Abstract
Purpose
To describe and validate a semi-automated targeted sampling (SATS) method for quantifying optic nerve axons in a feline glaucoma model.
Methods
Optic nerve cross sections were obtained from 15 cats, nine with mild to severe glaucoma and six with normal eyes. Optic nerves were dissected, fixed in paraformaldehyde and glutaraldehyde, and processed for light microscopy by resin embedding, sectioning, and staining of axon myelin sheaths with 1% p-phenylenediamine before axon quantification. Commercially available image analysis software was used as a semi-automated axon counting tool (SCT) and was first validated by comparison with a manual axon count (MAC). This counting tool was then used in a SATS method performed by three masked raters and in a semi-automated full count (SAFC) method performed by a single observer. Correlation was assessed between the SCT and MAC using a linear model and analysis of covariance (ANCOVA). Correlation between the SATS and SAFC methods was calculated and the bias, systematic errors, and variance component assessed. The intraclass correlation coefficient (ICC) was determined to establish inter-rater agreement. In addition, the time required to perform the SATS and SAFC methods was evaluated.
Results
Correlation between the axon counts obtained by the SCT and MAC was strong (r = 0.9985). There was evidence of an overcounting of axons by the SCT compared to the MAC with a percentage error rate of 13.0% (95% confidence interval [CI] 11.0%, 15.1%). Both the correlation of SATS count (average per rater) to SAFC (r = 0.9891) and inter-rater agreement (ICC = 0.986) were high. The SATS method presented an overall positive counting error (p<0.001) when compared to the SAFC, consistent with a fixed percentage overestimation of 11.2% (95% CI 8.3%, 14.2%) of the full count. The average time required to quantify axons by the SATS method was 10.9 min, only 27% of that required to conduct the SAFC.
Conclusions
Our data demonstrate that the SATS method provides a practical, rapid, and reliable means of estimating axon counts in the optic nerves of cats with glaucoma.
Introduction
Glaucoma is a leading cause of vision loss in the adult human population and although less prevalent, a devastating cause of vision loss in children. Loss of vision results from the death of retinal ganglion cells (RGCs) and the loss of their axons, which constitute the optic nerve. It is now widely accepted that the initial insult responsible for the characteristic axonopathy occurs within the lamina cribrosa region of the optic nerve head. We have established a colony of cats with a spontaneously arising, recessively inherited, primary congenital glaucoma (PCG) associated with a mutation in the latent transforming growth factor beta binding protein 2 (LTBP2; OMIM 602091) gene [1,2]. These animals represent an authentic homolog of human PCG at the Glaucoma 3, primary congenital, D (GLC3D; OMIM 613086) locus [3,4]. To facilitate the practical application of this model in translational research, an efficient, accurate, and reproducible method of quantification of RGC and/or optic nerve axon loss is required. Most conventional methods for quantifying RGCs are time intensive and costly, necessitating complex immune-labeling protocols or even requiring prior surgical intervention to back label RGCs with Fluoro-Gold (Fluorochrome, Denver, CO) or 1.1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI), which would be impractical and undesirable in our large animal model [5]. Quantification of RGCs in the feline retina is complicated by the substantial population of small (<10 µm) neurons, identified as displaced amacrine cells, that are estimated to account for approximately 80% of the cell population in the ganglion cell layer of the cat [6-9]. The histomorphometry of RGCs is further complicated in felines by the uneven distribution of RGCs throughout the fundus, with the greatest density in the area centralis [10,11]. We therefore elected to focus our efforts on the evaluation of optic nerve axon loss in this model. The most accurate method of quantifying optic nerve axons is the full axon count technique performed either manually using transmission electron microscopy (TEM) or semi-automatically by light microscopy. These techniques are extremely time consuming and not 100% accurate, requiring extrapolations to circumvent their inherent underestimation of axon count numbers [12].
Faster methods, such as random sampling of the optic nerve cross-sectional area [13] or semiquantitative grading [12], present unreliable results due to focal variability in the degree of nerve damage and subjectivity of grading schemes. In our feline model, as in human glaucoma, a regional pattern of axon loss is often observed. Therefore, we elected to modify a rapid method, initially developed for use in rodent models, that combines automated counting and targeted sampling of distinct regions of relatively uniform optic nerve damage [14]. The purpose of our study was to validate a semi-automated targeted sampling (SATS) method for quantifying optic nerve axons in cats.
