Abstract
Purpose
To identify disease-causing mutations in Chinese families who presented with retinitis pigmentosa (RP) and Leber congenital amaurosis (LCA).
Methods
The pathogenic variant in a Chinese family with autosomal dominant RP was investigated with a specific hereditary eye disease enrichment panel (HEDEP) based on targeted exome capture technology. The identified variant was confirmed with Sanger sequencing. CRB1 mutations in 67 patients with sporadic retinal dystrophy were examined with Sanger sequencing.
Results
Compound heterozygous mutations were identified in a family who had undergone HEDEP analysis. After complete sequence analysis of the CRB1 gene was performed in 67 patients with sporadic retinal dystrophy, other compound heterozygous mutations were detected in three families. The mutations included three novel heterozygous mutations: c.3059delT (p.M1020SfsX1), c.3460T>A (p.C1154S), and c.4207G>C (p.E1403Q). The mutation frequency of CRB1 in this study was 5.9% (8/136).
Conclusions
Our findings broaden the spectrum of CRB1 mutations and the phenotypic spectrum of the disease in Chinese patients. The results from this study show that patients with LCA carry CRB1 null mutations more frequently than patients with RP.
Introduction
Mutations in the Crumbs homolog 1 (CRB1) gene are known to cause retinitis pigmentosa-12 (RP-12) [1] and Leber congenital amaurosis 8 (LCA8) [2]. A CRB1 mutation was also reported in a family with possible autosomal dominant inherited pigmented paravenous chorioretinal atrophy (PPCRA) [3], but this was not verified by other studies. RP is caused by progressive loss of rod and cone photoreceptors. Typical symptoms include night blindness followed by decreasing visual fields, leading to tunnel vision and eventually blindness. With a prevalence of 1:3,784, RP is one of the most common types of retinal degeneration in China [4]. In contrast, Leber congenital amaurosis (LCA) is the most severe nonsyndromic retinal dystrophy, characterized by blindness or severe visual impairment, extinguished electroretinogram (ERG), and nystagmus before the age of 1 year [5]. There is clinical overlap between LCA and early onset RP, and in some cases, the diagnosis is ambiguous. According to a previous report, CRB1 mutations were a relatively frequent cause of autosomal recessive (ar) early onset retinal degeneration in Israeli and Palestinian populations (10% of families with LCA), and caused severe retinal degeneration at an early age [6].
CRB1 is a human homolog of the Drosophila transmembrane Crumbs protein, and is preferentially expressed in the inner segments of mammalian photoreceptors, also expressed in the brain [1,7]. The Crumbs protein has been implicated in mechanisms that control cell–cell adhesion, apicobasal cell polarity, and photoreceptor morphogenesis [7-10]. CRB1 is composed of 12 exons, and may give rise to four known isoforms. The longest isoform consists of 1,406 amino acids and contains an extracellular region, a transmembrane domain, and a cytoplasmic region. The extracellular region contains 19 epidermal growth factor (EGF)-like domains, three laminin A globular (AG)-like domains, and a signal peptide. The cytoplasmic region contains a conserved FERM binding domain, which is involved in localizing proteins to the plasma membrane, and a PDZ binding motif (PBM), used in the process of anchoring to the cytoskeleton [1,7,11]. To date, up to 194 mutations have been found in the CRB1 gene (among those, 59 were reported as non-disease-causing single nucleotide polymorphisms [SNPs]) with 189 occurring in the extracellular region, one in the transmembrane domain, and four in the cytoplasmic region.
In this study, four pairs of compound heterozygous mutations in the CRB1 gene were identified in four Chinese families presenting with RP and LCA. Among these mutations, three are first reported in this study. We discuss the clinical variability of patients harboring CRB1 mutations.
Methods
Patients
One Chinese family (family 1) of Han ethnicity with arRP was identified at the Beijing Tongren Eye Center. Family 1 has three affected individuals in the same generation; 11 individuals, including three affected and eight unaffected, participated in the study (Figure 1A). Medical and ophthalmic histories were obtained, and ophthalmological examinations were performed. Sixty-seven patients who presented with sporadic RP (45 cases), LCA (18 cases), and PPCRA (four cases) at the Department of Ophthalmology, Peking University Third Hospital were recruited for CRB1 gene mutation screening. One hundred general healthy individuals from the Chinese Han ethnic population were recruited to serve as controls. All procedures used in this study conformed to the tenets of the Declaration of Helsinki. All experiments involving DNA of the patients and their relatives were approved by Peking University Third Hospital Medical Ethics Committee. Informed consent was obtained from all participants.
