a glycosaminoglycanomic study of breast cancer showed similar results, suggesting overall content of GAGs and increased levels of 6S (CS-C) in cancer tissues (Weyers et al., 2012).
In the current study, several lipid structures were found to have significant changes in expression levels in cancerous tissues (Table 3). Among these, glycolipid structures (Fig. 3) appeared to be promising lipid structures to serve as biomarkers. Lipid structures that are less promising biomarkers (i.e., where <90% of patients show a corresponding increase in cancerous tissue) may still serve as important indicators of underlying shifts in lipid metabolism and signaling in cancerous tissue. The only phospholipid to display a consistent increase in cancerous tissue was the LPC lipid type (Fig. 4). While the observed increases in cancerous tissue LPC levels are not consistent across all patients, the increase in LPC may indicate underlying metabolic shifts in the cancerous tissue. Increased levels of LPC are thought to act as a growth factor, stimulating cell proliferation in both ovarian cancer (Fang et al., 2000) and endothelial cells (Sato et al., 1998). LPC, a metabolite of PC, has been shown to decrease in the plasma of cancer patients of several different cancer types (not including colon cancer) (Taylor et al., 2007). However, ras-transformed cell lines (associated with the cell proliferation and differentiation and with a variety of cancers, including colon cancer) (Yamamoto et al., 1999) exhibit an increased PC synthesis and degradation rate (Taylor et al., 2007) and ras-transformed cell lines consume PC and LPC at a higher rate than normal tissue (Baburina and Jackowski, 1999). The decreases in plasma levels are thus thought to be driven by the increased LPC consumption of the cancerous tissue (Yamamoto et al., 1999), which may result in increased tissue content in colon cancer. While some possible LPC receptors have been identified (Chang et al., 2008; Ishii et al., 2004), the mechanisms and signaling pathways have yet to be clearly established.
The sphingolipid SM was found to be significantly increased in cancerous tissue (Fig. 4); much like LPC, this increase was not seen in all patients, but may indicate an underlying metabolic shift in the tumor cells. While SM levels have been found to be lowered in some colon cancer studies (analyzed with magnetic resonance) (Merchant et al., 1991), several studies support the idea that SM is important in carcinogenesis, including an increase in SM in the presence of a known carcinogen in a rat model of colon cancer (Dahiya et al., 1987). SM is implicated as a cancer reducer in a number of studies, possibly acting to modulate important inflammatory responses.
Four of the glycolipids analyzed in this work (GlcCer, LacCer, GM3 16:0, and Gb4Cer) showed significant increases in cancerous tissue (Fig. 5 and Table 4) and showed promise as possible biomarkers. Several studies have linked glycolipids with colon cancer. Specific glycolipids structures have been identified as colon cancer biomarkers (Watanabe et al., 1976), apoptosis suppressants in colon cancer (Kakugawa et al., 2002), and as agents of metastasis in adenocarcinoma (Inufusa et al., 2001). In addition, certain forms of glycolipids have been implicated in increased E-selectin binding (important for metastasis) (Burdick et al., 2003; Miyazaki et al., 2004) and cell invasion and tumor budding (Oshiro et al., 2012). Of the lipid structures analyzed here, GM3 is the only structure to have been studied in depth in the context of cancer growth and metastasis. GM3 studies have shown higher expression levels in cancer (Devenica et al., 2011), GM3 influence tumor growth and phenotype (Taïeb et al., 2009), and potential use of GM3 as a melanoma vaccine target (Carr et al., 2001).
The observed significant increases in LPC, SM, GlcCer, LacCer, GM3 16:0, and Gb4Cer all have some preliminary literature studies supporting these findings. In particular, LacCer was shown to have cancerous lipid expression levels that were at least double that of normal tissue in all patients, making it the most promising colon cancer biomarker identified here (Fig. 6).
