Abstract
Cytokines and chemokines are key participants in pathways that drive inflammatory, immune, and other cellular responses to exogenous insults such as infection, trauma, and physiological stress. Persistent and aberrant expression of these factors has been linked to autoimmune, degenerative, and neoplastic diseases. Consequently, cytokine and chemokine expression is tightly governed at each level of gene regulation. Recent studies have demonstrated a role for KH-type splicing regulatory protein (KSRP) in curtailing cytokine and chemokine expression through transcriptional and post-transcriptional mechanisms, including promotion of microRNA maturation. Understanding the role of KSRP in cytokine mRNA metabolism should identify promising targets for the modulation of immune and inflammatory responses.
Introduction
Cytokines and chemokines help orchestrate finely tuned immune and inflammatory responses to infection, physical injury, and other assaults to the biological system. When left unchecked, these signaling molecules can drive or participate in many pathological processes ranging from neurodegenerative diseases (eg, amyotrophic lateral sclerosis and Alzheimer's) to cancer. A simple Pubmed inquiry using search terms “cytokine” and “disease” yields nearly 6,000 references, covering virtually every organ in the body. Thus, humans and other organisms have evolved an intricate regulatory system to keep cytokine and chemokine production in check. As a part of this system, cytokine gene expression is subjected to transcriptional as well as post-transcriptional control (Stoecklin and Anderson 2006; Hamilton and others 2007; Khabar 2007). For the latter, specific cis elements within the mRNA molecule govern stability and translational efficiency to regulate expression (Wilusz and others 2001; Wilusz and Wilusz 2004). The adenine and uridine-rich element (ARE) in the 3′ untranslated region (UTR) of inherently unstable mRNAs represents a major regulatory locus and is present, often in clusters, in many cytokine and chemokine mRNAs (Chen and Shyu 1995; Bakheet and others 2006). The AREs regulate mRNA decay and/or translational efficiency by recruiting ARE-binding proteins (ARE-BPs) (Bevilacqua and others 2003; Barreau and others 2005; Espel 2005). These proteins contain one or more RNA-binding domains that specifically bind AREs, and their role in post-transcriptional control of cytokine gene expression has been recently reviewed (Anderson 2008, 2010). KH-type splicing regulatory protein (KSRP), an ARE-BP, serves as one of the regulatory watchdogs of cytokine expression by negatively modulating a subset of cytokines and chemokines at multiple levels, including translational silencing, RNA instability, microRNA maturation, and transcriptional repression (Fig. 1). This review will primarily focus on recent findings on the role of KSRP in control of type I interferon (IFN) and cytokine expression.
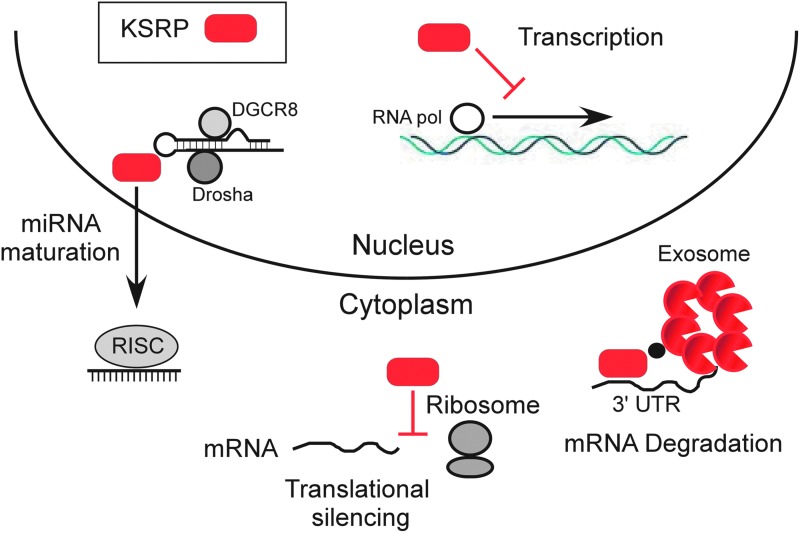
FIG. 1.
KH-type splicing regulatory protein (KSRP) negatively regulates cytokine and chemokine mRNA expression at multiple levels. KSRP promotes mRNA decay by binding to AU-rich elements in the 3′ untranslated region (UTR) and associating with the exosome (RNA degradation machine), and by binding to microRNA precursors and promoting their maturation. KSRP silences translation through dissociation of the mRNA from the polysome. KSRP also represses cytokine transcription by mechanisms that are not yet understood.