Methods
Subjects
We used fixed optic nerve tissues collected postmortem from cats in a research colony that had been established from a pedigree of cats with spontaneously occurring recessively inherited PCG. Samples were selected from animals ranging in age from 6 months to 6 years and representing a range of different stages in the progression of disease. Weekly intraocular pressure (IOP) data, as measured by rebound tonometry [15], were available for all cats in the study. All glaucomatous animals had persistently elevated IOP, a mean (standard deviation [SD]) IOP of 41.4 (14.6) mmHg, while mean (SD) IOP in normal cats was 18.3 (2.4) mmHg. The cup-to-disc ratio cannot be reliably assessed on ophthalmoscopy in cats, but all cats in the former group exhibited abnormalities consistent with glaucoma on electrophysiological testing (visual evoked potentials and pattern electroretinography) or optical coherence tomography. Single optic nerves were selected from normal cats (n = 6) and from cats that demonstrated mild to severe PCG due to a consistent mutation in LTBP2 (n = 9). All procedures were conducted with the approval of the University of Wisconsin-Madison institutional animal care and use committee.
Tissue processing
Animal were euthanized by an intravenous overdose of pentobarbital sodium (120-180 mg/kg). Immediately following euthanasia, animals were transcardially perfused with ice-cold 0.1 M PBS (100 mM phosphate, 154 mM sodium chloride, pH 7.4), followed by 4% paraformaldehyde solution in 0.1 M PBS (Electron Microscopy Sciences, Washington, PA). Eyes were enucleated and fixed overnight in 4% paraformaldehyde in 0.1 M PBS (Electron Microscopy Sciences) at 4 °C, then transferred to 0.4% paraformaldehyde in 0.1 M PBS for storage at 4 °C. An approximately 2-mm long segment of optic nerve was dissected 2 mm posterior to the globe. Shallow radial orientation cuts were made, one superiorly and two temporally, before postfixation in 2.5% glutaraldehyde/0.1 M PBS for 48 h at 4 °C. Nerve samples were then osmicated in 1% osmium tetroxide in 0.1 M PBS, rinsed, and dehydrated through an ascending series of alcohol treatments before routine epoxy resin embedding and sectioning. Semithin (1-µm) sections were stained with 1% p-phenylenediamine (PPD) for evaluation by light microscopy.
Optic nerve imaging and axon counting
Full optic nerve cross sections were analyzed from each optic nerve. Images from the PPD-stained optic nerve sections were obtained using a bright light microscope Olympus BX43 (Olympus Inc., Center Valley, PA) with attached Olympus DP72 digital camera (Olympus Inc.) and captured using commercially available image analysis software (cellSens Dimension®, Olympus Inc.). As described below, the software’s semi-automated axon counting tool (SCT) was compared to a manual axon count (MAC) for validation and then used in SATS and semi-automated full count (SAFC) methods. Comparisons were then made between the SATS and SAFC methods for axon counting. All counts were performed by raters masked to animal identity and disease status and to the other raters’ results.
