Abstract
We report a novel HIV-1 circulating recombinant form (CRF64_BC) that was isolated from five epidemiologically unlinked HIV-infected persons in Yunnan province. CRF64_BC was composed of subtype B and subtype C, with five short subtype B segments inserted into the subtype C backbone. Phylogenetic analysis demonstrated that the C subregion was correlated with the India C lineage, which was transmitted into China in the early 1990s. The evolutionary history of the B subregion was not as clear as the C subregion, as the short length of this region yielded poor phylogenetic results. Dehong is considered the epicenter of HIV-1 in China, and recombinant strains such as CRF07_BC and CRF08_BC, which also originated from this region, have spread widely in China. The newly emerged CRF64_BC increases the complexity of the HIV epidemic in China and complicates the development of subtype-specific tools against HIV transmission.
The M group of HIV-1 is the most widely epidemic form of HIV in the world, and a great number of pure subtypes and recombinant strains cocirculate globally. To date, nearly 20% of HIV-1 isolates are recombinant forms, which contain more than 60 circulating recombinant forms (CRFs).1 The analysis and surveillance for recombinant strains therefore play a major part in the prevention and control efforts for HIV. In China, three of the four most prevalent strains are CRF strains, including CRF01_AE, CRF07_BC, and CRF08_BC, which together consist of more than 90% of all cases of infection.2,3 Molecular evidence has traced the entry and origination of these CRFs to Yunnan province, typically considered to be China's epicenter for HIV,3,4,5 and the proportion and complexity of recombinant strains found here are much higher than anywhere else in China.6,7 In this study, we report a new CRF (CRF64_BC) identified in high-risk populations.
Blood plasma was collected from five HIV-positive patients, YNFL10, YNFL16, YNFL22, YNFL31, and YNFL33, in Dehong prefecture, Yunnan. Basic epidemiological information is shown in Table 1. These patients were recruited from a national HIV-1 drug resistance survey in Yunnan. The study was approved by the institutional review boards of the National Center for AIDS/STD Control and Prevention of China. An epidemiological survey suggested that there was no obvious transmission linkage between them. Near full-length genome (NFLG) amplification and sequencing were performed as previously reported.8
Table 1.
Sociodemographic Characteristics of Study Subjects
| Strain name* | Sampling year | Geographic origin (prefecture, province) | Sex | Age | Ethnic group | Risk factor | Accession number |
|---|---|---|---|---|---|---|---|
| YNFL10.1 | 2010 | Dehong, Yunnan | Male | 33 | Jingpo | Injecting drug user | KC870032 |
| YNFL10.2 | KC870033 | ||||||
| YNFL16 | 2010 | Dehong, Yunnan | Female | 47 | Han | Heterosexual | KC870036 |
| YNFL22 | 2010 | Dehong, Yunnan | Male | 29 | Jingpo | Heterosexual | KC870040 |
| YNFL31 | 2009 | Dehong, Yunnan | Female | 24 | Jingpo | Heterosexual | KC870042 |
| YNFL33 | 2009 | Dehong, Yunnan | Male | 27 | Han | Injecting drug user | KC870043 |
YNFL10.1 and YNFL10.2 were isolated from a single person.
Briefly, RNA was extracted using the QIAamp Viral Mini Kit (QIAGEN, Germany), then transcribed into cDNA using the Superscript III first-strand synthesis system (Invitrogen, USA). With the near-endpoint diluted cDNA template, the NFLGs were amplified with TaKaRa LA Taq (TaKaRa, Dalian, China) using the same nested polymerase chain reaction (PCR) amplification conditions. The positive PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germany) and sequenced by ABI 3730XL sequencer using BigDye terminators (Applied Biosystems, Foster City, CA). The chromatogram data were cleaned and assembled using Sequencher v4.9 (Gene Codes, Ann Arbor, MI). Recombination breakpoints were determined using RIP and jpHMM (www.hiv.lanl.gov) and SimPlot.8 Breakpoint confirmation and the origin of each segment were then analyzed by phylogenetic trees. The standard subtype reference alignment file including all HIV-1 group M, group P, CRF01_AE, CRF07_BC, and CRF08_BC was downloaded from the Los Alamos National Laboratory HIV Database (www.hiv.lanl.gov/content /sequence /NEWALIGN/align.html#ref). Nucleotide sequences were first aligned with the HXB2 standard reference strain using Clustal W, merged into the reference file, and adjusted manually using BioEdit.9 Phylogenetic and subregion tree analyses were performed using the neighbor-joining method based on a GTR model in MEGA.10 The reliability of tree branches was evaluated by 1,000 bootstrap replicates, and bootstrap values above 0.7 were considered stable.