Figure 1.
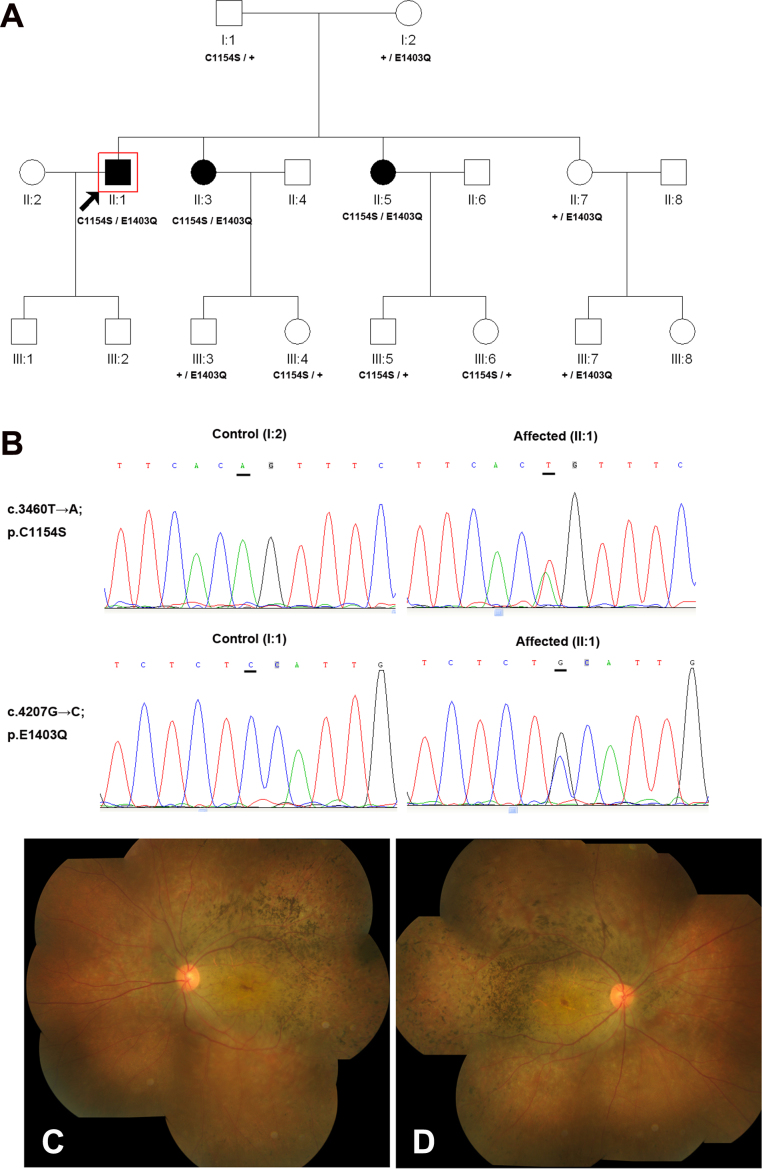
Family 1 with compound heterozygote for c.3460T>A (p.C1154S) and c.4207G>C (p.E1403Q) in the CRB1 gene. A: The pedigree of family 1. The filled symbols represent affected individuals and unfilled symbols unaffected individuals. Squares signify men, and circles women. An arrow marks the index patient; + means a normal allele. B: Representative sequence chromatograms for the proband (right) and his normal parents (left). C–D: Fundus examinations in the 35-year-old proband showed attenuation of retinal arterioles, numerous pigment deposits, and RPE degeneration mainly in the temporal quadrant and the posterior pole.