Post-Transcriptional Control of Type I Interferon Expression
Type I IFNs (IFN-α and IFN-β) play a critical role in the innate immune response against viral infection (Garcia-Sastre and Biron 2006; Stetson and Medzhitov 2006). Expression of type I IFNs is highly induced upon viral infection and rapidly shut off thereafter (Raj and Pitha 1981; Whittemore and Maniatis 1990). Although primarily controlled at the transcriptional level, rapid mRNA decay also contributes to the transient nature of IFN expression (Khabar and Young 2007). Although transcriptional regulation of type I IFN genes has been extensively studied (Honda and others 2006), the role of post-transcriptional regulation is less clear.
Since mRNAs encoding IFN-β and most of the IFN-α members (∼14 subtypes) contain AREs in their 3′ UTRs (Khabar and Young 2007), they are likely subjected to mRNA decay and/or translational control through an ARE-dependent mechanism. Indeed, human IFNB is subjected to mRNA stability control through an ARE in the 3′ UTR as well as an instability element in the coding region (Whittemore and Maniatis 1990; Paste and others 2003). Early studies also demonstrated that the 3′ UTR of human IFNB inhibited translation using in vitro translation systems (Kruys and others 1987; Grafi and others 1993) and that the murine Ifna 3′ UTR inhibited translation of a reporter gene (van Heuvel and others 1986). Subsequently, a 65-kDa protein was found to bind the ARE of IFNB, but its identity was not characterized (Raj and Pitha 1993). Although type I IFN mRNA stability and translation has been demonstrated, factors that modulate expression through an interaction with the 3′ UTR, and the underlying mechanism of regulation, remain largely unknown.
Role of KSRP in Control of Type I IFN Expression
KSRP was originally identified as a component of a multiprotein complex assembled on an intronic splicing enhancer element downstream of the neuron-specific c-src N1 exon (Min and others 1997). It was later purified as an ARE-BP and shown to promote mRNA decay by recruiting mRNA decay machinery (Chen and others 2001; Gherzi and others 2004). Subsequent to these studies, KSRP has been shown to facilitate the degradation of ARE-containing mRNAs in a variety of assay systems and models, and found to be required for the decay of reporter mRNAs containing various AREs from c-fos, TNF-α, and IL-8 (Chen and others 2001; Gherzi and others 2004; Winzen and others 2007) and endogenous mRNAs encoding cytokines and regulators for tissue development and cell growth (Briata and others 2003, 2005; Gherzi and others 2006; Ruggiero and others 2007; Winzen and others 2007; Nechama and others 2008; Graham and others 2010). Recent studies have also demonstrated that KSRP promotes the maturation of a subset of miRNAs (Ruggiero and others 2009; Trabucchi and others 2009; Zhang and others 2011; Briata and others 2012; Sundaram and others 2013; Wan and others 2013). These findings indicate that KSRP plays a pivotal role in controlling gene expression at multiple levels (Fig. 1).
The role of KSRP in controlling the stability of type I IFN mRNAs was recently revealed through the characterization of Ksrp-null mice (Lin and others 2011). Both Ifna and Ifnb were up-regulated in Ksrp−/− cells and Ksrp−/− mice in response to viral infection because of increased mRNA stability. KSRP physically interacted with Ifna4 and Ifnb transcripts through the ARE-containing 3′ UTRs. The AREs promoted mRNA decay and were necessary for KSRP-mediated regulation (Lin and others 2011). More importantly, the increased expression of type I IFNs contributed to resistance to herpes simplex virus I (HSV1) and vesicular stomatitis virus (VSV) infection. In the same study, KSRP was shown to target the mRNAs for decay in both stimulated and non-stimulated cells. Since Ifna and Ifnb are constantly expressed at very low levels in non-stimulated cells (Takaoka and others 2000; Gough and others 2012), these data imply that KSRP plays a critical role in maintaining low basal IFN expression in these cells. While the role of KSRP in regulating Ifna expression was established with Ifna4, most of other subtypes contain AREs, making them likely targets for decay by KSRP. Since Ksrp−/− cells are now available, this possibility can be easily tested. Altogether, these findings indicate that KSRP is a critical negative regulatory factor for type I IFN gene expression in innate immune responses and may serve as a therapeutic target for combating virus infection. Since type I IFNs also play a critical role in the pathogenesis of certain autoimmune diseases (Banchereau and Pascual 2006), elucidating the role of KSRP in restraining their expression in immune cells may also lead to the development of therapeutic strategies for these diseases.
