Abstract
Background and Purpose
(R,S)-ketamine produces rapid and significant antidepressant effects in approximately 65% of patients suffering from treatment-resistant bipolar depression (BD). The genetic, pharmacological and biochemical differences between ketamine responders and non-responders have not been identified. The purpose of this study was to employ a metabolomics approach, a global, non-targeted determination of endogenous metabolic patterns, to identify potential markers of ketamine response and non-response.
Experimental Approach
Plasma samples from 22 BD patients were analyzed to produce metabolomic patterns. The patients had received ketamine in a placebo-controlled crossover study and the samples were obtained 230 min post-administration at which time the patients were categorized as responders or non-responders. Matching plasma samples from the placebo arm of the study were also analysed. During the study, the patients were maintained on either lithium or valproate.
Key Results
The metabolomic patterns were significantly different between the patients maintained on lithium and those maintained on valproate, irrespective of response to ketamine. In the patients maintained on lithium, 18 biomarkers were identified. In responders, lysophosphatidylethanolamines (4) and lysophosphatidylcholines (9) were increased relative to non-responders.
Conclusions and Implications
The results indicate that the differences between patients who respond to ketamine and those who do not are due to alterations in the mitochondrial β-oxidation of fatty acids. These differences were not produced by ketamine administration. The data indicate that pretreatment metabolomics screening may be a guide to the prediction of response and a potential approach to the individualization of ketamine therapy.
Linked Articles
This article is part of a themed issue on Mitochondrial Pharmacology: Energy, Injury & Beyond. To view the other articles in this issue visit http://dx.doi.org/10.1111/bph.2014.171.issue-8
Keywords: mitochondrial function, fatty acid metabolism, pharmacometabolomics, lithium, valproate
Introduction
The majority of the mood stabilizers and antidepressants used to treat bipolar depression (BD) target the 5-hydroxytryptaminergic and noradrenergic systems (Machado-Vieira et al., 2008). These drugs are associated with a considerable lag in the onset of antidepressant action, high inter-individual variability in response and many ‘treatment-resistant’ patients (Machado-Vieira et al., 2008). However, recent data have indicated that there is an alternative approach to the treatment of BD based upon the targeting of the glutamatergic system using antagonists of the NMDA receptor (Zarate et al., 2006; 2012). A number of these studies have utilized a subanesthetic dose of the NMDA receptor antagonist (R,S)-ketamine and observed a rapid (within 4 h), and significant antidepressant effect in approximately 65% of previously treatment-resistant BD patients (Zarate et al., 2006, 2012; Dolgin, 2013). The genetic, pharmacological and biochemical differences between ketamine-responders (Rs) and non-responders (NRs) have not been identified. The objective of this pilot study is to employ a metabolomics approach to identify potential markers of ketamine response and non-response.
Metabolomics, a global, non-targeted approach to the study of biochemical processes and metabolic networks (Kaddurah-Daouk et al., 2008), has been used to identify disease-specific metabolic profiles and biomarkers of CNS disorders, including major depressive disorder (MDD) (Schwarz and Bahn, 2008; Kaddurah-Daouk and Krishnan, 2009; Quinones and Kaddurah-Daouk, 2009). Recent studies in MDD patients have also investigated changes in metabolomic patterns produced by administration of selective 5-hydroxytryptamine (5-HT) reuptake inhibitors (SSRI) and pharmacogenetic analysis to identify glycine and glycine dehydrogenase as citalopram/escitalopram response makers (Ji et al., 2011; Abo et al., 2012). Metabolomic studies have demonstrated that fatty acid metabolism is significantly lower and shifted from β-oxidation to ω-oxidation in depressed patients, compared with non-depressed controls (Maes et al., 1996; Paige et al., 2007; Steffens et al., 2010), indicating that mitochondrial function may be associated with the disease state. These results are consistent with the association of mitochondrial function with depression and anxiety (Burroughs and French, 2007) and with MDD and BD-related changes in phospholipid metabolism (Modica-Napolitano and Renshaw, 2004). In addition, stress-induced changes in mitochondria membrane potential have been suggested as a mechanism for hippocampus atrophy in posttraumatic stress disorder (PTSD) (Zhang et al., 2006). Postmortem studies in patients with PTSD identified 119 dysregulated genes, a number of these genes are associated with mitochondrial dysfunction and oxidative phosphorylation (Su et al., 2008).
In the current pilot study, a metabolomic analysis was performed using plasma samples obtained from 22 treatment-resistant patients with BD who had received ketamine in a placebo-controlled crossover study (Diazgranados et al., 2010; Zarate et al., 2012). The samples were obtained 230 min post-ketamine administration, at which time the patients were categorized as Rs or NRs. Metabolomic global profiling was also performed in 17 patients using matching plasma samples from the placebo arm of the study. During the study, the patients were maintained on either lithium or valproate. The effects of these mood stabilizers on ketamine response patterns were also examined as previous studies of post mortem brain tissues from BD patients and rat studies have indicated that lithium and valproate have different effects on the levels of excitatory/inhibitory neurotransmitters (Lan et al., 2009).
The results demonstrate that the metabolomic patterns observed in the patients receiving lithium (lithium-subgroup) were significantly different from those obtained with the valproate-subgroup, irrespective of the response to ketamine. Within the lithium-subgroup, 18 compounds were found to be significantly different between patients who responded to ketamine ( Rs ) and non-responders (NRs), with 16 of the 18 associated with the metabolism of fatty acids. Relative increases or decreases were observed in lysophosphatidylcholines (9 compounds), lysophosphatidylethanolamines (4), monoglycerides (2) and the carnitine precursor N6,N6,N6-trimethyl-L-lysine. Thus, the data suggest that key factors in the clinical response or non-response to ketamine in BD patients are differences in mitochondrial function reflected in fatty acid metabolism.
Methods
Patient selection and ketamine administration
The Combined Neuroscience Institutional Review Board of the National Institutes of Health approved the study. All subjects provided written informed consent and were studied as inpatients at the National Institute of Mental Health Clinical Research Center, Mood Disorders Research Unit in Bethesda, Maryland. Plasma samples were obtained from 22 BD patients enrolled in a placebo-controlled study of the effect of ketamine on depression (Diazgranados et al., 2010). At the time of the study, the patients were experiencing a major depressive episode without psychotic features and had been maintained on a mood stabilizer either lithium or valproate for 4 weeks before the trial and during the ketamine infusion. Lithium and valproate were administered twice a day (morning and evening) and the blood levels of the agents were within therapeutic range, serum lithium, 0.6–1.2 mEq·L−1, serum valproate 50–125 μg·mL−1, throughout the study. No other psychotropic medications were permitted in the 2 weeks before and during the study.
The patients received a single i.v. infusion of 0.5 mg·kg−1 of ketamine hydrochloride over the course of 40 min, and symptoms, including Montgomery Åsberg Depression Rating Scale (MADRS) scores and blood samples, were collected into heparinized tubes at 40 min (the end of the infusion), 80, 110 and 230 min post-infusion, and at day 1. The differential in MADRS scores from 0 to 230 min were used to delineate Rs and NRs with a relative improvement of ≥50% in MADRS score signifying a response to ketamine.