Mutation screening
Genomic DNA was prepared from venous leukocytes with standard protocols (D2492 Blood DNA Maxi Kit, Omega Bio-Tek, Norcross, GA). High-throughput DNA sequencing was applied for the mutation screening of family 1. Briefly, a specific hereditary eye disease enrichment panel (HEDEP) based on targeted exome capture technology was used to collect the protein-coding regions of the targeted genes (designed by MyGenostics, Baltimore, MD). This HEDEP captured 371 hereditary eye disease genes, which covered 35 arRP associated genes (ABCA4, BEST1, C2orf71, C8ORF37, CERKL, CLRN1, CNGA1, CNGB1, CRB1, DHDDS, EYS, FAM161A, IDH3B, IMPG2, LRAT, MAK, MERTK, NR2E3, NRL, PDE6A, PDE6B, PDE6G, PRCD, PROM1, RGR, RHO, RLBP1, RP1, RPE65, SAG, SPATA7, TTC8, TULP1, USH2A, ZNF513). Fifty μg of genomic DNA from the proband (II:1) and his parents (I:1 and I:2) were used for targeted exome capture. The exon-enriched DNA libraries were then prepared for high throughput sequencing with the Illumina HiSeq 2000 (Illumina, San Diego, CA) platform. The obtained mean exome coverage was more than 98%, with variants accuracy at more than 99%. The changes were filtered against exome data from ethnic Han Chinese Beijing available in the 1000 Genomes Project, and against the Han Chinese Beijing SNPs in the dbSNP131. We analyzed only the mutations that occurred in 35 genes related to arRP for family 1. Sanger sequencing was then used to validate the identified potential disease-causing variants in available family members.
CRB1 mutations in 67 patients with sporadic retinal dystrophies (including families 2–5) were examined with Sanger sequencing. All the exons (exons 1–12) and exon–intron boundaries of CRB1 (NM_201253.2) were amplified and sequenced with the primers listed in Table 1. Sequencing results were analyzed with Sequencher (Gene Codes, Ann Arbor, MI); the identified mutations were further evaluated for segregation in available family members and 100 controls. The possible pathogenicity of missense changes was evaluated using the PolyPhen-2 and SIFT prediction programs. Primer pairs for individual exons were designed using the Primer program.
Table 1. Primers used for CRB1 amplification and sequencing.
| Primer | Forward | Reverse | Product |
|---|---|---|---|
|
CRB1 Exon 1 |
TGAGACAGACAGGGATCAGGA |
AAGCCAGAAATAAACCAGGAA |
195 |
|
CRB1 Exon 2 |
TTTGGTTGAGGCAGCACAAA |
TCTGCTTCTGCCACTTAGAAA |
738 |
|
CRB1 Exon 3 |
AACAAAGCATTGTCAAATTGC |
CCGAGAACGTGAGAGCTCTAA |
371 |
|
CRB1 Exon 4 |
AAGATGATGCCATGGGTCTT |
ATTTTGTCACTCACCAGGCCA |
262 |
|
CRB1 Exon 5 |
CAGGCACATCAACTTGCTAAA |
AGCCATGGTCTGCCATAAAA |
358 |
|
CRB1 Exon 6 |
GCTATTCATGCACTTCTGCAA |
TTTGCTGTTTCTGCTCTGCT |
1122 |
|
CRB1 Exon 7 |
TGCTTGTGTGCATGTGTGTGT |
TGGTGGGTCAGTAACATCATC |
779 |
|
CRB1 Exon 8 |
CACCACTCTGCCCTTTTAGAA |
AGGCAAGAGGCCAGTCAGTAT |
529 |
|
CRB1 Exon 9 |
TTCTTCTTCCATAAAATGGGG |
CTTGAGGAGAGAGCTTTCCAA |
1205 |
|
CRB1 Exon 10 |
GCTCCTCCAGCCTGAGTACTT |
CGACAGCAACCATATTTGCA |
583 |
|
CRB1 Exon 11 |
GCTGTTCCAGAGAGATAAGGC |
AATCATAGTGATGGAGGGCAA |
710 |
| CRB1 Exon 12 | TGTCGCCTTGTTACTGATCCT | TCCAGTGTAATCCCAGTTGCA | 278 |
Results and Discussion
In this study, four pairs of compound heterozygote mutations in the CRB1 gene were identified in four Chinese families. These included c.3460T>A and c.4207G>C (family 1), c.1831T>C and c.3059delT (family 2), c.1576C>T and c.2234C>T (family 3), and c.1429G>A and c.1576C>T (family 4). Among these mutations, three are first reported in this study (c.3059delT, c.3460T>A, and c.4207G>C). The mutation frequency of CRB1 in this study was 5.9% (8/136).