Validation of the semi-automated counting tool
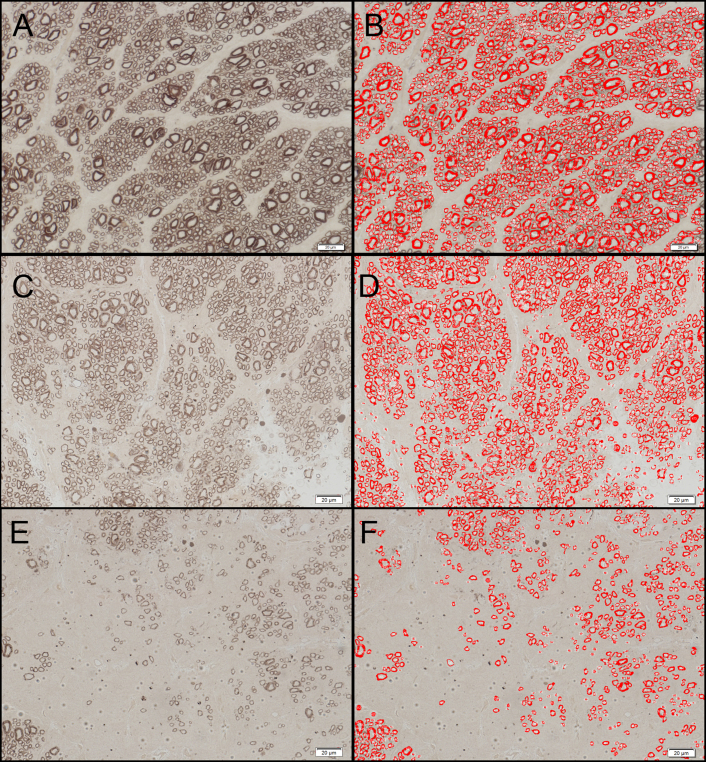
A semi-automated axon detection process using the built-in automatic object recognition tool of the image analysis software was compared to the MAC method by a single rater. Digital images from nine different fields were obtained under a 63X microscope objective lens. The images represented three subjectively distinct areas of the optic nerves from each of three different affected animals including those judged to be normal or densely populated areas (n = 3), moderately damaged areas (n = 3), and severely damaged areas (n = 3). Semi-automated axon detection was performed using cellSens Dimension® image analysis software. The images were analyzed under the “count and measure” menu. Using the adaptive threshold tool, the black PPD-stained myelin sheath of the axons was selected by visually adjusting the threshold values until all the axons in the field were selected, taking care not to obliterate the smaller axons (Figure 1). We established that a threshold with a minimum intensity value of −255 and maximum value of −11 was appropriate for our samples, and this threshold was used for all subsequent semi-automated axon counting (including SATS and SAFC methods described below). The “auto split objects” tool was applied to the whole image to separate contiguous axons, and the “count and measure” tool was used to count the axons. Nonaxonal darkly staining elements of the optic nerve tissue that were being counted by the software were manually selected and deleted. Axons at the margins of the image that were not entirely visible were automatically excluded from counting by the software. The final axon count for each image was recorded.
Figure 1.
Semi-automated counting tool used in axon detection and quantification. Histologic sections of the optic nerves from affected cats. The images represent three distinct areas of the optic nerves judged as normal or densely populated (A), moderately damaged (C), and severely damaged (E). Images B, D and F represent the corresponding images after semi-automated axon detection was performed using the image analysis software. The contour of the black-stained axon myelin sheath is highlighted in red and automatically counted by the software. The scale bar = 20 μm.
Manual axon counts
Manual axon counts were obtained for the same nine digital images by the same rater. Each and every axon in the image was visually identified and selected using the software’s manual count tool. The results obtained by the SCT were then compared to the MAC to determine the agreement between the techniques.
Semi-automated targeted sampling method
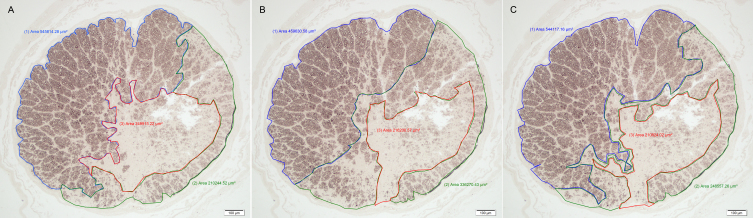
The SATS method was adapted from the method previously described by Marina et al. [14]. Briefly, up to three distinct relatively homogeneous regions were visually identified in low-power (10X objective) photomicrographs of the entire optic nerve cross sections, and the area of each region (in μm2) was recorded using the image analysis software (Figure 2).
Figure 2.
Selections of regions with homogeneous damage in an optic nerve cross section from a glaucomatous cat. Images A, B, and C depict region selection by three different raters in the same optic nerve section. The blue region labeled (1) represents a normal region, the green region labeled (2) a moderately damaged region, and the red region labeled (3) a severely damaged region. Note the variation of the selected areas by the different raters and the final area of each region in μm2. The scale bar = 100 μm.
Axon counts from up to five randomly selected high-power fields (63X objective) from each region were obtained using the SCT described earlier. The average axon density (axons/µm2) for each region was calculated by dividing the average axon count of the selected fields by the known high-power field area (242,611 µm2). Then, by multiplying the average axon density of that region by the total region area (µm2), we obtained the estimated axon count for each region. Estimated counts for each distinct region were subsequently summed, and an estimate of the total count over the entire cross section was thus obtained.