Metabolomics assay of plasma samples using LC-QTOF-MS
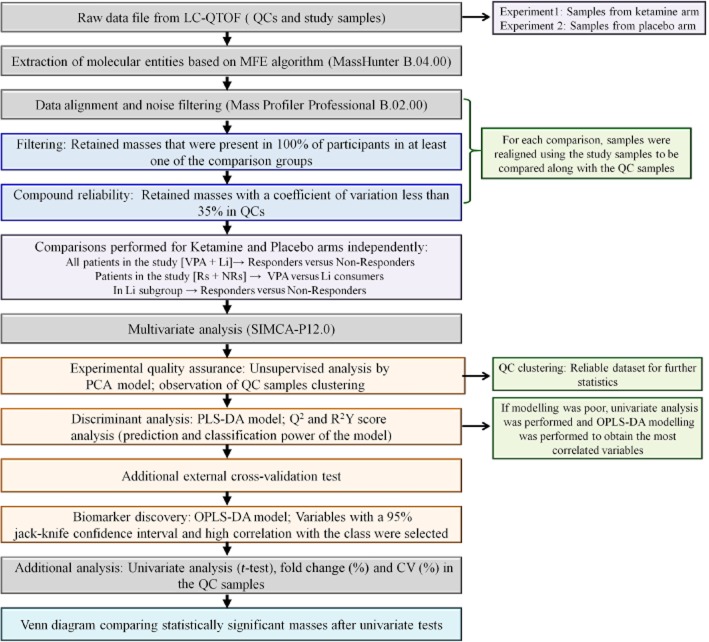
The experimental procedures utilized in the metabolomics study involved the sequential statistical treatment of data obtained from the analysis of the experimental and control samples. The samples were analyzed using liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer (LC-QTOF-MS), which initiated the detection, quantitation and identification of the unique biochemicals associated with the observed clinical response (Figure 1).
Figure 1.
Analysis of metabolomics data from plasma samples. A schematic representation of the sample analysis including detection, quantitation and identification of the unique biochemicals associated with response/non-response to ketamine therapy in bipolar depression.
In the initial step, a frozen plasma sample was thawed on ice, 3 volumes of ice-cold methanol : ethanol (1:1, v/v) was added to 1 volume of plasma, the mixture vortexed for 1 min, placed on ice for 5 min and then centrifuged at 15 700 g for 20 min at 4°C. The supernatant was collected and filtered through a 0.22 μm nylon filter. Quality Control (QC) samples were prepared by pooling an aliquot of each of the filtered supernatant. The samples were analysed by liquid chromatography following a previously described approach (Ciborowski et al., 2012). All chromatographic separations were performed using a 1200 series HPLC (Agilent Technologies, Waldbronn, Germany), Supelco Discovery HS C18 analytical column (15 cm × 2.1 mm, 3 μm) and guard column (2 cm × 2.1 mm, 3 μm; Sigma Aldrich, Steinheim, Germany), the autosampler and columns were maintained at 4 and 40°C, respectively, and the injection volume was set to 10 μL. Data were collected in positive electrospray ionization mode in separate runs on a QTOF (Agilent 6520) operated in full scan mode from 100 to 1000 m/z. During the analysis, two reference masses: 121.0509 m/z (C5H4N4) and 922.0098 m/z (C18H18O6N3P3F24), were continuously measured to allow constant mass correction. The capillary voltage was 3000 V with a scan rate of 1.02 scan per second and the nebulizer gas flow rate was 10.5 L·min−1. LC-MS grade acetonitrile, methanol, ethanol, formic acid and standards used were purchased from Sigma-Aldrich. Ultra-pure water Milli-Q Water System (Millipore, Billerica, MA, USA) was used for mobile phase and all standard solutions.
LC-QTOF-MS data processing and analysis
Data were re-processed using molecular feature extraction (MFE) tool using the Mass Hunter Qualitative Analysis software B.04.00 (Agilent Technologies) which allowed subtraction of background noise and data reduction. The MFE file gave a list of each mass and retention time pairs with associated intensities for all detected peaks. Alignment and filtering of the primary data were performed on Mass Profiler Professional B.02.00 (Agilent Technologies) software. Masses in the samples that were not present in 100% of participants in at least one group and that had a coefficient of variation above 35% in QC samples were filtered out.
Biomarker discovery
The masses were exported to SIMCA-P+ 12.0 (Umetrics, Umeå, Sweden) for multivariate statistical analysis. Principal component analysis (PCA), partial least square discriminant analysis (PLS-DA) and orthogonal PLS-DA (OPLS-DA) models were used to discriminate the samples. Potential biomarkers were selected through an OPLS-DA model. Differences in metabolites were assessed through the loading column plot where an error bar (jack-knife) was calculated for each variable. Variables with a 95% jack-knife confidence level were selected for further identification.
An external cross-validation test was used to verify the predictability of the PLS-DA model and avoid the risk of overfitting (Rubingh et al., 2006). The sample set was randomized and split in three groups. In each prediction set, one group was excluded and predicted by the remaining groups. The procedure was repeated until the three groups were predicted and the global percentage of samples classified correctly was calculated.
Compound identification
The databases METLIN, LIPID MAPS, MASSTRIX and HMDB were searched for hits against the identified discriminant accurate masses. For each hit, the proposed formula was compared with the experimental isotopic pattern distribution. To confirm the identity of statistically significant compounds, LC-MS/MS analyses were performed on a QTOF (model 6520, Agilent Technologies) using the initial chromatographic conditions. Ions were targeted based on previously determined mass and retention time, nitrogen was used as the collision gas, and collision energy was adjustable with slope of 3.6 V/100 Da and offset 4.8 V for fragmentation. Compound identification was performed as previously described (Lin et al., 2010) – in brief, the corresponding molecular ion (m/z) was searched as extracted ion chromatogram and according to its retention time. Elemental composition of the peak (chemical formula) based on the exact mass and isotope pattern recognition was compared with the database hit considering the probability score. If available, the MS/MS spectra were compared with spectra in the MS/MS spectra library (METLIN). For compounds whose fragmentation pattern was not present in METLIN database, the patterns were predicted using ACD/ChemSketch software v.12.01 (ACD/Labs, Toronto, ON, Canada). When possible, the identity of the compound was confirmed using a commercially available reagent.
Results
Demographics and treatment characteristics of the patient population
The patient samples (n = 22) analysed in the study were obtained at 230 min post-initiation of ketamine or placebo infusion (Diazgranados et al., 2010). The MADRS score at 230 min was compared with the pre-infusion scores and patients with a ≥50% reduction were classified as Rs, and, using the criteria, this study included 13 Rs (67 ± 12% reduction) and 9 NRs (13 ± 13% reduction). Sixteen of the patients were maintained on lithium and 6 on valproate (Table 1).
Table 1.
Clinical characteristics and demographics of the BD patients included in this study
| Characteristics | Combined | Patients taking lithium | Patients taking valproate | |||
|---|---|---|---|---|---|---|
| Responders | Non-responders | Responders | Non-responders | Responders | Non-responders | |
| Sample size (N) | 13 | 9 | 9 | 7 | 4 | 2 |
| Age (years) mean ± SD |
42.3 ± 10.6 | 50.9 ± 10.2 | 40.6 ± 11.1 | 50.6 ± 10.2 | 46.3 ± 9.5 | 52 ± 14.1 |
| Gender (% female) |
69.2 | 66.7 | 77.8 | 57.1 | 50 | 100 |
| Race (% Caucasian) |
100 | 88.9 | 100 | 85.7 | 100 | 100 |
| MADRS score at 230 min mean ± SD |
11 ± 4 | 29 ± 7 | 12 ± 4 | 28 ± 7 | 8 ± 5 | 32 ± 1 |
| Percentage reduction in MADRS mean ± SD |
67 ± 12 | 13 ± 13 | 64 ± 11 | 14 ± 15 | 75 ± 13 | 9 |
BD, bipolar depression; MADRS, Montgomery Åsberg Depression Rating Scale.