Family 1
We selected three individuals in family 1 for targeted exome capture. We generated an average of 0.77 Gb of sequence with 228× average coverage for each individual with paired 100 bp reads. The generated sequence covered average 99.0% of the targeted bases with the accuracy of a variant call more than 99%. About 450 variants were identified in each patient with HEDEP analysis. After filtering against exome data from ethnic Han Chinese Beijing available in the 1000 Genomes Project, and against the Han Chinese Beijing SNPs in the dbSNP131, about 20 variants were left. The heterozygous compound changes or homozygous changes in the affected individual but heterozygous changes in his parents were identified as the potential disease-causing mutation. Novel compound heterozygosity for c.3460T>A (p.C1154S) and c.4207G>C (p.E1403Q) in the CRB1 gene was found. Sanger sequencing validation and segregation analysis were performed, which demonstrated that compound heterozygosity for c.3460T>A (p.C1154S) and c.4207G>C (p.E1403Q) was cosegregated with the disease phenotype in this family, and the unaffected family members carried only one mutant allele (Figure 1A–B). These two mutations were absent in the 100 normal controls. The CRB1 mutations found in this study are summarized in Table 2. The two novel missense mutations were perfectly conserved among mammals and yielded a high score for pathogenicity using the online SIFT and PolyPhen-2 prediction programs (Figure 2). The proband and other affected members in the family had complained of night blindness since 20 years old, followed by progressive loss of visual acuity from the third decade of life. Fundus examinations in the 35-year-old proband (II:1) showed attenuation of retinal arterioles, numerous pigment deposits, and RPE degeneration mainly in the temporal quadrant and the posterior pole (Figure 1C–D).
Table 2. CRB1 mutations identified in four families with retinitis pigmentosa and Leber congenital amaurosis.
| Family Number | Clinical Diagnosis | Allele 1 Exon Nucleotide 1 Protein 1 | Allele 2 Exon Nucleotide 2 Protein 2 | Sex | Age(y) | Onset age (yrs) | Visual acuity (corrected) R eye L eye | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 |
RP |
Exon 9 |
c.3460T>A* |
p.C1154S* |
Exon 12 |
c.4207G>C* |
p.E1403Q* |
M |
35 |
25 |
0.3 |
0.1 |
| Family 2 |
LCA |
Exon 6 |
c.1831T>C |
p.S611P |
Exon 9 |
c.3059delT* |
p.M1020SfsX1* |
M |
7 |
<1 |
0.01 |
0.01 |
| Family 3 |
LCA |
Exon 6 |
c.1576C>T |
p.R526X |
Exon 7 |
c.2234C>T |
p.T745M |
F |
24 |
1 |
0.05 |
FC |
| Family 4 | LCA | Exon 6 | c.1429G>A | p.G477R | Exon 6 | c.1576C>T | p.R526X | M | 22 | childhood | 0.02 | 0.02 |
* Novel mutation FC means finger counting.
Figure 2.

Evaluation of two novel CRB1 missense mutations. Multiple alignments using Clustal W and amino acid conservation of three novel missense sequence variants were performed. The alignment results showed that cysteine at codon 1154 and glutamic acid at codon 1403 were fully conserved through all species. The predicted effect of the mutation on the protein was estimated with the online prediction programs SIFT and PolyPhen-2, which are shown at the bottom. For each mutation, a high pathogenicity score for the altered amino acid was obtained.
The mutation p.C1154S localized to the third cysteine residues of the 15th EGF-like domain. EGF-like domains typically consist of six cysteine residues that interact with each other by forming disulfide bridges. These stabilize the native fold, which comprises a major and minor β-sheet. Disulfide bridges are formed between the first and third cysteine residues, the second and fourth residues, and the fifth and sixth residues [12]. The mutation p.C1154S affected the formation of disulfide bridges and therefore caused disruption of the protein secondary structure. The mutation p.E1403Q happened in the cytoplasmic region of the CRB1 protein. Previous reports have identified only four mutations in the cytoplasmic region of the CRB1 gene: p.P1381L [13], p.R1383H [14], p.R1390X [15], and c.4121_30del [16]. The cytoplasmic domain was of crucial importance since it has been shown to link CRB homologs to several cytoplasmic proteins, for example, the conserved C-terminal ERLI motif (codon 1403–06) bound to PDZ domains [11]. The novel p.E1403Q amino acid substitution affected a residue that is highly conserved in the ERLI motif, and therefore affected its binding properties with PDZ domains. The heterozygous mutation of p.C1154S and p.E1403Q alone was not disease-causing. When they coexisted, RP developed. It was assumed that these two mutations disrupted the appropriate assembly and localization of the CRB1 protein.