The key in defining a molecule as biomarker or drug target is a full understanding of its signaling and/or metabolic pathways. As part of these efforts, glycomics approach is being successfully used to examine the structure and function of glycans, glycolipids, glycoproteins, and glycan-binding proteins (Ly et al., 2010; Nairn et al., 2007). In the current study, we have emphasized the consequences of structure and composition of glycolipids in colon cancer. The data presented suggest that glycolipids, GAG, and PG can be potential diagnostic and therapeutic biomarkers in colon cancer.
Supplementary Material
Abbreviations Used
- CEA
carcinoembryonic antigen
- CM
ceramide, N-acyl-sphing-4-enine
- CS
chondroitin sulfate
- DS
dermatan sulfate
- EGFR
epidermal growth factor receptor
- HILIC
hydrophilic interaction liquid chromatography
- HS
heparan sulfate
- LPC
lysophosphatidylcholine, 1-acyl-sn-glycero-3-phosphocholine
- Gb4Cer
globoside 4, GalNAcβ1-3Galα1-4Galβ1-4Glcβ-ceramide
- Gb5Cer
globoside 5, Galβ1-3GalNAcβ1-3Galα1-3Galβ1-4Glcβ-ceramide
- GlcCer
glucosylceramide, Glcβ-ceramide
- GM3
monosialic ganglioside, NeuAcα2-3Galβ1-4Glcβ-ceramide
- LacCer
lactosylceramide, Galβ1,4Glcβ-ceramide
- PC
phosphatidylcholine, 1,2-diacyl-sn-glycero-3-phosphocholine
- PE
phosphoethanolamine, 1,2-diacyl-sn-glycero-3-phosphoethanolamine
- PG
proteoglycan
- SM
sphingomyelin, N-acyl-sphing-4-enine-1-phosphocholine
Acknowledgments
The authors acknowledge support from the National Institutes of Health in the form of grants GM38060, ES020903, and HL62244. This work was supported in part by Konyang University Myunggok Research Fund of 2013.
Author Disclosure Statement
No competing financial interests exist.
References
- Baburina I, and Jackowski S. (1999). Cellular responses to excess phospholipid. J Biol Chem 274, 9400–9408 [DOI] [PubMed] [Google Scholar]
- Beauvais DM, and Rapraeger AC. (2004). Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick MM, McCaffery JM, Kim YS, Bochner BS, and Konstantopoulos K. (2003). Colon carcinoma cell glycolipids, integrins, and other glycoproteins mediate adhesion to HUVECs under flow. Am J Physiol Cell Physiol 284, C977–C987 [DOI] [PubMed] [Google Scholar]
- Capila I, and Linhardt RJ. (2002). Heparin-protein interactions. Angew Chem Int Ed Engl 41, 391–412 [DOI] [PubMed] [Google Scholar]
- Carr A, Mazorra Z, Alonso DF, et al. (2001). A purified GM3 ganglioside conjugated vaccine induces specific, adjuvant-dependent and non-transient antitumour activity against B16 mouse melanoma in vitro and in vivo. Melanoma Res 11, 219–227 [DOI] [PubMed] [Google Scholar]
- Cartwright TH. (2012). Treatment decisions after diagnosis of metastatic colorectal cancer. Clin Colorectal Cancer 11, 155–166 [DOI] [PubMed] [Google Scholar]
- Castro NP, Osório CA, Torres C, et al. (2008). Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res 10, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DH, Deng H, Matthews P, et al. (2008). Inflammation-associated lysophospholipids as ligands for CD1d-restricted T cells in human cancer. Blood 112, 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R, Dudeja PK, and Brasitus TA. (1987). Premalignant alterations in the glycosphingolipid composition of colonic epithelial cells of rats treated with 1,2-dimethylhydrazine. Cancer Res 47, 1031–1035 [PubMed] [Google Scholar]
- Deepa SS, Umehara Y, Higashiyama S, Itoh N, and Sugahara K. (2002). Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. Implications as a physiological binding partner in the brain and other tissues. J Biol Chem 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- Đevenica D, Čikeš Čulić V, Vuica A, and Markotić A. (2011). Biochemical, pathological and oncological relevance of Gb3Cer receptor. Med Oncol 28, S675–S684 [DOI] [PubMed] [Google Scholar]