Global pharmacometabolomic profiles
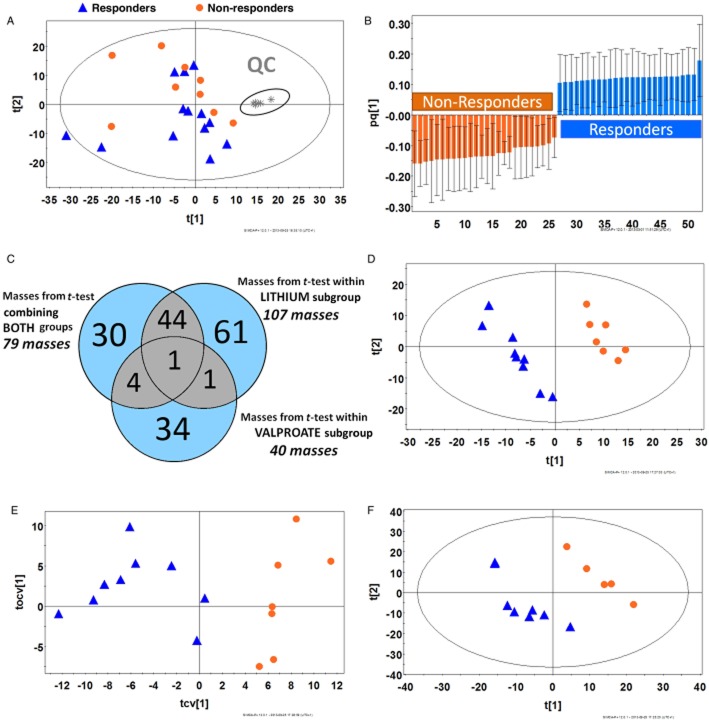
Chromatograms from the analysis of the samples obtained from 22 patients post-ketamine infusion and 6 QC samples were multi-aligned and data were filtered to remove noise. The quality of the global profiling analysis was evaluated using PCA method. The results indicated that the QCs clustered tightly (Figure 2A), suggesting that the dataset can be used for further statistical analysis (Milne et al., 2006). Samples were categorized as coming from R or NRs as described above. The dataset was analysed by PLS-DA and OPLS-DA. Modelling the whole dataset presented both reasonable and poor predictive power (Q2 > 0.4 and 0.2 respectively; data not shown). The t-test analysis indicated that 75 metabolites were statistically significant (P < 0.05). Using these metabolites, an OPLS-DA model was generated. The OPLS-DA was jack-knifed (Figure 2B) and the result suggests that the 52 masses that were selected predicted the metabolic variability observed between the Rs and NRs groups. Putative metabolite identification indicated that 11 metabolites were up-/down-regulated in NRs, with the majority of the compounds identified as lipids and fatty acid acyl derivatives (Table 2). Global profiling was also performed on 17 plasma samples obtained from the placebo arm of the study. The placebo samples were classified as obtained from Rs (n = 10) or NRs (n = 7) based on the observed response in the ketamine arm of the study. Multivariate analysis of the placebo samples did not show any model fitting. However, univariate analysis revealed that in the patients categorized as NRs, octanoyl-and decanoylcarnitines were increased while carnitine and two glycerophospholipids (16:1 and 19:1) were decreased (Supporting Information Table S1).
Figure 2.
Statistical analysis of global profiles derived from bipolar patients treated with ketamine or placebo. (A) PCA model of LC-MS dataset after filtration (970 out of 18926 features) from 22 patients with BD after 230 min post-infusion of ketamine showing the clustering of QC samples (shown in the gray oval) [R2Y = 0.34; Q2 = 0.09]. (B) Covariance plot from OPLS-DA model for metabolites different between Rs and NRs after t-test including jack-knife (confidence interval >95%) [R2Y = 0.98; Q2 = 0.85; n = 75 features]. (C) Venn diagram displaying overlapping metabolites after t-test for the comparison of Rs versus NRs to ketamine therapy in lithium and valproate subgroups individually and when combined together. (D) PLS-DA plot of plasma samples in patients taking lithium in the ketamine arm of the therapy, Rs (n = 9) against NRs (n = 7) using the filtered dataset [R2Y = 0.997; Q2 = 0.672; n = 885 features]. (E) OPLS-DA cross-validated plot of plasma samples in patients taking lithium in the ketamine arm of the therapy, Rs (n = 9) against NRs (n = 7) using the filtered dataset [R2Y = 1.000; Q2 = 0.779; n = 885 features]. (F) PLS-DA model of Rs (n = 8) against NRs (n = 5) from patients taking lithium in the placebo arm [R2Y = 0.949; Q2 = 0.0.539; n = 1325 features]. UV scaling was used for modelling.
Table 2.
Tentative identification of significant metabolites in patients undergoing ketamine treatment
| Compound | RT (min) | Measured mass (Da) | Mass error (p.p.m.) | Molecular formula | FC (%) in NRs | P value | CV (%) for QCs | Biochemical category | Subcategory | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Phenyllactic acid§ | 13.58 | 166.0624 | −3.5 | C9H10O3 | 62.7 | 0.049 | 5.58 | Phenolic, benzoyl, and phenyl derivatives | – |
| 2 | Phenylvaleric acid | 10.78 | 178.0988 | −3.3 | C11H14O2 | 70.6 | 0.015 | 3.53 | Phenolic, benzoyl, and phenyl derivatives | – |
| 3 | LPC (16:1)§ | 16.93 | 493.3162 | −1.3 | C24H48NO7P | −21.2 | 0.048 | 5.51 | Glycerophos-pholipids | Monoacyl-glycerophos-phocholines |
| 4 | Deoxytetradecasphingenine | 21.81 | 227.2240 | −4.0 | C14H29NO | −38.0 | 0.022 | 1.53 | Sphingolipids | Sphingoid base |
| 5 | Deoxytetradecasphinganine | 11.27 | 229.2394 | −5.1 | C14H31NO | 17.4 | 0.029 | 7.99 | Sphingolipids | Sphingoid base |
| 6 | Decanamide | 9.23 | 171.1613 | −5.9 | C10H21NO | −46.3 | 0.045 | 2.20 | Fatty acyls | Primary amides |
| 7 | Pentadecatetraenal | 5.65 | 218.1663 | −3.5 | C15H22O | −42.1 | 0.021 | 14.80 | Fatty acyls | Fatty aldehydes |
| 8 | Dimethyldioxododecatrienal | 24.45 | 234.1269 | 5.6 | C14H18O3 | 33.8 | 0.058 | 9.12 | Fatty acyls | Fatty aldehydes |
| 9 | Hexadienoic acid | 8.82 | 112.0523 | −1.1 | C6H8O2 | 143.4 | 0.030 | 8.37 | Fatty acyls | Unsaturated FAs |
| Hexenedial | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Oxohexenal | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Hydroxy-hexadienal | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Dihydrobenzenediol | C6H8O2 | Phenolic, benzoyl, and phenyl derivatives | ||||||||
| 10 | Hexadienoic acid | 9.22 | 112.0523 | −1.1 | C6H8O2 | 145.1 | 0.029 | 7.69 | Fatty acyls | Unsaturated FAs |
| Hexenedial | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Oxohexenal | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Hydroxy-hexadienal | C6H8O2 | Fatty acyls | Fatty aldehydes | |||||||
| Dihydrobenzenediol | C6H8O2 | Phenolic, benzoyl, and phenyl derivatives | – | |||||||
| 11 | Unknown§ | 7.35 | 162.1038 | C11H14O | 53.9 | 0.024 | 4.47 | – |
Tentative identification of metabolites significantly associated with response to ketamine treatment in 22 BD patients taking either lithium or VPA.