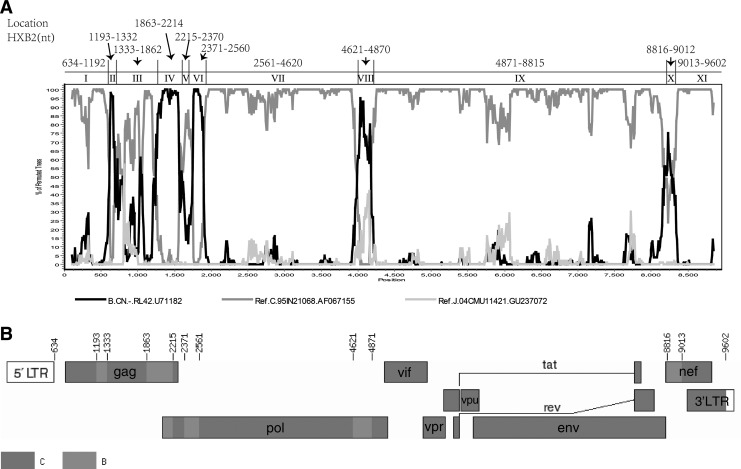
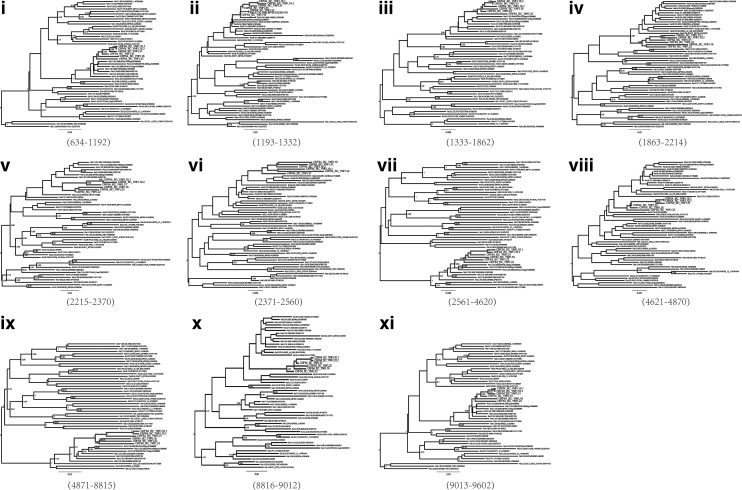
In total, six NFLG sequences were amplified, with two from YNFL10 named YNFL10.1 and YNFL10.2. The six sequences formed a distinct monophyletic cluster, distantly related to all known HIV-1 subtype/CRFs (Fig. 1). Recombination analysis showed that the sequences were composed of subtypes B and C, with five regions (II, IV, VI, VIII, and X) of subtype B inserted into the subtype C backbone (I, III, V, VII, IX, and XI) (Fig. 2). The breakpoint positions refer to HXB2 coordinates, and were located by HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE /locate.html). Therefore, the map of the recombinant genome is as follows: IC (634–1,192 nt), IIB (1,193–1,332 nt), IIIC (1,333–1,862 nt), IVB (1,863–2,214 nt), VC (2,215–2,370 nt), VIB (2,371–2,560 nt), VIIC (2,561–4,620 nt), VIIIB (4,621–4,870 nt), IXC (4,871–8,815 nt), XB (8,816–9,612 nt), and XIC (9,013–9,602 nt). All subtype C segments were verified by phylogenetic analysis, while the trees of B segments were not as stable as the C segment except for IVB (bootstrap value 0.97) (Fig. 3). The main reason may be that the B segments were shorter and that the low bootstrap value of reference strains is the best evidence. The analysis also demonstrated that the parental origin of the subtype B regions was most likely from the Thai B lineage, and the subtype C regions originated from the India C lineage (Fig. 3). The recombinant form is distinct from any known CRFs to date. Therefore, these new recombinants are now designated CRF64_BC.
FIG. 1.
Phylogenetic tree analysis of the near full-length genome (NFLG) sequences of the six isolates. All HIV-1 group M reference sequences were used to construct the phylogenetic tree (neighbor-joining method). Bootstrap values above 0.7 are shown at the corresponding nodes, and the CRF64_BC sequences are marked in bold gray.