- Fang X, Gaudette D, Furui T, et al. (2000). Lysophospholipid growth factors in the initiation, progression, metastases, and management of ovarian cancer. Ann NY Acad Sci 905, 188–208 [DOI] [PubMed] [Google Scholar]
- Furuya H, Shimizu Y, and Kawamori T. (2011). Sphingolipids in cancer. Cancer Metastasis Rev. 30, 567–576 [DOI] [PubMed] [Google Scholar]
- Fuster MM, and Esko JD. (2005). The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- Goldoni S, Humphries A, Nyström A, et al. (2009). Decorin is a novel antagonistic ligand of the Met receptor. J Cell Biol 185, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AD, Kaya M, Shi W, et al. (1998). OCI-5/GPC3, a glypican encoded by a gene that is mutated in the Simpson-Golabi-Behmel overgrowth syndrome, induces apoptosis in a cell line-specific manner. J Cell Biol 141, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I, Park H, and Oh ES. (2004). New insights into syndecan-2 expression and tumourigenic activity in colon carcinoma cells. J Mol Histol 35, 319–326 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Skacel M, and Adams JC. (2008). Association of loss of epithelial syndecan-1 with stage and local metastasis of colorectal adenocarcinomas: An immunohistochemical study of clinically annotated tumors. BMC Cancer 8, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inufusa H, Adachi T, Kiyokawa T, et al. (2001). Ley glycolipid-recognizing monoclonal antibody inhibits procoagulant activity and metastasis of human adenocarcinoma. Int J Oncol 19, 941–946 [DOI] [PubMed] [Google Scholar]
- Ishii I, Fukushima N, Ye X, and Chun J. (2004). Lysophospholipid receptors: Signaling and biology. Annu Rev Biochem 73, 321–354 [DOI] [PubMed] [Google Scholar]
- Kakugawa Y, Wada T, Yamaguchi K, et al. (2002). Up-regulation of plasma membrane-associated ganglioside sialidase (Neu3) in human colon cancer and its involvement in apoptosis suppression. Proc Natl Acad Sci USA 99, 10718–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Ogawa I, Kitajima S, et al. (2006). Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res 66, 6928–6935 [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Bettencourt-Dias M, Hoffmann I, Arnold M, and Sandvig L. (2009). γ-Tubulin-containing abnormal centrioles are induced by insufficient Plk4 in human HCT116 colorectal cancer cells. J Cell Sci 122, 2014–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Yu C, et al. (2008). Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47, 1211–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Huber R, Schlessinger D, and Morin PJ. (1999). Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res 59, 807–810 [PubMed] [Google Scholar]
- Linhardt RJ, and Toida T. (2004). Role of glycosaminoglycans in cellular communication. Acc Chem Res 37, 431–438 [DOI] [PubMed] [Google Scholar]
- Ly M, Laremore TN, and Linhardt RJ. (2010). Proteoglycomics: Recent progress and future challenges. OMICS 14, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant TE, Kasimos JN, de Graaf PW, Minsky BD, Gierke LW, and Glonek T. (1991). Phospholipid profiles of human colon cancer using 31P magnetic resonance spectroscopy. Int J Colorectal Dis 6, 121–126 [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Ohmori K, Izawa M, et al. (2004). Loss of disialyl Lewis(a), the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (Siglec-7) associated with increased sialyl Lewis(a) expression on human colon cancers. Cancer Res. 64, 4498–4505 [DOI] [PubMed] [Google Scholar]
- Mizumoto S, and Sugahara K. (2013). Glycosaminoglycans are functional ligands for receptor for advanced glycation end-products in tumors. FEBS J. 280, 2462–2470 [DOI] [PubMed] [Google Scholar]
- Moertel CG, O'Fallon JR, Ho LW, O'Connell MJ, and Thynne GS. (1986). The preoperative carcinoembryonic antigen test in the diagnosis, staginh, and prognosis of colorectal cancer. Cancer 58, 603–610 [DOI] [PubMed] [Google Scholar]