Families 2, 3, and 4
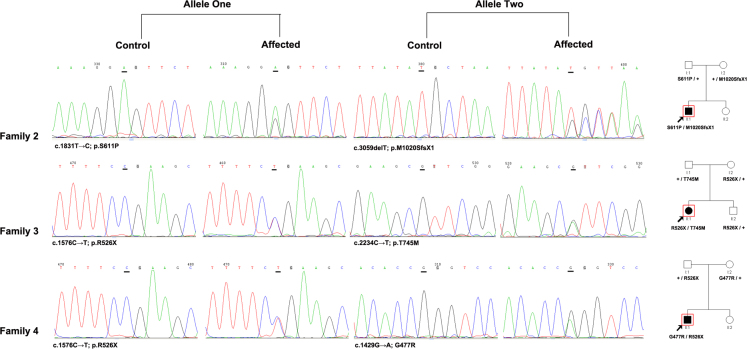
Upon complete sequence analysis of the coding and adjacent intronic region of the CRB1 gene in 67 patients with sporadic retinal dystrophy, novel compound heterozygous mutations for c.1831T>C (p.S611P) and c.3059delT (p.M1020SfsX1) were identified in family 2, c.1576C>T (p.R526X) and c.2234C>T (p.T745M) were identified in family 3, and c.1429G>A (p.G477R) and c.1576C>T (p.R526X) were identified in family 4 (Figure 3). The patients inherited the two heterozygous mutations from their father and mother, and these mutations were absent in the 100 normal controls. The 7-year-old proband of family 2 had complained of poor vision, photophobia, and nystagmus since he was 1 year old, with severe visual loss (0.01) at this visit. Fundus examination showed widespread nummular pigment clumps and macula atrophy (data not shown). The 24-year-old proband of family 3 had complained of poor vision, color vision defect, and nystagmus since she was 1 year old, with severe visual loss (0.05 in the right eye and finger counting in the left eye) at this visit. Fundus examination showed widespread nummular pigment clumps admixed with small white spots, but the optic disc appeared relatively normal (data not shown). The 22-year-old proband of family 4 had complained of poor vision since childhood, with severe visual loss (0.02) at this visit. Fundus examination showed numerous nummular pigments admixed with white spots throughout the fundus, and macular degeneration was also observed (Figure 4).
Figure 3.
Representative sequence chromatograms for the probands and normal controls in families 2–4.
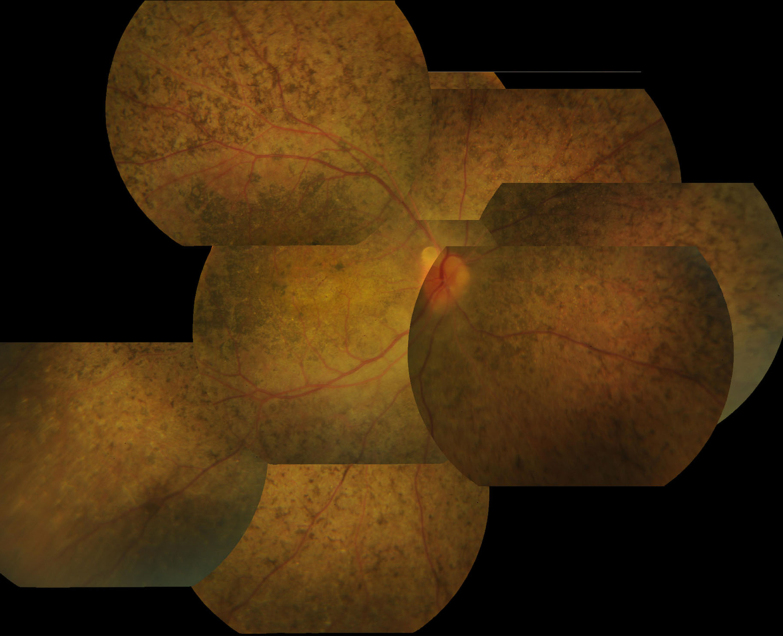
Figure 4.