FIG. 2.
Bootscan plots and schematic representation of the near full-length genome's mosaic structure. (A) Bootscan plots of CRF64_BC (YNFL31) with subtype B (U71182), subtype C (AF067155), and subtype J (GU237072) as reference genotypes. The parameters of SimPlot bootscan analysis were set as the default except for a window size of 200 and step size of 10. The breakpoint positions refer to HXB2 coordinates located by HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/ LOCATE/locate.html). (B) Genomic map of CRF64_BC. The mosaic map was generated using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content /sequence/ DRAW_CRF/recom_mapper.html).
FIG. 3.
Phylogenetic analyses of 11 mosaic segments defined by bootscanning. Phylogenetic trees were constructed using the neighbor-joining method based on the GTR model in MEGA. The reliability of tree branches was evaluated by 1,000 bootstrap replicates, and bootstrap values above 0.7 were considered stable. The sequences of CRF64_BC isolates are marked in bold gray.
Dehong prefecture, a hotspot of China's AIDS epidemic, is considered the place of origin for HIV-1 subtype B, CRF07_BC, and CRF08_BC in China.6,11 Multiple BC recombinants are now circulating in this region and pose a high risk of spreading to other places of China. In this study, we characterized a novel HIV-1 circulating recombinant form CRF64_BC from five donors without obvious epidemiological linkage in Dehong. Phylogenetic analysis showed that the subtype C segments likely originated from the Indian subtype C lineage, which was transmitted into China in the 1990s. The other regions of the genome were most likely derived from subtype B; these segments may be too short to provide enough signal for accurate inference of their evolutionary history, but nonetheless appeared most closely related to Thai subtype B strains. The emergence of a new CRF may further complicate the development of effective vaccines and other subtype-specific molecular tools for controlling the HIV-1 epidemic in China. Further study is also needed to assess whether the new CRF's phenotypic traits, such as fitness and virulence, are higher than the presently dominant strains in China.
Sequence Data
The nucleotide sequences of the six isolates YNFL10.1, YNFL10.2, YNFL16, YNFL22, YNFL31, and YNFL33 have been submitted to GenBank with the following accession numbers: KC870032, KC870033, KC870036, KC870040, KC870042, and KC870043, respectively.
Acknowledgments
This work was supported by the National Major Projects for Infectious Diseases Control and Prevention Grants (2012ZX10001-008 and 2012ZX10001-002), the National Natural Science Foundation of China Grants (81020108030 and 81261120393), a State Key Laboratory for Infectious Disease Development Grant (2012SKLID103), and the International Development Research Center of Canada Grant (104519-010). We would like to thank Brian Foley from the Los Alamos National Laboratory for advice on CRF nomenclature, and professor Yutaka Takebe, AIDS Research Center, National Institute of Infectious Diseases of Japan, for suggestions on recombinant analysis.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Zhang M, Foley B, Schultz AK, et al. : The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 2010;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X, Xing H, Ruan Y, et al. : A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 2012;7(10):e47289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su L, Graf M, Zhang Y, et al. : Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B') recombinant strain in China. J Virol 2000;74(23):11367–11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graf M, Shao Y, Zhao Q, et al. : Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B'-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses 1998;14(3):285–288 [DOI] [PubMed] [Google Scholar]
- 5.Li Z, He X, Wang Z, et al. : Tracing the origin and history of HIV-1 subtype B' epidemic by near full-length genome analyses. AIDS 2012;26(7):877–884 [DOI] [PubMed] [Google Scholar]
- 6.Yang R, Kusagawa S, Zhang C, et al. : Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J Virol 2003;77(1):685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Chen L, Yang S, et al. : Recombination form and epidemiology of HIV-1 unique recombinant strains identified in Yunnan, China. PLoS One 2012;7(10):e46777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei H, Su L, Feng Y, et al. : Near full-length genomic characterization of a novel HIV type 1 CRF07_ BC/01_AE recombinant in men who have sex with men from Sichuan, China. AIDS Res Hum Retroviruses 2013;29(8):1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall TA: BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper presented at the Nucleic Acids Symposium Series1999 [Google Scholar]
- 10.Tamura K, Dudley J, Nei M, and Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24(8):1596–1599 [DOI] [PubMed] [Google Scholar]
- 11.Tee KK, Pybus OG, Li XJ, et al. : Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J Virol 2008;82(18):9206–9215 [DOI] [PMC free article] [PubMed] [Google Scholar]