Note: For putative identification, all the compounds with a score >80% were first formula matched with the experimental isotopic pattern distribution on Mass Hunter software.
Found in the comparison of Rs and NRs for lithium-subgroup in the same direction.
FC, fold change; FC was calculated as follows: (Average [NRs] – Average [Rs])/Average [Rs] × 100).
+/–, increase/decrease in NRs when compared with Rs.
LPC, lysophosphatidylcholine; this entity has been named with the number of carbon of the fatty acid attached to the backbone and the number of unsaturation, for example, LPC (16:1).
BD, bipolar depression; KET, ketamine; Li, lithium; NR, non-responders; R, responders; VPA, valproate.
Influence of lithium and valproate on global pharmacometabolomic profiles
In order to determine the influence of mood stabilizer on the response to ketamine therapy, the Rs and NRs within lithium and valproate subgroups were compared independently by univariate t-test analysis. Significant masses from this analysis were compared with masses identified from t-test analysis of the combined (lithium + valproate) group. The results indicate that in the ketamine arm of the study, 28 metabolites were significantly different between patients taking lithium and those receiving valproate (Supporting Information Table S2). Statistically significant masses (P < 0.05) from each of the comparison are displayed in a Venn diagram (Figure 2C). Only two masses were common between the lithium-and the valproate-subgroups, and only one mass was common among the three comparisons, suggesting that the mood stabilizers influenced the metabolomic patterns. These differences were also observed in the samples obtained from the placebo arm of the study (Supporting Information Table S3), indicating that the differences in the metabolomic patterns were associated with the administration of valproate and lithium rather than ketamine. In both the ketamine and placebo arms of the study, the concentrations of acyl-carnitines, octenoic acid and its analogue metabolites were increased in the valproate-subgroup relative to the lithium-subgroup. In addition, there were relative increases in phenylalanine and tryptophan in the lithium-subgroup, which appears to be due to the action of ketamine, independent of patient response. R2Y and Q2 parameters for these models showed high-quality sample classification and good predictive power (Supporting Information Figure S1).
Metabolomic profile of patients taking lithium as the mood stabilizer
The initial objective of the study was to determine the global metabolomic markers associated with the clinical response of patients with BD to treatment with ketamine. Thus, all of the patients were included in the analysis irrespective of the administered mood stabilizer. However, the presence of large differences in the pharmacometabolomic profiles associated with the valproate-subgroup and lithium-subgroup required an independent analysis of the two subgroups. Unfortunately, the number of patients in the valproate-subgroup (n = 6) was not large enough to achieve a significant statistical analysis of the data and only the data from the lithium-subgroup (n = 16) were re-analysed. Therefore, the chromatograms from the lithium-subgroup and QCs were re-aligned and filtered. A PLS-DA model was able to effectively (99.7%) classify the samples from the ketamine arm of the study with a prediction capacity of over 60% (Figure 2D). This indicated that there was an inherent metabolic change between Rs and NRs, which was not observed when the valproate-subgroup was included in the analysis. This model was subjected to an external cross-validation test that demonstrated that 87.5% of the samples were classified correctly.
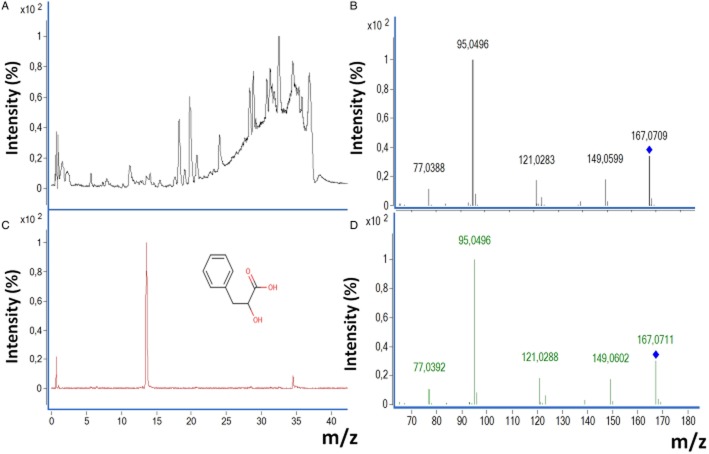
The OPLS-DA model from Rs and NRs was used to select the statistically significant (jack-knife P < 0.05) biomarkers (Figure 2E) and 165 biomarker candidates were selected for further analysis. The identification process included total ion chromatogram, extracted ion chromatogram with retention time, MS/MS experimental spectrum and, when available, MS/MS spectrum of commercial analytical standard. An example of biomarker identification is presented using phenyl lactic acid (PLA), Figure 3. This process identified 18 compounds, 6 of which were increased and 12 were decreased in NRs relative to Rs. The identified metabolites are summarized in Table 3. Of these metabolites, lysophospholipids were confirmed through their characteristic fragments as described in the literature (Milne et al., 2006). For lysophosphatidylcholines (LPCs), fragments 184.07, 104.11 and 86.1 m/z, and for lysophosphatidylethanolamines (LPEs), a fragment of ([M + H]-141.02 m/z) were observed. N6,N6,N6-trimethyl-L-lysine (TML) and a majority of the LPCs (7/9) and LPEs (3/4) were decreased in NRs, relative to Rs, while monoglycerides (MG) and PLA were increased.
Figure 3.
Identification of a selected biomarker using the example of PLA. (A) Total ion chromatogram from plasma sample of a BD patient was selected. (B) Extracted ion chromatogram of 167.0696 (m/z), retention time of 13.6 min. (C) MS/MS experimental spectrum from the ion [167.0696 (m/z), RT = 13.6 min]. (D) MS/MS spectrum of commercial analytical standard PLA reagent (collision energy = 175 V) was also compared with the experimental fragmentation.
Table 3.