Fundus images of a patient with Leber congenital amaurosis (from family 4). The patient was a 22-year-old man who had had poor vision since childhood, with severe visual loss (0.02) at this visit. Fundus examination showed numerous nummular pigments admixed with white spots throughout the fundus; macular degeneration was also observed.
Among the compound heterozygous mutations, one heterozygote was a missense mutation, and the other heterozygote was a mutation resulting in premature termination codons (PTCs). The c.1831T>C (p.S611P) mutation identified in family 2 was first reported by Li et al. in a patient with LCA [17]. The c.2234C>T (p.T745M) mutation identified in family 3 was first reported by den Hollander et al. in a patient with RP [1], and was further verified by Henderson et al. in patients with early onset retinal dystrophy [13]. The c.1429G>A (p.G477R) mutation identified in family 4 was first reported by Abu-Safieh et al. in a patient with RP recently [18]. The p.S611P and p.T745M mutations localized to the laminin AG-like domain, which have been identified in various proteins and served as protein interaction modules. These mutations have also been predicted to affect calcium binding and protein folding [11,19,20]. The mutation p.G477R localized to the 11th EGF-like domain, which typically consists of six cysteine residues that interact with each other by forming disulfide bridges. The nonsense mutation p.R526X (identified in families 3 and 5) was first reported by Seong in Korean patients with LCA [21]. The nonsense mutation p.R526X and the novel small deletion c.3059delT (p.M1020SfsX1) resulted in truncation proteins without the transmembrane and cytoplasmic domains. According to a previous report, CRB1 mutations bearing PTCs before the last exon behaved as a null allele, resulted in haploinsufficiency as their corresponding mRNA was degraded by nonsense-mediated mRNA decay (NMD) [22]. Therefore, the function loss of CRB1 was the most probable reason in these patients. All patients who carried p.R526X and c.3059delT mutations were diagnosed with LCA. Den Hollander et al. showed that patients with LCA carry CRB1 null mutations more frequently than patients with RP [14].
In summary, we report compound heterozygous mutations of the CRB1 gene in four of 67 Chinese families and showed that mutations in CRB1 accounted for 5.8% of the Chinese patients with retinal dystrophy in this study. Our findings broaden the spectrum of CRB1 mutations and the phenotypic spectrum of the disease in Chinese patients.
Acknowledgments
The authors thank the patients and all family members for their participation in this study. This study was supported by the National Natural Science Foundation of China (Grant 81,170,877). Dr. Zhizhong Ma (mazzpuh3@163.com) and Dr. Genlin Li (ligenglin@263.net) are co-corresponding authors for this paper.
References
- 1.den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, van de Pol DJ, Payne AM, Bhattacharya SS, Kellner U, Hoyng CB, Westerveld A, Brunner HG, Bleeker-Wagemakers EM, Deutman AF, Heckenlively JR, Cremers FP, Bergen AA. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat Genet. 1999;23:217–21. doi: 10.1038/13848. [DOI] [PubMed] [Google Scholar]
- 2.Lotery AJ, Jacobson SG, Fishman GA, Weleber RG, Fulton AB, Namperumalsamy P, Héon E, Levin AV, Grover S, Rosenow JR, Kopp KK, Sheffield VC, Stone EM. Mutations in the CRB1 gene cause Leber congenital amaurosis. Arch Ophthalmol. 2001;119:415–20. doi: 10.1001/archopht.119.3.415. [DOI] [PubMed] [Google Scholar]
- 3.McKay GJ, Clarke S, Davis JA, Simpson DA, Silvestri G. Pigmented paravenous chorioretinal atrophy is associated with a mutation within the crumbs homolog 1 (CRB1) gene. Invest Ophthalmol Vis Sci. 2005;46:322–8. doi: 10.1167/iovs.04-0734. [DOI] [PubMed] [Google Scholar]
