Abstract
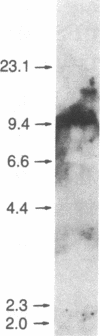
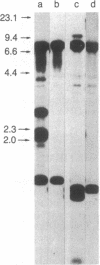
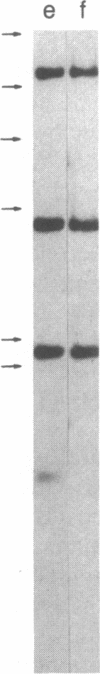
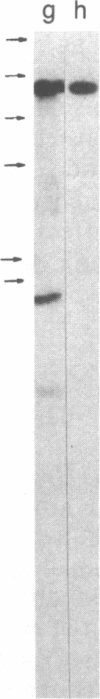
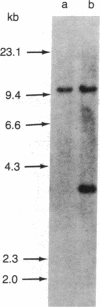
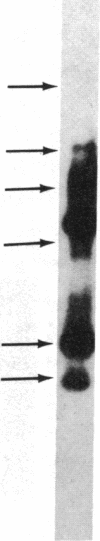
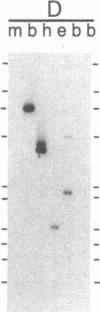
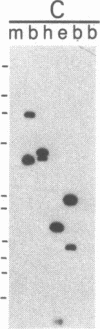
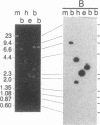
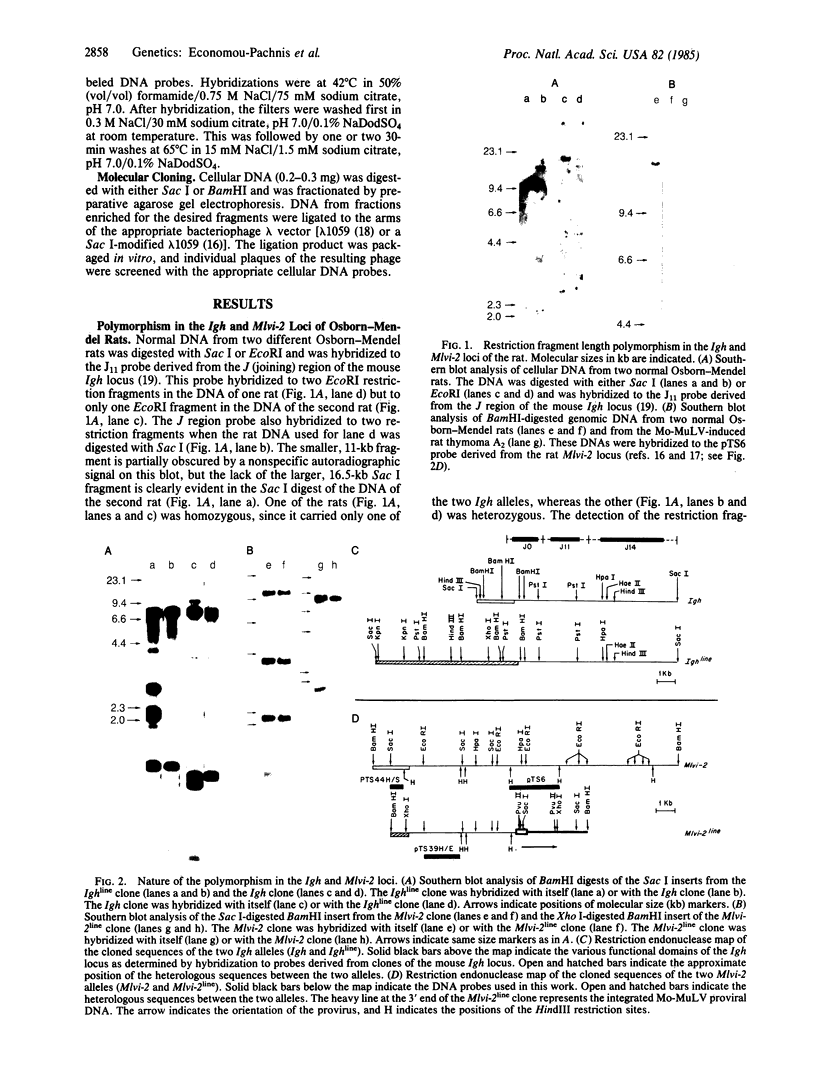
Restriction enzyme analysis of normal DNA derived from individual rats of the National Institutes of Health outbred Osborn-Mendel colony revealed that two independent single-copy loci, the Igh (immunoglobulin heavy chain) locus and the Mlvi-2 (Moloney leukemia virus integration 2) locus, a putative oncogene, are polymorphic (i.e., exhibit allelic variation). The polymorphism at both loci was due to the presence or absence of a long interspersed repeated DNA element (LINE). The LINE insertion in the Igh locus occurred in the joining (J) region, which is involved in the physiological rearrangement of this locus. The LINE insertion in the Mlvi-2 locus has occurred approximately 6 kilobases from the region of provirus integration in Moloney murine leukemia virus-induced rat thymomas. The two inserts are colinear with each other and with other randomly selected cloned copies of the rat LINE family, the general characteristics of which we also present. LINE insertion in the Mlvi-2 locus was observed in several rat strains, established from independent rat colonies, suggesting that LINE-containing Mlvi-2 alleles may be widespread in the rat population. LINE insertion in the Igh locus was observed in 1 of 27 rats. The detection of a LINE-related polymorphism at two nonselected loci indicates that LINEs are transposable. The presence or absence of these long (greater than 5 kilobases), highly transcribed elements at single-copy loci could have profound effects on gene activity. Furthermore, LINE-containing single-copy loci could be affected by homologous interaction between the resident LINE and any of the other 50,000 or so copies of these elements in the rat genome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. W., Kaufman R. E., Kretschmer P. J., Harrison M., Nienhuis A. W. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980 Dec 20;8(24):6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971 Jun;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Fanning T. G. Size and structure of the highly repetitive BAM HI element in mice. Nucleic Acids Res. 1983 Aug 11;11(15):5073–5091. doi: 10.1093/nar/11.15.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuff E. L., Feenstra A., Lueders K., Smith L., Hawley R., Hozumi N., Shulman M. Intracisternal A-particle genes as movable elements in the mouse genome. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1992–1996. doi: 10.1073/pnas.80.7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R. B., Stanton L. W., Marcu K. B. On immunoglobulin heavy chain gene switching: two gamma 2b genes are rearranged via switch sequences in MPC-11 cells but only one is expressed. Nucleic Acids Res. 1982 Jan 22;10(2):611–630. doi: 10.1093/nar/10.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemischka I., Sharp P. A. The sequences of an expressed rat alpha-tubulin gene and a pseudogene with an inserted repetitive element. Nature. 1982 Nov 25;300(5890):330–335. doi: 10.1038/300330a0. [DOI] [PubMed] [Google Scholar]