Metabolites significantly different between Rs and NRs taking lithium in the ketamine arm of the therapy
| Compound | RT (min) | Measured mass (Da) | Mass error (p.p.m.) | Molecular formula | MS/MS fragments | FC (%) in NRs | P value | CV (%) for QCs | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Phenyllactic acid¶, § | 13.58 | 166.0623 | −4.2 | C9H10O3 | 167.07, 149.06, 121.03, 95.05, 77.04 | 47.9 | 0.028 | 5.1 |
| 2 | N6,N6,N6-Trimethyl-L-lysine | 0.61 | 188.1509 | −10.1 | C9H20N2O2 | 189.16, 173.02, 130.09, 84.08 | −22.9 | JK | 19.7 |
| 3 | MG (16:0) | 27.67 | 330.2765 | −1.5 | C19H38O4 | 331.28,313.27, 257.24, 239.24, 123.11, 109.10, 95.08, 85.10, 83.08, 81.07, 71.09, 57.07 | 12.4 | JK | 12.1 |
| 4 | MG (16:0) | 28.38 | 330.2772 | 0.6 | C19H38O4 | 331.28,313.27, 257.24, 239.24, 123.11, 109.10, 95.08, 85.10, 71.09, 57.07 | 12.9 | JK | 7.9 |
| 5 | LPE (18:2) | 17.46 | 477.2854 | −0.2 | C23H44NO7P | 478.30, 337.27, 81.07 | −39.1 | JK | 8.1 |
| 6 | LPE (18:2) | 18.12 | 477.2852 | −0.7 | C23H44NO7P | 478.29, 338.28, 337.27, 306.28, 135.11, 95.08, 81.07, 62.06 | −46.1 | 0.029 | 3.2 |
| 7 | LPE (18:1) | 20.64 | 479.3006 | −1.2 | C23H46NO7P | 480.31, 340.30, 339.29, 308.29, 155.01, 62.06 | −19.8 | JK | 2.8 |
| 8 | LPE (22:6) | 18.13 | 525.2848 | −1.4 | C27H44NO7P | 526.28, 385.27, 93.10, 79.06, 62.06 | 25.2 | JK | 3.3 |
| 9 | LPC (16:1)§ | 16.94 | 493.3163 | −1.1 | C24H48NO7P | 494.32, 476.31, 184.07, 104.11, 86.10 | −39.8 | 0.031 | 6.5 |
| 10 | LPC (16:0) | 19.06 | 495.3331 | 1.2 | C24H50NO7P | 496.33, 184.07, 86.10 | −14.2 | JK | 7.6 |
| 11 | LPC (16:0) | 19.87 | 495.3337 | 2.4 | C24H50NO7P | 496.34, 184.07, 104.11, 86.10 | −6.3 | JK | 4.1 |
| 12 | LPC (18:2) | 17.63 | 519.3325 | 0.0 | C23H48NO7P | 520.34, 184.07, 86.10 | −16.8 | JK | 11.2 |
| 13 | LPC (18:2) | 18.29 | 519.3334 | 1.8 | C23H48NO7P | 520.34, 502.33, 184.07, 104.11, 86.10 | −19.8 | JK | 10.8 |
| 14 | LPC (20:3) | 19.77 | 545.3479 | −0.4 | C28H52NO7P | 546.35, 184.07, 104.11, 86.10 | −56.0 | JK | 16.7 |
| 15 | LPC (20:2) | 21.88 | 547.3622 | −2.9 | C28H54NO7P | 548.37, 184.07, 104.11, 86.10 | −22.3 | JK | 17.9 |
| 16 | LPC (22:6) | 17.75 | 567.3311 | −2.5 | C30H50NO7P | 568.34, 184.07, 86.10 | 36.0 | 0.042 | 13.8 |
| 17 | LPC (22:6) | 18.27 | 567.3320 | −0.9 | C30H50NO7P | 568.34, 184.07, 104.11, 86.10 | 33.1 | JK | 19.2 |
| 18 | Unknown§ | 7.35 | 162.1038 | – | C11H15O | 163.11, 135.08, 107.05 | 41.5 | 0.026 | 5.1 |
Note: FC, fold change; FC was calculated as follows: (Average [NRs] – Average [Rs])/Average [Rs] × 100); +/–-increase/decrease in NRs compared with Rs; Unknown, compound which MS/MS spectra was not interpretable or not informative.
Confirmed also by standard
Found in the comparison of Rs and NRs in combined subgroups (Li + VPA) in the same direction;
In MS/MS fragments the underlined number refers to the most abundant fragment observed.
MG, monoglyceride; LPE, lysophosphatidylethanolamine; LPC, lysophosphatidylcholine, these entities has been named with the number of carbon of the fatty acid attached to the backbone and the number of unsaturation, for example, LPC (16:1); JK, jack-knife analysis.
Li, lithium; NR, non-responders; R, responders; VPA, valproate.
A good sample separation was obtained with plasma samples from the lithium-subgroup from the placebo arm of the study using the PLS-DA model (Figure 2F, Table 4). The comparison of the data from Rs and NRs in the placebo arm indicated that the same acyl-carnitines and glycerophospholipid compounds were significantly different and followed the same pattern as observed in the combined (lithium + valproate) placebo group. The metabolites from placebo and ketamine arms did not match, suggesting that the metabolites identified in Table 3 are associated with response to ketamine therapy in the lithium-subgroup.
Table 4.
Tentative identification of metabolites significantly different between Rs and NRs taking lithium in the placebo arm of the therapy
| Compound | RT (min) | Measured mass (Da) | Mass error (p.p.m.) | Molecular formula | FC (%) in NRs | P value | CV (%) for QCs | Biochemical category | Subcategory | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1a | Octanoylcarnitine | 4.14 | 287.2090 | −2.3 | C15H29NO4 | 65.0 | 0.028 | 8.92 | Fatty acyls | Fatty esters |
| 2a | Decanoylcarnitine | 8.18 | 315.2403 | −2.1 | C17H33NO4 | 66.6 | 0.024 | 7.64 | Fatty acyls | Fatty esters |
| 3 | Dodecanoylcarnitine | 12.10 | 343.2716 | −1.9 | C19H37NO4 | 40.7 | 0.049 | 5.59 | Fatty acyls | Fatty esters |
| N-palmitoyl serine | C19H37NO4 | Fatty acyls | N-acyl amines | |||||||
| 4a | LPC (16:1) | 16.41 | 493.3166 | −0.5 | C24H48NO7P | −34.9 | 0.025 | 8.21 | Glycerophospholipids | Monoacylglycerophosphocholines |
| LPE (19:1) | C24H48NO7P | Glycerophospholipids | Monoacylglycerophosphoethanolamines | |||||||
| 5 | Trp Phe | 1.03 | 351.1588 | 1.4 | C20H21N3O3 | −40.8 | 0.030 | 27.88 | Dipeptide |
Note: For putative identification, all the compounds with a score >80% were first formula matched with the experimental isotopic pattern distribution on Mass Hunter software.
Also found changed in the same direction in the comparison of Rs and NRs in the ketamine arm infusion.
FC, fold change; FC was calculated as follows: (Average [NRs] – Average [Rs])/Average [Rs] × 100).
+/–, increase/decrease in NRs when compared with Rs.
LPC, lysophosphatidylcholine; this entity has been named with the number of carbon of the fatty acid attached to the backbone and the number of unsaturation, for example, LPC (16:1).
KET, ketamine; NR, non-responders; R, responders.
Discussion
Over the past five decades, a considerable number of antidepressants have been developed, representing a wide range of molecular and therapeutic classes. The primary pharmacological activities associated with these agents are either based on 5-HT (SSRI) or on noradrenaline (5-HT-noradrenaline reuptake inhibitors;SNRI). However, there are a substantial number of patients with major depression who do not respond to current antidepressant therapy and none of these agents have demonstrated a significant advantage in terms of clinical efficacy. In addition, there is a considerable latency in the onset of antidepressant effects, which typically take 6 weeks or more. Due to these limitations, new therapeutic targets are being explored, with the hopes of developing more effective and rapidly acting treatments. A primary focus in this effort is the development of glutamatergic modulators, particularly those acting at the NMDA receptor.