Role of Tristetraprolin in Control of Type I IFN Expression
The tristetraprolin (TTP) family of CCCH tandem zinc-finger proteins is composed of 3 known members in mammals, including TTP/ZFP36, butyrate response factor 1 (BRF1/ZFP36L1), and butyrate response factor 2 (BRF2/ZFP36L2), and facilitates the decay of ARE-containing cytokine mRNAs (Blackshear 2002; Sanduja and others 2011). In a previous study, the stability of Ifnb and Ifna4 mRNA was increased by 2- to 4-fold in the absence of KSRP, but the transcripts were still degraded (Lin and others 2011), suggesting that additional decay-promoting ARE-BPs may be involved in the decay. Indeed, the down-regulation of TTP decreased Ifnb and Ifna mRNA decay rate and simultaneously, the down-regulation of TTP and KSRP additively increased mRNA stability (W.-J. Lin and C.-Y. Chen, unpublished data), indicating that KSRP and TTP play a redundant, yet independent, role in controlling IFN mRNA decay. Thus, we suggest that other decay-promoting ARE-BPs, such as BRF1, BRF2, and AUF1, may also regulate the decay of Ifna and/or Ifnb mRNAs. Since knockout mouse models for these ARE-BPs are available, it will be of interest to determine their roles in post-transcriptional control of type I IFN gene expression.
While TTP is expressed at very low levels in non-stimulated cells, it is rapidly induced by a variety of stimuli (Taylor and others 1995; Mahtani and others 2001; Brook and others 2006; Hitti and others 2006). TTP was found to be induced in HSV1-infected cells (Esclatine and others 2004). Treatment of bone marrow-derived macrophages (BMMs) with lipopolysaccharide (LPS) increased Ttp expression in a biphasic manner with an initial peak at 1 h and a second peak at 6 h after treatment (Sauer and others 2006). The 6 h peak was significantly attenuated in Ifnb-deficient BMMs, indicating that LPS induction of IFN-β at the early phase is essential for the late phase induction of Ttp. From these studies, we propose an integrated model for controlling type I IFN mRNA decay by KSRP and TTP (Fig. 2). In uninfected or non-stimulated cells, Ifnb and perhaps Ifna4, the first α subtype induced in response to viral infection (Marie and others 1998), are expressed by low-level transcription. KSRP, which is constitutively expressed in these cells, targets the mRNAs for decay and restrains their expression. In the early phase of viral infection, Ifnb and Ifna4 are induced through the activation of IFN regulatory factors, IRF3 and IRF7 (Sato and others 2000; Honda and others 2006). Meanwhile, Ttp is induced by the virus. The induced Ifna4/b mRNAs are then targeted by both KSRP and TTP for decay. In the late phase of viral infection, enhanced levels of IFN-β and IFN-α4 act in an autocrine and a paracrine fashion to boost transcription of IRF7, resulting in further amplification of Ifnb and other Ifna transcription. IFN-β also induces Ttp transcription. Thus, Ifnb mRNA and perhaps mRNAs encoding other Ifna subtypes are subjected to decay control by both TTP and KSRP to prevent an uncontrollable overexpression that could lead to apoptosis and/or dysregulated immune responses (Gough and others 2012).
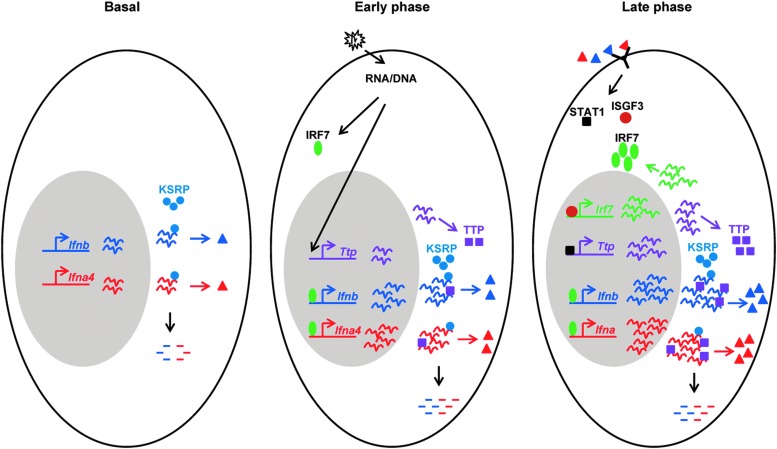
FIG. 2.