- 4.Hu DN. Prevalence and mode of inheritance of major genetic eye diseases in China. J Med Genet. 1987;24:584–8. doi: 10.1136/jmg.24.10.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Beryozkin A, Zelinger L, Bandah-Rozenfeld D, Harel A, Strom TA, Merin S, Chowers I, Banin E, Sharon D. Mutations in CRB1 are a relatively common cause of autosomal recessive early-onset retinal degeneration in the Israeli and Palestinian populations. Invest Ophthalmol Vis Sci. 2013;54:2068–75. doi: 10.1167/iovs.12-11419. [DOI] [PubMed] [Google Scholar]
- 7.Richard M, Roepman R, Aartsen WM, van Rossum AG, den Hollander AI, Knust E, Wijnholds J, Cremers FP. Towards understanding CRUMBS function in retinal dystrophies. Hum Mol Genet. 2006;15:R235–43. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- 8.Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12 is essential for photoreceptor morphogenesis. Nature. 2002;416:143–9. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- 9.Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–83. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- 10.Chartier FJ, Hardy ÉJ, Laprise P. Crumbs limits oxidase-dependent signaling to maintain epithelial integrity and prevent photoreceptor cell death. J Cell Biol. 2012;198:991–8. doi: 10.1083/jcb.201203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosens I, den Hollander AI, Cremers FP, Roepman R. Composition and function of the Crumbs protein complex in the mammalian retina. Exp Eye Res. 2008;86:713–26. doi: 10.1016/j.exer.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Cooke RM, Wilkinson AJ, Baron M, Pastore A, Tappin MJ, Campbell ID, Gregory H, Sheard B. The solution structure of human epidermal growth factor. Nature. 1987;327:339–41. doi: 10.1038/327339a0. [DOI] [PubMed] [Google Scholar]
- 13.Henderson RH, Mackay DS, Li Z, Moradi P, Sergouniotis P, Russell-Eggitt I, Thompson DA, Robson AG, Holder GE, Webster AR, Moore AT. Phenotypic variability in patients with retinal dystrophies due to mutations in CRB1. Br J Ophthalmol. 2011;95:811–7. doi: 10.1136/bjo.2010.186882. [DOI] [PubMed] [Google Scholar]
- 14.den Hollander AI, Davis J, van der Velde-Visser SD, Zonneveld MN, Pierrottet CO, Koenekoop RK, Kellner U, van den Born LI, Heckenlively JR, Hoyng CB, Handford PA, Roepman R, Cremers FP. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat. 2004;24:355–69. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- 15.Corton M, Tatu SD, Avila-Fernandez A, Vallespín E, Tapias I, Cantalapiedra D, Blanco-Kelly F, Riveiro-Alvarez R, Bernal S, García-Sandoval B, Baiget M, Ayuso C. High frequency of CRB1 mutations as cause of Early-Onset Retinal Dystrophies in the Spanish population. Orphanet J Rare Dis. 2013;8:20. doi: 10.1186/1750-1172-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber S, Perrault I, Hanein S, Shalev S, Zlotogora J, Barbet F, Ducroq D, Dufier J, Munnich A, Rozet J, Kaplan J. A novel mutation disrupting the cytoplasmic domain of CRB1 in a large consanguineous family of Palestinian origin affected with Leber congenital amaurosis. Ophthalmic Genet. 2002;23:225–35. doi: 10.1076/opge.23.4.225.13879. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Xiao X, Li S, Jia X, Wang P, Guo X, Jiao X, Zhang Q, Hejtmancik JF. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PLoS ONE. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, Al-Zahrani J, Al-Abdi L, Hashem M, Al-Tarimi S, Sebai MA, Shamia A, Ray-Zack MD, Nassan M, Al-Hassnan ZN, Rahbeeni Z, Waheeb S, Alkharashi A, Abboud E, Al-Hazzaa SA, Alkuraya FS. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–47. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JA, Handford PA, Redfield C. The N1317H substitution associated with Leber congenital amaurosis results in impaired interdomain packing in human CRB1 epidermal growth factor-like (EGF) domains. J Biol Chem. 2007;282:28807–14. doi: 10.1074/jbc.M704015200. [DOI] [PubMed] [Google Scholar]
- 20.Tepass U, Theres C, Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–99. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- 21.Seong MW, Kim SY, Yu YS, Hwang JM, Kim JY, Park SS. Molecular characterization of Leber congenital amaurosis in Koreans. Mol Vis. 2008;14:1429–36. PMID:18682808. [PMC free article] [PubMed] [Google Scholar]
- 22.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]