To estimate inter-rater variability, three different masked raters applied the SATS method to the same set of 15 optic nerve cross sections (nine affected cats and six controls). Raters selected regions and representative fields within regions independently. In addition, one rater recorded the time required for image acquisition and counting using both the SATS and the SAFC methods (see below).
Semi-automated full count method
SAFCs were obtained by a single masked rater for the same set of 15 optic nerve cross sections in which the SATS method was applied. A consecutive series of images (up to 40 images) representing the entire optic nerve cross section was captured under a 63X microscope objective. To ensure the whole tissue section was captured without duplications or omissions, anatomic and other tissue landmarks, such as blood vessels or indentations, were used as reference points while manually navigating across each slide. All axons in each image were then counted using the SCT, as described previously, and the sum of the counts from all photomicrographs provided a total count of axons for the entire nerve cross section. The time taken to acquire images and axon counts by this method was recorded.
Statistical analyses
To validate the SCT, we calculated the correlation between the axon counts obtained by the SCT and by the MAC techniques in the nine different images and we fit a linear model of the counting error (SCT less MAC) to the MAC. An analysis of covariance (ANCOVA) was performed to determine if this linear relationship varied by slide, and we performed tests for non-zero intercept and non-zero slope.
To compare the axon counts obtained by the SATS method performed by three raters for the 15 optic nerves to the SAFC for the same nerves, we fit a linear mixed-effects model of the counting error (SATS count less SAFC count) that included a fixed-effect linear relationship to the SAFC count and random effects for slide and rater. We tested the SATS method for an overall bias using a Wilcoxon signed rank test of the differences between the average SATS count (averaged over raters) and the SAFC count for each slide, and we fit a two-stage linear model of the average counting error (average SATS count less SAFC) versus SAFC to test for non-zero slope and non-zero intercept and to estimate constant absolute error and constant percentage error models for the counting error. We also used the linear mixed-effects model to calculate variance component estimates. We determined an ICC for inter-rater agreement, and we estimated the correlation of the SATS method to the SAFC method for a typical rater by averaging the correlations between SAFC and SATS counts for each rater.