Ketamine, an NMDA receptor modulator, is currently under investigation for use in the treatment of depression and neuropathic pain. We have demonstrated that a subanesthetic dose of ketamine produces rapid antidepressant effects in patients diagnosed with BD (Diazgranados et al., 2010) and MDD (Zarate et al., 2006), and these effects can last up to 7 days (Zarate et al., 2012). A simultaneous population pharmacokinetic model for ketamine and three of its major metabolites in BD patients has been developed, utilizing plasma samples collected up to 3 days after ketamine administration (Zhao et al., 2012), and a pharmacodynamic study in a combined cohort of patients with MDD (n = 45) and BD (n = 22) has also been conducted (Zarate et al., 2012). The data from these studies indicate that at 230 min after infusion, the clinical effect of ketamine robustly separates from placebo (Zarate et al., 2006; 2012; Diazgranados et al., 2010). In addition, 76% (25/33) of Rs at 230 min continued to meet response criteria at day 1 and only 10% (5/50) of NRs at 230 min were classified as Rs at a later time point. Based upon these data, the 230 min time point was chosen as the sampling point to capture clinical response and non-response. Thus, the preliminary metabolomic comparison of ketamine Rs and NRs in the treatment of BD was limited to the analysis of the 230 min plasma samples.
The initial analysis of 22 BD patients included in the study indicated that there were response-related differences in metabolomic patterns (Table 2), which was confirmed by PLS-DA and OPLS-DA models. After compound selection (Figure 2B) and database identification, 11 compounds were increased or decreased in Rs relative to NRs. A majority of the compounds were from the fatty acyl family. The observation of differences in fatty acyl metabolism between ketamine Rs and NRs was supported by analysis of plasma samples from the placebo arm in which acyl-carnitines were increased and L-carnitine decreased in NRs compared with Rs (Supporting Information Table S1).
While the differences between NRs and Rs were statistically significant in univariate analysis, in multivatiare analysis the differences were small and the quality of the models poor. A potential source of this result was the lack of homogeneity within the BD cohort produced by maintenance on either lithium or valproate. Therefore, the plasma samples from the ketamine arm of the study were divided into lithium – and valproate-subgroups based upon the mood stabilizers administered to the BD patients. The metabolomic pattern of each subgroup was determined and then compared in a Venn diagram, with the data from the analysis of the combined (lithium + valproate) group (Table 1). The patterns were significantly different with only two common compounds (Figure 2C). A similar result was obtained from the analysis of the samples from the placebo arm of the study, in which there was a greater than twofold increase in acyl-carnitines in the valproate-subgroup compared with lithium-subgroup (Supporting Information Table S3). This result is consistent with valproate-associated depletion of free carnitine due to formation of valproyl carnitine and decreased tubular reabsorption of acetylcarnitine (Lheureux and Hantson, 2009) and with the observations that valproate affects mitochondrial fatty acid oxidation (Silva et al., 2008).
In the ketamine arm of the study, the relative concentrations of phenylalanine, tryptophan and bilirubin were increased in the lithium-subgroup and amino-octanoic acid, and arachidonoyl-serine was increased in the valproate-subgroup, independent of response to treatment with ketamine. Increased plasma levels of phenylalanine have been previously observed in heart failure patients diagnosed with MDD, relative to matched controls, suggesting a potential alteration in the phenylalanine metabolic pathway (Steffens et al., 2010); and tyrosine plasma concentrations, a product of another metabolic pathway of phenylalanine, are also affected in the depressed patients (Nordin, 1988; Kaddurah-Daouk et al., 2012). In addition, data from a proton nuclear magnetic resonance spectroscopy-based metabolonmic analysis using post mortem brain tissue from patients with a history of BD and rat brains obtained after chronic treatment with lithium or valproate indicate that valproate treatment increased glutamate levels and decreased glutamate/glutamine ratios, while lithium treatment increased GABA levels (Lan et al., 2009). The results indicate that even though the concomitant administration of lithium or valproate does not produce a difference in the clinical response to ketamine therapy, it does produce a difference in the background metabolomic patterns. Based upon these differences, the analysis of the combined subgroups was not continued and only the plasma samples from the lithium-subgroup were subjected to additional analysis. It is important to note that these differences do not negate the use of a pretreatment metabolomics screen, but only highlights the challenges in application.
The metabolomic patterns in BD patients maintained on lithium were significantly different between Rs and NRs (Figure 2D and E). Eighteen of the 165 biomarker candidates selected for further analysis were identified, and the majority (15/18) of these were LPEs (4), LPCs (9) and MGs (2). The signals associated with 3/4 LPEs and 7/9 LPCs were increased in Rs relative to NRs, while the signals associated with both of the identified MGs were decreased in Rs (Table 2). The data indicate that there are differences in mitochondrial fatty acid metabolism between the two groups.
The analysis of the plasma samples also revealed that TML was increased in Rs, compared with levels in NRs. TML is a precursor of L-carnitine, a key factor in the mitochondrial β-oxidation of fatty acids. The differences in TML plasma level in Rs and NRs are consistent with the dissimilarities in fatty acid metabolism between the two groups. In addition, in the samples from the placebo arm of the study, octanoyl-and decanoyl-carnitines were up-regulated in NRs compared with Rs. While acyl-carnitines have antidepressant effects in elderly (Pettegrew et al., 2000; 2002) and mice (Di Cesare Mannelli et al., 2011), increased concentrations in NRs may reflect a dysfunction in signalling of the mitochondrial acyl-carnitine receptor. It is of interest to note that TML is synthesized from lysine by S-adenosyl-methionine (SAM) catalyzed N-methylation and that SAM is in turn synthesized from L-methionine. Higher levels of methionine were observed in MDD patients who were in remission (Kaddurah-Daouk et al., 2012), which may reflect an alteration in the conversion of methionine to SAM affecting LPC synthesis.
In contrast to the increased levels of LPCs, LPEs and TML, the signals associated with PLA were decreased in Rs, compared with NRs. PLA is a product of the NADH-mediated reduction of phenylpyruvate and an indication of a disruption in phenylalanine metabolism. The potential alteration in phenylalanine metabolic pathways is in accord with previous observations that phenylalanine plasma levels are higher in heart failure patients diagnosed with MDD, relative to matched controls (Steffens et al., 2010) and that the tyrosine pathway is compromised in depressed patients (Nordin, 1988; Kaddurah-Daouk et al., 2012). Tyrosine is produced by the phenylalanine metabolic pathway. Moreover PLA and LPC (16:1) followed similar pattern of changes observed in plasma samples obtained from the combined group (lithium + valproate), suggesting that the differences in fatty acid metabolism and phenylalanine pathway were involved in the response or /non-response to ketamine, independent of the co-medication.
The results of this study indicate that in BD patients an underlying basis for a positive response or a non-response to treatment with ketamine is a difference in the mitochondrial metabolism of fatty acids. While the experimental approach used in this study, metabolomics, relies on an unbiased analysis of data, the identification of markers associated with mitochondrial function is not surprising. Mitochondrial function or dysfunction has been associated with mood disorders including BD (Horrobin and Bennett, 1999; Hroudova and Fisar, 2012; Tang and Wang, 2012). In a recent review of data from brain imaging studies and mitochondrial functional studies, the authors concluded that the results support the hypothesis of mitochondrial dysfunction in BD and suggest that BD is associated with decreased energy production and a shift towards anaerobic glycolysis (Minuzzi et al., 2011). A key marker of changes in disease-related mitochondrial function is fatty acid and phospholipid metabolism (Horrobin and Bennett, 1999) and metabolomic studies have demonstrated that fatty acid metabolism is significantly lower and shifted from β-oxidation to ω-oxidation in depressed patients, compared with that in non-depressed controls (Maes et al., 1996; Paige et al., 2007; Steffens et al., 2010) and BD-related changes in phospholipid metabolism (Modica-Napolitano and Renshaw, 2004).