Integrated model for type I interferon (IFN) mRNA decay by KSRP and tristetraprolin (TTP) during the multistage induction. In non-infected cells, Ifnb and perhaps Ifna4 are expressed at very low levels. Constitutively expressed KSRP targets the mRNAs for decay. In the early phase of viral infection, activation of IFN regulatory factors, IRF3 and IRF7, leads to elevated expression of Ifnb, Ifna4, and Ttp. These induced Ifna4/b mRNAs are targeted for decay by both KSRP and TTP. In the late phase of viral infection, increased production of IFN-β and IFN-α4 acting in an autocrine and a paracrine fashion leads to a strong production of IRF7 by activated ISGF3. Activation of the newly synthesized IRF7 results in further amplification of Ifnb and other Ifna transcription. IFN-β induces STAT1-dependent Ttp transcription, in which Ifnb mRNA and perhaps mRNAs encoding other Ifna subtypes are subjected to decay by both TTP and KSRP.
Role of KSRP in Control of Cytokine Expression
Control of cytokine expression through transcriptional repression
KSRP, also known as fuse binding protein 2 (FBP2), was first characterized as a homologue of FBP1, a single-strand DNA binding protein with both transactivating and repressor domains capable of binding the c-myc promoter and modulating its activity (Davis-Smyth and others 1996; Duncan and others 1996). We recently reported that KSRP suppresses TNF-α promoter activity in LPS-stimulated primary astrocytes and RAW 264.7 macrophage cells (Li and others 2012). The former are immunocompetent cells that are located in the central nervous system which can be induced to express a number of cytokines, including TNF-α, IL-6, and TGF-β (Dong and Benveniste 2001; Sofroniew and Vinters 2010). Astrocytes can promote inflammation and disease progression in a wide range of disorders, including neurodegeneration (eg, Alzheimers, ALS), autoimmunity (eg, multiple sclerosis), and infection (Sofroniew and Vinters 2010). Mapping studies indicate that KSRP-induced suppression of TNF-α promoter activity, in part, localizes to a region containing the third NF-κB binding site. We also observed suppression of the triggering receptor expressed on myeloid cells (TREM)-1 promoter. This gene encodes a cell surface receptor that amplifies inflammatory cytokine release, including IL-1β and TNF-α, especially during infection and septic shock (Bouchon and others 2001; Klesney-Tait and others 2006). Thus, KSRP also regulates cytokines indirectly by blocking pathways that amplify their release. For TREM-1, the mRNA increased 4-fold in Ksrp−/− astrocytes without any change in RNA half life, suggesting that transcriptional suppression is the main mode of regulation. For both TNF-α and TREM-1, however, we were unable to demonstrate any direct DNA-protein interaction by chromatin immunoprecipitation or gel shift analysis, leaving the actual mechanism for promoter suppression unexplained. In addition to a direct inhibitory effect on DNA, possibilities include suppressed expression of key transactivators or increased expression of silencers through its regulation of miRNAs and mRNA stability/translational efficiency (see next 3 sections).
Control of cytokine expression through miRNA maturation
The role of KSRP in miRNA maturation was first described by Trabucchi and others (2009). They found that KSRP is essential for the maturation of a subset of miRNAs by binding to the terminal loop of the pre-miRNA and interacting with Drosha and Dicer complexes (Fig. 1). A link to cytokine regulation was later observed when KSRP was shown to regulate miR-155 in primary murine and RAW 264.7 macrophages (Ruggiero and others 2009). In macrophages stimulated with LPS, the induction of miR-155 was reversed on KSRP knockdown with a concomitant accumulation of pri-miRNA forms. Microarray analysis of cells transfected with anti-miR-155 showed that a broad subset of cytokine and chemokine mRNAs, including IL-1β, IL-12b, and CXCL11, was significantly affected (Table 1). Targets with delayed induction (>4 h post LPS stimulation) were most impacted, and this time frame paralleled the miR-155 maturation kinetics. Some of the mRNA targets may have indirectly been affected through changes in NF-κB signaling, as the noncanonical NF-κB kinase, IKK-ɛ, was significantly altered. More recently, KSRP has been shown to regulate mature miR-155 in lung epithelial cells from patients with cystic fibrosis (Bhattacharyya and others 2013). MiR-155 drives a hyper-inflammatory response in these cells that are characterized by high levels of IL-8 (Bhattacharyya and others 2011). Investigators found that TTP antagonizes miR-155 expression in these cells. Interestingly, KSRP itself was shown to be regulated by miR-27 in gut epithelial cells after cryptosporidium parvum infection (Zhou and others 2012). In a TLR4/NF-κB-dependent manner, INOS mRNA becomes stabilized and up-regulated, because KSRP is translationally silenced by miR-27. Ectopic KSRP expression without its 3′ UTR reverses this suppression and leads to increased infection. The extent of direct or indirect cytokine regulation by KSRP through miRNAs is potentially far reaching based on in silico analyses of transcripts that contain binding sites for miRNAs linked to KSRP (Asirvatham and others 2009).