We also performed a comparison of the time required for one reviewer to count with the SATS and SAFC methods using a Wilcoxon signed rank test. Data analyses were performed using R version 3.0.1 [16] and the lme4 package version 1.0–4 [17].
Results
Comparison of counts of semi-automated axon counting tool to manual axon counts
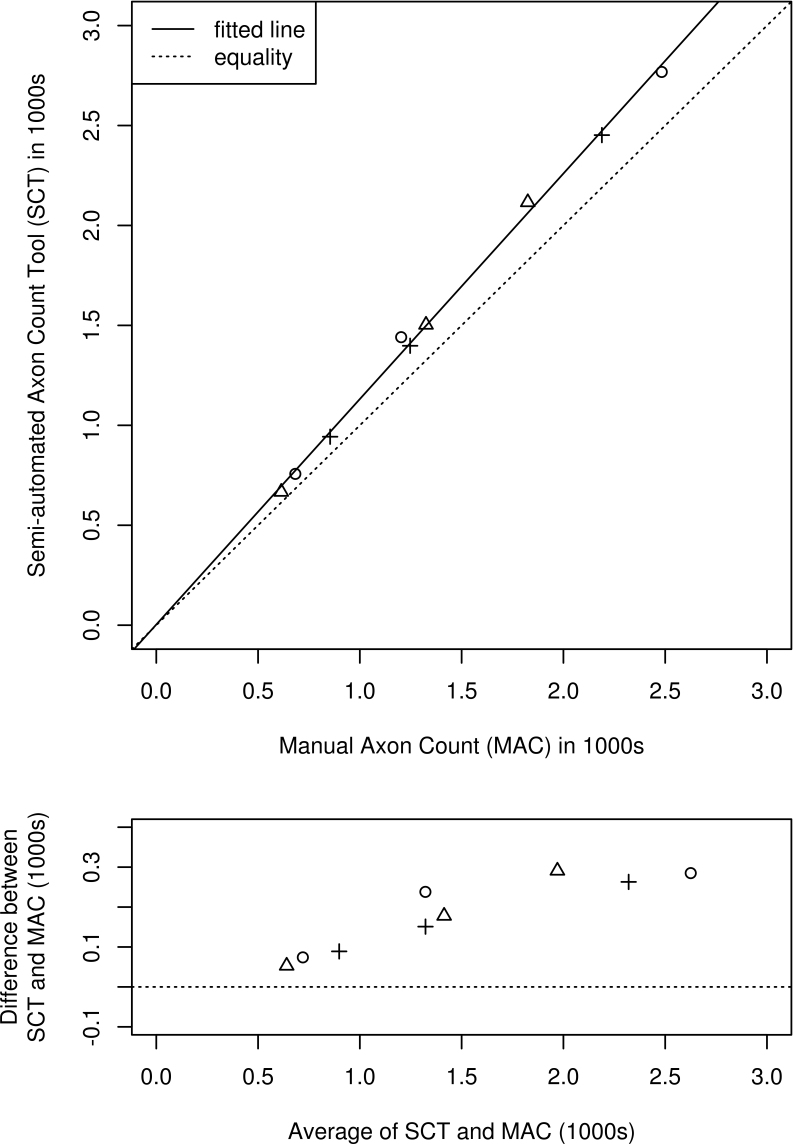
Figure 3 shows a scatter plot of axon counts obtained by SCT versus MAC for the nine fields with the least-squares linear fit (solid) and a line representing equality of the counts (dotted) for comparison. The correlation between SCT and MAC was high (r = 0.9985).
Figure 3.
Degree of agreement between semi-automated axon count tool (SCT) and manual axon count (MAC) methods. The top panel shows a scatter plot of SCT versus MAC counts for n=9 fields, one each of low, medium, and high density selected from each of three animals (designated by three different plotting symbols). The dotted line represents equality of counting methods, and the solid line represents the best linear fit. The bottom panel shows a Bland-Altman plot of the difference versus the average of the two counting methods.
An ANCOVA analysis showed no significant differences by slide in the fitted intercept or slope. Also, there was no evidence of a non-zero intercept, consistent with a constant percentage error rate over the large range of per-field counts sampled (614 to 2,483 axons). A model with zero intercept gave an estimated constant percentage error rate of 13.0% (95% CI 11.0%, 15.1%), representing overcounting of axons by the SCT compared to the MAC technique. Individual raters selected areas and representative fields independently and subjectively, and although choice of areas and fields varied considerably from rater to rater (Figure 2), both inter-rater agreement (ICC = 0.986) and the average per-rater correlation of SATS count to SAFC (r = 0.9891) were high.
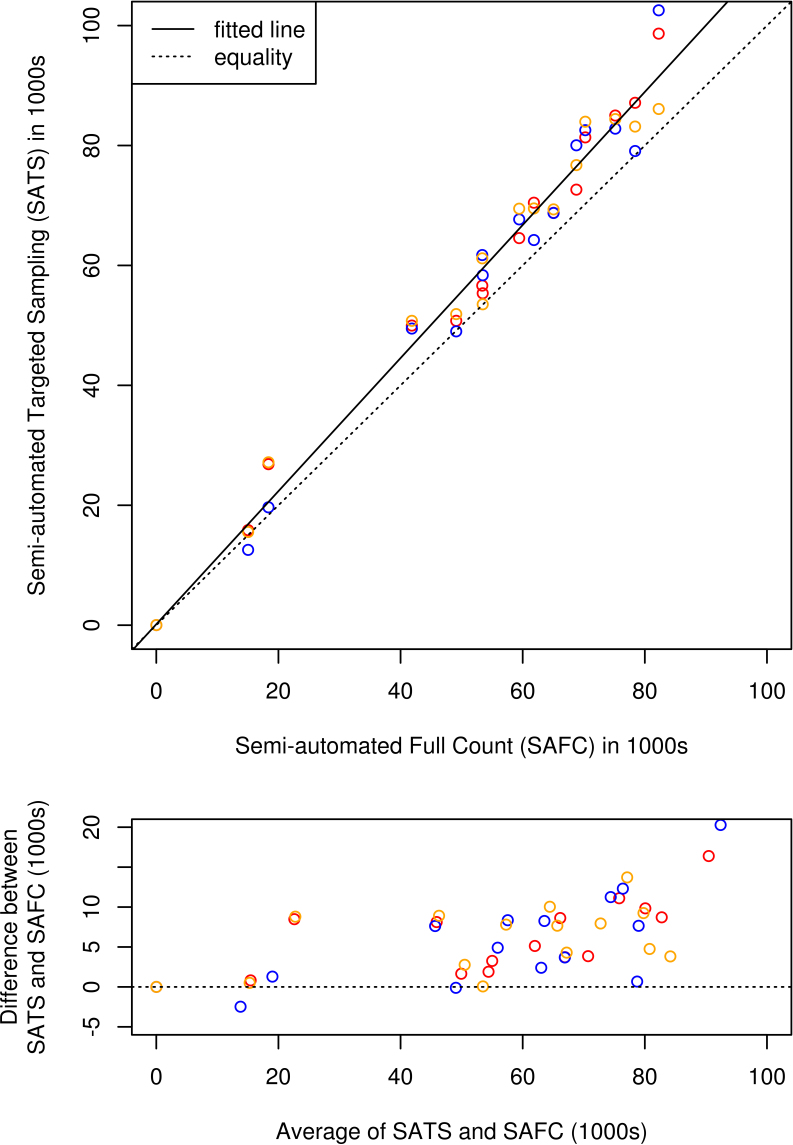