In summary, this is the first study of the plasma metabolomic patterns in patients receiving ketamine for the treatment of BD. The results indicate that there are distinct biochemical differences between patients who respond to treatment (Rs) and those who do not (NRs), and that the differences appear to be due to alterations in the mitochondrial metabolism of fatty acids. The major observation is that the differences in the metabolomics patterns observed between R and NRs were not produced by ketamine administration. Instead, they appear to set up the biochemical basis for the pharmacological response to ketamine. Thus, pretreatment metabolomics screening may be a guide to the prediction of response and a potential approach to the individualization of ketamine therapy in BD. In addition, these differences appear to be associated with disease-related dysregulation of mitochondrial function and networks. While the source of these differences is not clear, previous studies have identified genetic links between BD and variants in mitochondrial DNA (Hroudova and Fisar, 2012). Additional prospective studies will be required to better understand these observations.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (NIH) and the National Institute of Mental Health, NIH. The authors also acknowledge the funding from Spanish Ministerio de Economía y Competitividad (MEC) grant CTQ2011-23562 and A. V. acknowledges her fellowship to EADS CASA.
Glossary
- 5-HT
5-hydroxytryptamine
- BD
bipolar depression
- LC-QTOF-MS
liquid chromatography coupled to QTOF-MS
- LPC
lysophosphatidylcholine
- LPE
lysophosphatidylethanolamine
- MADRS
Montgomery Åsberg Depression Rating Scale
- MDD
major depressive disorder
- MFE
molecular feature extraction algorithm
- MG
monoglycerides
- NR
non-responder
- OPLS-DA
orthogonal partial least square discriminant analysis
- PCA
principal component analysis
- PLA
phenyl lactic acid
- PLS-DA
partial least square discriminant analysis
- PTSD
post-traumatic stress disorder
- Q2
prediction power score
- QC
quality control
- QTOF
quadrupole time-of-flight mass spectrometer
- R
responder
- R2Y
classification score
- SAM
S-adenosyl-methionine
- SNRI
serotonin-noradrenaline reuptake inhibitors
- SSRI
selective 5-HT reuptake inhibitors
- TML
N6,N6,N6-trimethyl-L-lysine
Conflict of interest
C. Z. and I. W. W. have submitted a patent for the use of ketamine metabolites in the treatment of bipolar disorder and major depression. They have assigned their rights in the patent to the U.S. government, but will share a percentage of any royalties that may be received by the government. A. R., M. S., G. L., C. B., M. P. L. report no potential conflicts of interest.
EADS CASA (who provided a Fellowship for A. V.) is an aeronautical company with no interest or property in the results.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site: http://dx.doi.org/10.1111/bph.12494
Figure S1 Statistical analysis of global profiles of patients taking lithium or valproate with ketamine /placebo. (A) PLS-DA and (B) OPLS-DA cross-validated plot of lithium (n = 10) versus valproate (n = 6) in ketamine arm of the therapy (Rs and NRs are combined within each group) [for (A), R2Y = 0.99; Q2 = 0.51 and for (B), R2Y = 1.00; Q2 = 0.80; n = 1080 features]. (C) PLS-DA and (D) OPLS-DA crossvalidated plot of lithium (n = 7) versus valproate (n = 4) in placebo arm of the therapy (Rs and NRs were classified according to their response to ketamine therapy [for (A), R2Y = 0.99; Q2 = 0.85 and for (B), R2Y = 1.00; Q2 = 0.81; n = 1325 features]. Key: – Li,
– Li, – VPA; UV scaling was used for modelling.
– VPA; UV scaling was used for modelling.
Table S1 Tentative identification of metabolites in the placebo arm of the study after global profiling of the entire cohort.
Table S2 Tentative identification of metabolites significantly different between patients taking lithium or valproate in the ketamine arm of the therapy.
Table S3 Tentative identification of metabolites significantly different between patients taking lithium or valproate in the placebo arm of the therapy.
References
- Abo R, Hebbring S, Ji Y, Zhu H, Zeng Z-B, Batzler A, et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes’ single-nucleotide polymorphism imputation: selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenet Genomics. 2012;22:247–253. doi: 10.1097/FPC.0b013e32835001c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs S, French D. Depression and anxiety: role of mitochondria. Curr Anaesth Crit Care. 2007;18:34–41. [Google Scholar]
- Ciborowski M, Teul J, Martin-Ventura JL, Egido J, Barbas C. Metabolomics with LC-QTOF-MS permits the prediction of disease stage in aortic abdominal aneurysm based on plasma metabolic fingerprint. PLoS ONE. 2012;7:e31982. doi: 10.1371/journal.pone.0031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare Mannelli L, Vivoli E, Salvicchi A, Schiovone N, Koverechi A, Messano M, et al. Antidepressant-like effect of artemin in mice: a mechanism for acetyl-L-carnitine activity on depression. Psychopharmacology (Berl) 2011;218:347–356. doi: 10.1007/s00213-011-2326-0. [DOI] [PubMed] [Google Scholar]
- Diazgranados N, Ibrahim L, Brutsche NE, Newberg A, Kronstein P, Khalife S, et al. A randomized add-on trial of an N-methyl-D-aspartate antagonist in treatment-resistant bipolar depression. Arch Gen Psychiatry. 2010;67:793–802. doi: 10.1001/archgenpsychiatry.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. Rapid antidepressant effects of ketamine ignite drug discovery. Nat Med. 2013;19:8. doi: 10.1038/nm0113-8. [DOI] [PubMed] [Google Scholar]
- Horrobin DF, Bennett CN. Depression and bipolar disorder: relationships to impaired fatty acid and phospholipid metabolism and to diabetes, cardiovascular disease, immunological abnormalities, cancer, ageing and osteoporosis. Possible candidate genes. Prostaglandins Leukot Essent Fatty Acids. 1999;60:217–234. doi: 10.1054/plef.1999.0037. [DOI] [PubMed] [Google Scholar]
- Hroudova J, Fisar Z. In vitro inhibition of mitochondrial respiratory rate by antidepressants. Toxicol Lett. 2012;213:345–352. doi: 10.1016/j.toxlet.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Krishnan K. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology. 2009;34:173–186. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R, Yuan P, Boyle SH, Matson W, Wang Z, Zeng ZB, et al. Cerebrospinal fluid metabolome in mood disorders-remission state has a unique metabolic profile. Sci Rep. 2012;2:667. doi: 10.1038/srep00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan MJ, McLoughlin GA, Griffin JL, Tsang TM, Huang JTJ, Yuan P, et al. Metabonomic analysis identifies molecular changes associated with the pathophysiology and drug treatment of bipolar disorder. Mol Psychiatry. 2009;14:269–279. doi: 10.1038/sj.mp.4002130. [DOI] [PubMed] [Google Scholar]