- Murthy SS, Shen T, De Rienzo A, et al. (2000). Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene 19, 410–416 [DOI] [PubMed] [Google Scholar]
- Nairn AV, Kinoshita-Toyoda A, Toyoda H, et al. (2007). Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res 6, 4374–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro R, Yamamoto H, Takahashi H, et al. (2012). C4.4A is associated with tumor budding and epithelial-mesenchymal transition of colorectal cancer. Cancer Sci 103, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CR, and Schulze A. (2012). Lipid metabolism in cancer. FEBS J 279, 2610–2623 [DOI] [PubMed] [Google Scholar]
- Sato N, Kokame K, Shimokado K, Kato H, and Miyata T. (1998). Changes of gene expression by lysophosphatidylcholine in vascular endothelial cells: 12 up-regulated distinct genes including 5 cell growth-related, 3 thrombosis-related, and 4 others. J Biochem 123, 1119–1126 [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, and Jemal A. (2012). Cancer statistics. CA: Cancer J ClinicianS 62, 10–29 [DOI] [PubMed] [Google Scholar]
- Taïeb N, Maresc M, Guo XJ, Garmy N, Fantini J, and Yahi N. (2009). The first extracellular domain of the tumour stem cell marker CD133 contains an antigenic ganglioside-binding motif. Cancer Lett 278, 164–173 [DOI] [PubMed] [Google Scholar]
- Taylor LA, Arends J, Hodina AK, Unger C, and Massing U. (2007). Plasma lyso-phosphatidylcholine concentration is decreased in cancer patients with weight loss and activated inflammatory status. Lipids Health Dis 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis AD, Skandalis SS, Tzanakakis GN, and Karamanos NK. (2010). Proteoglycans in health and disease: Novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J 277, 3904–3923 [DOI] [PubMed] [Google Scholar]
- Tsonis AI, Afratis N, Gialeli C, et al. (2013). Evaluation of the coordinated actions of estrogen receptors with epidermal growth factor receptor and insulin-like growth factor receptor in the expression of cell surface heparan sulfate proteoglycans and cell motility in breast cancer cells. FEBS J 280, 2248–2259 [DOI] [PubMed] [Google Scholar]
- Vicente CM, Ricci R, Nader HB, and Toma L. (2013). Syndecan-2 is upregulated in colorectal cancer cells through interactions with extracellular matrix produced by stromal fibroblasts. BMC Cell Biol 14, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Matsubara T, and Hakomori S. (1976). Alpha-L-fucopyranosylceramide, a novel glycolipid accumulated in some of the human colon tumors. J Biol Chem 251, 2385–2387 [PubMed] [Google Scholar]
- Weyers A, Yang B, Yoon DS, et al. (2012). A structural analysis of glycosaminoglycans from lethal and nonlethal breast cancer tissues: Toward a novel class of theragnostics for personalized medicine in oncology? OMICS 16, 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Mitsui Y, Kakoi N, Kinoshita M, Hayakawa T, and Kakehi K. (2012). One-pot characterization of cancer cells by the analysis of mucin-type glycans and glycosaminoglycans. Anal Biochem 421, 595–606 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Taya S, and Kaibuchi K. (1999). Ras-induced transformation and signaling pathway. J Biochem 126, 799–803 [DOI] [PubMed] [Google Scholar]
- Yang B, Chang Y, Weyers AM, Sterner E, and Linhardt RJ. (2012). Disaccharide analysis of glycosaminoglycan mixtures by ultra-high-performance liquid chromatography-mass spectrometry. J Chromatogr A 1225, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang B, Solakylidirim K., et al. (2013). Sequence analysis and domain motifs in the porcine skin decorin glycosaminoglycan chain. J Biol Chem 288, 9226–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JX, Goldoni S, Bix G, et al. (2005). Decorin evokes protacted internalization and degradation of the epidermal growth factor receptor via caveolar endocytosis. J Biol Chem 280, 32468–32479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.