Table 1.
Cytokine/Chemokine mRNAs Regulated by KSRP
| mRNA target | mRNA degradation | Translational silencinga | miRNA-mediated | Cell type | Reference(s) |
|---|---|---|---|---|---|
| Chemokines | |||||
| CXCL2 | x | T98G, HeLa | Herjan and others (2013) | ||
| CXCL3 | x | T98G, HeLa | Graham and others (2010) and Herjan and others (2013) | ||
| CXCL8 | x | HeLa | Winzen and others (2007) | ||
| CXCL10 | x | HeLa | Dhamija and others (2011) | ||
| miR-155 | Macrophage | Ruggiero and others (2009) | |||
| CXCL11 | miR-155 | Macrophage | Ruggiero and others (2009) | ||
| CCL2 | x | T98G | Graham and others (2010) | ||
| miR-155 | Macrophage | Ruggiero and others (2009) | |||
| CCL5 | miR-155 | Macrophage | Ruggiero and others (2009) | ||
| CCL20 | x | HeLa | Dhamija and others (2011) | ||
| Cytokines | |||||
| CSF2 | x | HeLa | Winzen and others (2007) | ||
| IFNB1 | x | HeLa | Dhamija and others (2011) | ||
| IL-1α | x | HeLa | Dhamija and others (2011) | ||
| IL-1β | x | ? | Astrocytes | Li and others (2012) | |
| RAW264.7 | |||||
| miR-155 | Macrophage | Ruggiero and others (2009) | |||
| IL-6 | x | x | HeLa | Dhamija and others (2011) and Winzen and others (2007) | |
| IL-10 | miR-155 | Macrophage | Dhamija and others (2011) and Winzen and others (2007) | ||
| IL-12 | miR-155 | Macrophage | Dhamija and others (2011) | ||
| IL-23A | x | HeLa | Dhamija and others (2011) | ||
| TNF-α | x | x | Astrocytes | Li and others (2012), Dhamija and others (2011) and Winzen and others (2007) | |
| RAW264.7 HeLa | |||||
| TNF-α | Transcriptional repression | Astrocytes | Li and others (2012) | ||
| RAW264.7 | |||||
| Related | |||||
| ATF3 | x | HeLa | Dhamija and others (2011) | ||
| IRF1 | x | HeLa | Dhamija and others (2011) | ||
| IL-16R | x | HeLa | Dhamija and others (2011) | ||
| INOS | x | DLD-1 | Linker and others (2005), Schmidt and others (2010), and Zhou and others (2012) | ||
| Chondrocytes | |||||
| Gut epithelial cells | |||||
| SOCS3 | miR-155 | Macrophage | Ruggiero and others (2009) | ||
| TREM-1 | Transcriptional repression | Astrocytes | Li and others (2012) | ||
As defined by polysome redistribution with KSRP knockdown.
KSRP, KH-type splicing regulatory protein.