- Lheureux PE, Hantson P. Carnitine in the treatment of valproic acid-induced toxicity. Clin Toxicol (Phila) 2009;47:101–111. doi: 10.1080/15563650902752376. [DOI] [PubMed] [Google Scholar]
- Lin L, Yu Q, Yan X, Hang W, Zheng J, Xing J, et al. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst. 2010;135:2970–2978. doi: 10.1039/c0an00265h. [DOI] [PubMed] [Google Scholar]
- Machado-Vieira R, Salvadore G, Luckenbaugh DA, Manji HK, Zarate CA., Jr Rapid onset of antidepressant action: a new paradigm in the research and treatment of major depressive disorder. J Clin Psychiatry. 2008;69:946–958. doi: 10.4088/jcp.v69n0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Smith R, Christophe A, Cosyns P, Desnyder R, Meltzer HY. Fatty acid composition in major depression: decreased ω3 fractions in cholesteryl esters and increased c20:4ω6/c20:5ω3 ratio in cholesteryl esters and phospholipids. J Affect Disord. 1996;38:35–46. doi: 10.1016/0165-0327(95)00092-5. [DOI] [PubMed] [Google Scholar]
- Milne S, Ivanova P, Forrester J, Alex Brown H. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 2006;39:92–103. doi: 10.1016/j.ymeth.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Minuzzi L, Behr GA, Moreira JCF, Frey BN. Mitochondrial dysfunction in bipolar disorder: lessons from brain imaging and molecular markers. Rev Colomb Psiquiatr. 2011;40:166S–182S. [Google Scholar]
- Modica-Napolitano JS, Renshaw PF. Ethanolamine and phosphoethanolamine inhibit mitochondrial function in vitro: implications for mitochondrial dysfunction hypothesis in depression and bipolar disorder. Biol Psychiatry. 2004;55:273–277. doi: 10.1016/s0006-3223(03)00784-4. [DOI] [PubMed] [Google Scholar]
- Nordin C. Relationships between clinical symptoms and monoamine metabolite concentrations in biochemically defined subgroups of depressed patients. Acta Psychiatr Scand. 1988;78:720–729. doi: 10.1111/j.1600-0447.1988.tb06411.x. [DOI] [PubMed] [Google Scholar]
- Paige LA, Mitchell MW, Krishnan KR, Kaddurah-Daouk R, Steffens DC. A preliminary metabolomics analysis of older adults with and without depression. Int J Geriatr Psychiatry. 2007;22:418–423. doi: 10.1002/gps.1690. [DOI] [PubMed] [Google Scholar]
- Pettegrew JW, Levine J, McClure RJ. Acetyl-L-carnitine physical-chemical, metabolic, and therapeutic properties: relevance for its mode of action in Alzheimer's disease and geriatric depression. Mol Psychiatry. 2000;5:616–632. doi: 10.1038/sj.mp.4000805. [DOI] [PubMed] [Google Scholar]
- Pettegrew JW, Levine J, Gershon S, Stanley JA, Servan-Schreiber D, Panchalingam K, et al. 31P-MRS study of acetyl-L-carnitine treatment in geriatric depression: preliminary results. Bipolar Disord. 2002;4:61–66. doi: 10.1034/j.1399-5618.2002.01180.x. [DOI] [PubMed] [Google Scholar]
- Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–176. doi: 10.1016/j.nbd.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Rubingh CM, Bijlsma S, Derks EPPA, Bobeldijk I, Verheij ER, Kachar S, et al. Assessing the performance of statistical validation tools for megavariate metabolomics data. Metabolomics. 2006;2:53–61. doi: 10.1007/s11306-006-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Bahn S. The utility of biomarker discovery approaches for the detection of disease mechanisms in psychiatric disorders. Br J Pharmacol. 2008;153:S133–S136. doi: 10.1038/sj.bjp.0707658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MFB, Aires CCP, Luis PB, Ruiter JPN, Ijlst L, Duran M, et al. Valproic acid metabolism and its effects on mitochondrial fatty acid oxidation: a review. J Inherit Metab Dis. 2008;31:205–216. doi: 10.1007/s10545-008-0841-x. [DOI] [PubMed] [Google Scholar]
- Steffens DC, Jiang W, Krishnan KRR, Karoly ED, Mitchell MW, O'Connor CM, et al. Metabolomic differences in heart failure patients with and without major depression. J Geriatr Psychiatry Neurol. 2010;23:138–146. doi: 10.1177/0891988709358592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YA, Wu J, Zhang L, Zhang Q, Su DM, He P, et al. Dysregulated mitochondrial genes and networks with drug targets in postmortem brain of patients with posttraumatic stress disorder (PTSD) revealed by human mitochondria-focused cDNA microarrays. Int J Biol Sci. 2008;4:223–235. doi: 10.7150/ijbs.4.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang V, Wang JF. Oxidative stress in bipolar disorder. Biochem Anal Biochem. 2012:S2–002. doi: 10.4172/2161-1009.S2-002. [Google Scholar]
- Zarate CA, Jr, Singh JB, Carlson PJ, Brutsche ME, Ameli R, Luckenbaugh DA, et al. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry. 2006;63:856–864. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- Zarate CA, Jr, Brutsche N, Laje G, Luckenbaugh DA, Venkata SLV, Ramamoorthy A, et al. Relationship of ketamine's plasma metabolites with efficacy, diagnosis, and psychotomimetic effects in patients with treatment-resistant depression. Biol Psychiatry. 2012;72:331–338. doi: 10.1016/j.biopsych.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou R, Li X, Ursano RJ, Li H. Stress-induced change of mitochondria membrane potential regulated by genomic and non-genomic GR signaling: a possible mechanism for hippocampus atrophy in PTSD. Med Hypotheses. 2006;66:1205–1208. doi: 10.1016/j.mehy.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Zhao X, Swarajya LV, Moaddel R, Luckenbaugh DA, Brutsche NE, Ibrahim L, et al. Simultaneous population pharmacokinetic modeling of ketamine and three major metabolites in patients with treatment-resistant bipolar depression. Br J Clin Pharmacol. 2012;74:304–314. doi: 10.1111/j.1365-2125.2012.04198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Statistical analysis of global profiles of patients taking lithium or valproate with ketamine /placebo. (A) PLS-DA and (B) OPLS-DA cross-validated plot of lithium (n = 10) versus valproate (n = 6) in ketamine arm of the therapy (Rs and NRs are combined within each group) [for (A), R2Y = 0.99; Q2 = 0.51 and for (B), R2Y = 1.00; Q2 = 0.80; n = 1080 features]. (C) PLS-DA and (D) OPLS-DA crossvalidated plot of lithium (n = 7) versus valproate (n = 4) in placebo arm of the therapy (Rs and NRs were classified according to their response to ketamine therapy [for (A), R2Y = 0.99; Q2 = 0.85 and for (B), R2Y = 1.00; Q2 = 0.81; n = 1325 features]. Key: – Li,
– Li, – VPA; UV scaling was used for modelling.
– VPA; UV scaling was used for modelling.
Table S1 Tentative identification of metabolites in the placebo arm of the study after global profiling of the entire cohort.
Table S2 Tentative identification of metabolites significantly different between patients taking lithium or valproate in the ketamine arm of the therapy.
Table S3 Tentative identification of metabolites significantly different between patients taking lithium or valproate in the placebo arm of the therapy.