The mean of the SAFC for the 15 optic nerves sampled was 52,827 axons with an SD of 24,483 axons. Figure 4 shows a scatter plot of SATS counts by three raters for each of the 15 optic nerves versus the SAFC for those nerves performed by one of the raters. A solid line giving the best linear fit is shown as is a dotted line representing equality of the counts for comparison (line of equivalence). The fitted linear relationship is
Figure 4.
Degree of agreement between semi-automated targeted sampling (SATS) and semi-automated full count (SAFC) methods. The top panel shows a scatter plot of SATS counts performed by three raters versus the SAFC count for each of n=15 optic nerve cross sections. Raters are identified by plot symbol color, the dotted line represents equality of the counting methods, and the solid line represents the best linear fit. The bottom panel shows a Bland-Altman plot of the differences versus the average of the two counting methods.
or equivalently:
representing strong evidence of an overall positive counting error (p<0.001) with overcounting of the SATS method compared to the SAFC method. There was no evidence of a non-zero intercept, consistent with a fixed percentage error rate of 11.2% of the total count (95% CI 8.3%, 14.2%).
In the fitted linear mixed-effects model, the random rater effect had zero variance, indicating consistency of the observed error pattern across raters (i.e., no systematic tendency of a rater to over- or undercount axons compared to other raters). The estimated residual error SD was 3,287 axons per slide (95% CI 2,603; 4,333), which represents the rater-to-rater variation observed in SATS counts performed by different raters of a single optic nerve slide. The estimated SD of the slide random effect was 2,572 axons per slide (95% CI 862; 4,054), which represents additional slide-to-slide variation attributable to features of a particular optic nerve cross section that make it more subject to over- or undercounting by the SATS method compared to other sections with similar axon density.
Comparison of time spent performing semi-automated targeted sampling and semi-automated full count methods
For the rater who applied both the SATS and SAFC methods to the 15 optic nerves, the SATS method took significantly less time, on average only 27% as much time, as the SAFC method: SATS mean (SD) 10.9 (4.9) min versus SAFC mean 45.3 (10.7) min; (p<0.001, n=15, paired Wilcoxon signed rank test). This included time saved in both image acquisition (mean 6.3 versus 21.1 min) and actual counting time (mean 5.7 versus 24.3 min).