Control of cytokine expression through mRNA destabilization
Many cytokine and chemokine mRNAs have AREs in the 3′ UTR that govern their stability and translational efficiency. The scope of targets for potential mRNA regulation broadened after a report by Winzen and others (2007) identified more than 1,600 endogenous mRNA targets bound to KSRP in a pull-down assay. One hundred of those targets were significantly up-regulated with KSRP knockdown, with some showing RNA stabilization. A number of cytokines and chemokines, including IL-6 and IL-8, were identified among these targets (Table 1). Interestingly, some chemokines and cytokine mRNAs were up-regulated but not identified in the pull-down assay, including IL-1α, IL-23, -24, -29, CXCL10, 11, and CCL20, suggesting indirect effects on cytokine regulation through KSRP. A more extensive analysis of IL-8 showed that KSRP destabilized the mRNA through the AREs in the 3′ UTR. When cells were stimulated with IL-1α, IL-8 mRNA became stable with a concomitant loss of KSRP binding. This phenotype could be reversed with inhibition of p38 but not MK2. Interestingly, there was a functional overlap between TTP and KSRP in the destabilization of IL-8 mRNA with TTP being regulated by MK2. Overlap of destabilizers BRF1 and KSRP was also observed for IL-3 and TNF-α AREs, and the contribution of each RBP varied based on cell type (Gherzi and others 2004). We looked at TNF-α and IL-1β mRNA stability in astrocytes from Ksrp−/− mice and observed stabilization of both transcripts with LPS but not TNF-α treatment, suggesting that the mode of KSRP-mediated RNA down-regulation varies depending on stimulus type (Li and others 2012). Another study looked at CXCL2 and 3 chemokine regulation in HeLa cells and found that IL-1α stimulation induced mRNA stabilization but did not affect KSRP binding and was independent of the ARE (Herjan and others 2013). KSRP, however, exerted a destabilizing effect on these targets in non-stimulated cells. These studies indicate differential stabilization of cytokine mRNAs depending on the nature of the stimulus and the cell type. Although not a cytokine, iNOS is a signaling molecule activated by cytokines and is an important component of the inflammatory response pathway. KSRP destabilizes iNOS mRNA through an ARE in the 3′ UTR (Linker and others 2005; Schmidt and others 2010; Zhou and others 2012). Interestingly, TTP was shown to enhance iNOS mRNA stability by physically interacting with KSRP and likely sequestering the KSRP-exosome complex away from the mRNA (Fechir and others 2005; Linker and others 2005). Thus, KSRP negatively affects different levels of cytokine signaling in inflammation, and TTP and KSRP can coordinately regulate target cytokine mRNA stability.
Control of cytokine expression through translational silencing
Translational silencing through the ARE represents another level of post-transcriptional regulation, which is distinct but intimately associated with RNA stability (Wilusz and Wilusz 2004; Barreau and others 2005; Abdelmohsen and others 2008). Features suggestive of this level of regulation are high RNA-to-protein levels, dissociation of the mRNA from the polysome, and low translation rates as determined by pulse labeling. A role for KSRP and silencing cytokine mRNA translation was shown for IL-6 and IL-1α (Dhamija and others 2011). An in-depth analysis of IL-6 showed that the cis elements responsible for silencing localized to the ARE, where KSRP binds the mRNA. Stimulation of cells with IL-1, a strong inducer of IL-6, reduced KSRP binding to IL-6 mRNA and increased mRNA association with polysomes. In that same study, investigators used microarray to identify 50 mRNA targets that were enriched in polysomes after KSRP knockdown. Additional cytokine or related mRNAs were identified among these targets, including IL-16R, IL-23A, and TNF-α (Table 1). The polysome enrichment of some targets (which lack AREs) may have resulted from indirect effects of KSRP related to maturation of miRNAs. Moreover, 2 of the 50 identified mRNA targets, ATF3 and IRF1, are transcription factors linked to cytokine and IFN expression (Romeo and others 2002; Thompson and others 2009), thus reflecting another indirect level of cytokine regulation. In Ksrp−/− astrocytes, translational silencing of TNF-α and IL-β was suspected based on large discrepancies between RNA and protein changes compared with controls (eg, 15-fold change in protein vs. a ∼2-fold change in mRNA) (Li and others 2012).
Perspectives
Since the initial characterization, KSRP has been demonstrated to negatively modulate gene expression at multiple levels. KSRP should be viewed as a part of an armamentarium of factors that serve to fine tune type I IFN and cytokine/chemokine regulation and to prevent the deleterious consequences of persistent expression. TTP and AUF1, for example, are 2 RNA binding proteins that also exert down-modulating effects on cytokine mRNAs, sometimes in concert with KSRP, by targeting AREs in the mRNA. KSRP and these RBPs, thus, have overlapping and also distinct cytokine mRNA targets that vary with cellular context and type of stressor. The transcriptional repressive function of KSRP and its role in microRNA maturation, however, extend the regulatory “reach” over cytokine expression beyond ARE-containing mRNAs. The complexity of cytokine and chemokine mRNA post-transcriptional regulation, as alluded to in this review, underscores the intricacies of cellular responses to the vast array of insults and physiological stressors. The list of KSRP mRNA targets has grown over the past decade. However, KSRP has not yet been linked to a disease model, and the associated phenotype(s) in vivo resulting from its deletion remains unclear. The availability of KSRP knockout mice or cells should foster our understanding of its in vivo function and the identification of other target mRNAs in a physiological setting.
Acknowledgments
P.H.K. is supported by the National Institutes of Health Grant RO1 NS064133, R21NS081743, and a Merit Review grant from the Department of Veterans Affairs. C.Y.C. is supported by the National Institutes of Health Grant RO1 GM68758.