- Lerman M. I., Thayer R. E., Singer M. F. Kpn I family of long interspersed repeated DNA sequences in primates: polymorphism of family members and evidence for transcription. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3966–3970. doi: 10.1073/pnas.80.13.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier-Rotival M., Bernardi G. The Bam repeats of the mouse genome belong in several superfamilies the longest of which is over 9 kb in size. Nucleic Acids Res. 1984 Feb 10;12(3):1593–1608. doi: 10.1093/nar/12.3.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier-Rotival M., Soriano P., Cuny G., Strauss F., Bernardi G. Sequence organization and genomic distribution of the major family of interspersed repeats of mouse DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(2):355–359. doi: 10.1073/pnas.79.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder A., Leder P. Unusual alpha-globin-like gene that has cleanly lost both globin intervening sequences. Proc Natl Acad Sci U S A. 1980 May;77(5):2806–2809. doi: 10.1073/pnas.77.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H., Tsuzuki T., Wakasugi S., Fukuda M., Shimada K. Interruption of a human nuclear sequence homologous to mitochondrial DNA by a member of the KpnI 1.8 kb family. Nucleic Acids Res. 1984 Jul 11;12(13):5225–5234. doi: 10.1093/nar/12.13.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Wu J. R., Bonner J. Analysis of rat repetitive DNA sequences. Biochemistry. 1978 Jan 10;17(1):51–59. doi: 10.1021/bi00594a008. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Siebenlist U., Korsmeyer S., Waldmann T., Leder P. Structure of the human immunoglobulin mu locus: characterization of embryonic and rearranged J and D genes. Cell. 1981 Dec;27(3 Pt 2):583–591. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980 Aug 14;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Shafit-Zagardo B., Brown F. L., Maio J. J., Adams J. W. KpnI families of long, interspersed repetitive DNAs associated with the human beta-globin gene cluster. Gene. 1982 Dec;20(3):397–407. doi: 10.1016/0378-1119(82)90208-6. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Singer M. F., Thayer R. E., Grimaldi G., Lerman M. I., Fanning T. G. Homology between the KpnI primate and BamH1 (M1F-1) rodent families of long interspersed repeated sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5739–5745. doi: 10.1093/nar/11.16.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer R. E., Singer M. F. Interruption of an alpha-satellite array by a short member of the KpnI family of interspersed, highly repeated monkey DNA sequences. Mol Cell Biol. 1983 Jun;3(6):967–973. doi: 10.1128/mcb.3.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Kozak C. A. Cellular DNA region involved in induction of thymic lymphomas (Mlvi-2) maps to mouse chromosome 15. Mol Cell Biol. 1984 May;4(5):997–1000. doi: 10.1128/mcb.4.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S. W., Denison R. A., Bernstein L. B., Weiner A. M., Manser T., Gesteland R. F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981 Oct;26(1 Pt 1):11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Witney F. R., Furano A. V. Highly repeated DNA families in the rat. J Biol Chem. 1984 Aug 25;259(16):10481–10492. [PubMed] [Google Scholar]
- Yamada S., Masuko K., Ito M., Nagayo T. Spontaneous thyoma in Buffalo rats. Gan. 1973 Jun;64(3):287–291. [PubMed] [Google Scholar]