Discussion
To validate the SCT, we analyzed nine fields of optic nerve cross sections with variable density and compared the obtained axon counts to MACs of the same fields. The correlation between the two approaches was high (r = 0.9985), but there was a consistent bias—an overestimation of axons by 13% when using the SCT compared to the MAC. The most likely explanation is a misidentification of axon-like structures by the software’s automatic detection system. On any given optic nerve cross section, and particularly in moderate to severely affected areas, myelin or cellular debris from degenerating axons are stained black by PPD and can be recognized and counted by the software. Furthermore, splitting of myelin fibers can cause a discontinuity of the circumferential myelin sheath surrounding an axon, causing the software to recognize the axon as two separate objects. To circumvent these problems we established an additional step in the protocol that includes manually selecting and deleting the detected nonaxonal structures and double-counted axons. Despite this extra effort, some of the artifacts erroneously counted by the software could not be easily recognized and excluded by the rater. The addition of this step caused a slight increase in the overall time spent analyzing a slide, but even with its flaws it proved to be important in creating a homogeneous set of data, a fact that can be appreciated by the high correlation between the SCT and MAC.
Our ANCOVA analysis showed that this 13% overestimation did not vary significantly across the three slides (that is, there was no tendency for any slides to be more or less prone to overestimation than the others). This suggests that it would be possible to develop an accurate calibration curve to correct for this bias in the SCT. Such a correction would need to be developed separately for a particular slide preparation and counting protocol using a larger sample of slides than we used in our analysis. However, our ANCOVA results provided confidence that even uncorrected counts obtained by the SCT are comparable from slide to slide because of the consistency of counting bias.
Full optic nerve axon counts in normal cats have been reported in the literature with values ranging from 86,000 axons by light microscopy [18] to 193,000 axons by TEM [19]. Control cats from our study presented mean axon counts of 73,751 axons with the SAFC method and 78,380 for the SATS method (average of three raters). It has been reported that light microscopy techniques tend to underestimate total axon counts in the order of 20%–30% [14]. This could be explained by the capacity of TEM to detect small axons that were most likely not detected by the SCT. Approximately 48%–52% of cat axons present diameters between 0.3 to 1.5 μm [20], and those with diameters in the lower end on this spectrum might not be detected by our method. In addition, the mesh grid employed in TEM procedures occupies 55% of the image area, leaving only 45% of the tissue available for an axon count; thus the total axon count must be extrapolated, with the potential for overestimation of axon counts. The cat optic nerve presents marked fascicular organization of axon fibers [21] as well as size grouping and regional variations in axonal density [22]. These factors could impact the final axon counts using TEM since the extrapolations assume that the remaining 55% of the tissue is of similar composition, introducing the potential for overestimation of axon numbers. However, it was not the intent of our study to determine reference ranges for axon counts in cats but to develop a reproducible and reliable method to quantify axons in our model. These counts, in turn, serve as a basis for validation in individual cats of the noninvasive structural and functional measures of glaucomatous damage (acquired by optical coherence tomography and electrophysiology, respectively) that are the focus of our ongoing longitudinal studies. Analysis of the optic nerves of a much larger number of subjects is now underway and might allow us to better characterize a reference range for feline axons counts obtained by light microscopy.
Both the correlation between the SATS and SAFC methods (r = 0.9891) and the inter-rater agreement (ICC = 0.986) were high, confirming the accuracy, validity, and reproducibility of the SATS method in our animal model.
The SATS method presented an overall positive counting error when compared to the SAFC method, consistent with a fixed percentage overestimation (11.2% of the full count). One explanation for the overestimation of axon counts using the SATS method is that the mean axon count of the five selected fields is multiplied by the region’s area (µm2). This calculation assumes that each field is composed only of axons. In reality, optic nerve sections of normal cats included in this study demonstrated that approximately 15% of the area of a high power (63X) field is composed of nonaxonal tissue (data not shown), and thus overestimation of axon numbers is to be expected. This problem is hard to address since to correct for this source of overestimation the nonaxonal area needs to be extracted from the images of each selected field and region. This process is time consuming and would defeat our initial purpose of creating a more rapid efficient method. Another alternative would be to apply a correction factor to the final counts. However, this correction factor would be hard to establish given the heterogeneity in different optic nerve regions and the fact that in the most affected areas it is hard to distinguish areas that represent optic nerve native connective tissue from those that represent areas with genuine axon loss.