Author Disclosure Statement
No competing financial interests exist.
References
- Abdelmohsen K, Yuki K, Kim HH, Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389(3):243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9(4):353–359 [DOI] [PubMed] [Google Scholar]
- Anderson P. 2010. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10(1):24–35 [DOI] [PubMed] [Google Scholar]
- Asirvatham AJ, Magner WJ, Tomasi TB. 2009. miRNA regulation of cytokine genes. Cytokine 45(2):58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. 2006. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res 34(Database issue):D111–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Pascual V. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25(3):383–392 [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33(22):7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. 2003. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol 195(3):356–372 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Balakathiresan NS, Dalgard C, Gutti U, Armistead D, Jozwik C, Srivastava M, Pollard HB, Biswas R. 2011. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J Biol Chem 286(13):11604–11615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Kumar P, Tsuchiya M, Bhattacharyya A, Biswas R. 2013. Regulation of miR-155 biogenesis in cystic fibrosis lung epithelial cells: antagonistic role of two mRNA-destabilizing proteins, KSRP and TTP. Biochem Biophys Res Commun 433(4):484–488 [DOI] [PubMed] [Google Scholar]
- Blackshear PJ. 2002. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans 30(Pt 6):945–952 [DOI] [PubMed] [Google Scholar]
- Bouchon A, Facchetti F, Weigand MA, Colonna M. 2001. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410(6832):1103–1107 [DOI] [PubMed] [Google Scholar]
- Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. 2005. p38-Dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell 20(6):891–903 [DOI] [PubMed] [Google Scholar]
- Briata P, Ilengo C, Corte G, Moroni C, Rosenfeld MG, Chen CY, Gherzi R. 2003. The Wnt/beta-catenin—>Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell 12(5):1201–1211 [DOI] [PubMed] [Google Scholar]
- Briata P, Lin WJ, Giovarelli M, Pasero M, Chou CF, Trabucchi M, Rosenfeld MG, Chen CY, Gherzi R. 2012. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ 19(3):478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol 26(6):2408–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107(4):451–464 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20(11):465–470 [DOI] [PubMed] [Google Scholar]
- Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. 1996. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem 271(49):31679–31687 [DOI] [PubMed] [Google Scholar]
- Dhamija S, Kuehne N, Winzen R, Doerrie A, Dittrich-Breiholz O, Thakur BK, Kracht M, Holtmann H. 2011. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J Biol Chem 286(38):33279–33288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Benveniste EN. 2001. Immune function of astrocytes. Glia 36(2):180–190 [DOI] [PubMed] [Google Scholar]
- Duncan R, Collins I, Tomonaga T, Zhang T, Levens D. 1996. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol Cell Biol 16(5):2274–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclatine A, Taddeo B, Roizman B. 2004. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J Virol 78(16):8582–8592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espel E. 2005. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol 16(1):59–67 [DOI] [PubMed] [Google Scholar]
- Fechir M, Linker K, Pautz A, Hubrich T, Forstermann U, Rodriguez-Pascual F, Kleinert H. 2005. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol Pharmacol 67(6):2148–2161 [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312(5775):879–882 [DOI] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell 14(5):571–583 [DOI] [PubMed] [Google Scholar]
- Gherzi R, Trabucchi M, Ponassi M, Ruggiero T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS, Briata P. 2006. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol 5(1):e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36(2):166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G, Sela I, Galili G. 1993. Translational regulation of human beta interferon mRNA: association of the 3′ AU-rich sequence with the poly(A) tail reduces translation efficiency in vitro. Mol Cell Biol 13(6):3487–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JR, Hendershott MC, Terragni J, Cooper GM. 2010. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol Cell Biol 30(22):5295–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TA, Novotny M, Datta S, Mandal P, Hartupee J, Tebo J, Li X. 2007. Chemokine and chemoattractant receptor expression: post-transcriptional regulation. J Leukoc Biol 82(2):213–219 [DOI] [PubMed] [Google Scholar]
- Herjan T, Novotny M, Hamilton TA. 2013. Diversity in sequence-dependent control of GRO chemokine mRNA half-life. J Leukoc Biol 93(6):895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol 26(6):2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25(3):349–360 [DOI] [PubMed] [Google Scholar]
- Khabar KS. 2007. Rapid transit in the immune cells: the role of mRNA turnover regulation. J Leukoc Biol 81(6):1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS, Young HA. 2007. Post-transcriptional control of the interferon system. Biochimie 89(6–7):761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesney-Tait J, Turnbull IR, Colonna M. 2006. The TREM receptor family and signal integration. Nat Immunol 7(12):1266–1273 [DOI] [PubMed] [Google Scholar]
- Kruys V, Wathelet M, Poupart P, Contreras R, Fiers W, Content J, Huez G. 1987. The 3′ untranslated region of the human interferon-beta mRNA has an inhibitory effect on translation. Proc Natl Acad Sci U S A 84(17):6030–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin WJ, Chen CY, Si Y, Zhang X, Lu L, Suswam E, Zheng L, King PH. 2012. KSRP: a checkpoint for inflammatory cytokine production in astrocytes. Glia 60(11):1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, Parker JN, Chen CY. 2011. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol 31(16):3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. 2005. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res 33(15):4813–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol 21(19):6461–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17(22):6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev 11(8):1023–1036 [DOI] [PubMed] [Google Scholar]
- Nechama M, Ben-Dov IZ, Briata P, Gherzi R, Naveh-Many T. 2008. The mRNA decay promoting factor K-homology splicing regulator protein post-transcriptionally determines parathyroid hormone mRNA levels. FASEB J 22(10):3458–3468 [DOI] [PubMed] [Google Scholar]
- Paste M, Huez G, Kruys V. 2003. Deadenylation of interferon-beta mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur J Biochem 270(7):1590–1597 [DOI] [PubMed] [Google Scholar]
- Raj NB, Pitha PM. 1981. Analysis of interferon mRNA in human fibroblast cells induced to produce interferon. Proc Natl Acad Sci U S A 78(12):7426–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj NB, Pitha PM. 1993. 65-kDa protein binds to destabilizing sequences in the IFN-beta mRNA coding and 3′ UTR. FASEB J 7(8):702–710 [DOI] [PubMed] [Google Scholar]
- Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. 2002. IRF-1 as a negative regulator of cell proliferation. J Interferon Cytokine Res 22(1):39–47 [DOI] [PubMed] [Google Scholar]
- Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. 2009. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J 23(9):2898–2908 [DOI] [PubMed] [Google Scholar]
- Ruggiero T, Trabucchi M, Ponassi M, Corte G, Chen CY, al-Haj L, Khabar KS, Briata P, Gherzi R. 2007. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol Biol 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2(1):42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13(4):539–548 [DOI] [PubMed] [Google Scholar]
- Sauer I, Schaljo B, Vogl C, Gattermeier I, Kolbe T, Muller M, Blackshear PJ, Kovarik P. 2006. Interferons limit inflammatory responses by induction of tristetraprolin. Blood 107(12):4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N, Pautz A, Art J, Rauschkolb P, Jung M, Erkel G, Goldring MB, Kleinert H. 2010. Transcriptional and post-transcriptional regulation of iNOS expression in human chondrocytes. Biochem Pharmacol 79(5):722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, Vinters HV. 2010. Astrocytes: biology and pathology. Acta Neuropathol 119(1):7–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. 2006. Type I interferons in host defense. Immunity 25(3):373–381 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Anderson P. 2006. Posttranscriptional mechanisms regulating the inflammatory response. Adv Immunol 89:1–37 [DOI] [PubMed] [Google Scholar]
- Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P. 2013. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 495(7439):103–106 [DOI] [PubMed] [Google Scholar]
- Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, Tanaka N, Taniguchi T. 2000. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science 288(5475):2357–2360 [DOI] [PubMed] [Google Scholar]
- Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. 1995. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem 270(22):13341–13347 [DOI] [PubMed] [Google Scholar]
- Thompson MR, Xu D, Williams BR. 2009. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 87(11):1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459(7249):1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heuvel M, Bosveld IJ, Luyten W, Trapman J, Zwarthoff EC. 1986. Transient expression of murine interferon-alpha genes in mouse and monkey cells. Gene 45(2):159–165 [DOI] [PubMed] [Google Scholar]
- Wan G, Zhang X, Langley RR, Liu Y, Hu X, Han C, Peng G, Ellis LM, Jones SN, Lu X. 2013. DNA-damage-induced nuclear export of precursor microRNAs is regulated by the ATM-AKT pathway. Cell Rep 3(6):2100–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore LA, Maniatis T. 1990. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol 10(4):1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 20(10):491–497 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. 2001. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2(4):237–246 [DOI] [PubMed] [Google Scholar]
- Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. 2007. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol 27(23):8388–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Berger FG, He X, Lu X. 2011. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41(4):371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Gong A-Y, Eischeid AN, Chen X-M. 2012. miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PLoS Pathog 8(5):e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]