When using time-efficient approaches, like semiquantitative grading schemes [12,23,24], random sampling [25,26], targeted [14,27], or automated full counts [28], to assess optic nerve damage, the subjectivity of important steps in the protocol can introduce significant variability in the data. Many of the published studies using these counting methods and grading schemes have not addressed variance between raters and optic nerve slides or attempted to evaluate sources of bias in counting. In our study, raters selected homogeneous optic nerve regions and fields independently and subjectively, and there was marked variability in the selections made by our raters (Figure 2). However, despite this subjective step, the inter-rater agreement (ICC = 0.986) and correlation of axon counts obtained by SATS (average per rater) to those obtained by SAFC (r = 0.9891) were high. The data analysis also found no significant random rater effect, meaning no tendency for a given rater to systematically over- or undercount axons. This suggests that, assuming adequate training in its application, the SATS method might be usefully employed in a large study extending over several years where it is not feasible for a single rater (or the exact same set of multiple raters) to perform every SATS count.
In experiments that generate a large number of optic nerves for analysis, time becomes an important consideration. Manual counting of axons on TEM and light microscopy images can be time consuming to the point of becoming impractical. In the current study, the average time spent acquiring images, selecting regions and fields, and counting axons for the SATS method was less than 30% of that required to conduct SAFC. These time savings were facilitated by the commercially available image analysis system used that, after appropriate thresholds were set, delivered axon counts in seconds. Alternative image analysis software packages, whether commercially or freely available, present similar capabilities and could be used with appropriate adaptations and validation.
Our method has several limitations. First, as for any other image analysis protocol, the results depend largely on the quality of the images analyzed. Image quality is influenced by proper processing, sectioning, mounting, and staining of the optic nerve tissue. Second, while we have provided characterizations of and models for the observed counting error when comparing SCT with MAC and SATS with SAFC methods, these errors were observed under a specific and strict slide preparation and analysis protocol that included, among other things, selection of specific values for the software’s detection “threshold” for the SCT. Counting errors are almost certainly dependent on details of the counting protocol, including the selected threshold value, and so our error models must be considered specific to the protocol and detection settings used. In particular, our models of counting error are unlikely to be useful to “recalibrate” SATS counts to match manual counts in a more general setting. Third, because our results were based on a single set of slides prepared and counted over a relatively short period of time according to a specific strict protocol (including specific threshold value), we were not able to fully characterize errors that might be observed across separate slide preparations at different time points, as might occur in a large multiyear study. With respect to the SCT and MAC comparison, while the correlation between the two methods was high and we found no evidence of slide to slide variation in the error rate, the sample number was small, encompassing only three animals and nine fields.
In summary, our data indicated that the SATS method provides a practical, rapid, and reliable means of estimating axon counts in the optic nerves of normal cats and cats with glaucoma. Despite the relative subjectivity of certain aspects of the method, it provided reproducible axon counts that were comparable to those obtained by the SAFC method while reducing the time taken to acquire axon counts by more than two-thirds.
Acknowledgments
This study was previously presented, in part at the 2012 Annual Meeting of the Association for Vision and Ophthalmology, Fort Lauderdale, Florida. The study was supported by NIH grants K08EY018609 and P30EY0016665; the Clinical and Translational Science Award (CTSA) program through the NIH National Center for Advancing Translational Sciences (NCATS) grant UL1TR000427; and an unrestricted award to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness. Glaucomatous subjects were derived from a breeding colony maintained at Iowa State University, supported in part by the Center for Integrated Animal Genomics, Iowa State University and a Battelle Platform Project Grant from the State of Iowa, and expertly managed by Dr N Matthew Ellinwood, DVM, PhD.
References
- 1.Kuehn MH, McLellan GJ, Pfleging A, Snella EM, Ellinwood NM. Spontaneous mutations in LTBP2 are associated with congenital glaucoma in cats. ARVO Annual Meeting; 2011 May 1-5; Fort Lauderdale, FL [Google Scholar]
- 2.Ellinwood NM, Deckman KH, Zhao Z, Rutz-Mendicino MM, Jens JK, David VA, Kuehn MH, O'Brien SJ, Menotti-Raymond MA, McLellan GJ. Candidate gene analysis of a feline model of primary congenital glaucoma implicates LTBP2 as the causative locus. ARVO Annual Meeting; 2010 May 1-5; Fort Lauderdale, FL [Google Scholar]
- 3.Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, Gilmour DF, Towns K, Murphy AL, Azmanov D, Tournev I, Cherninkova S, Jafri H, Raashid Y, Toomes C, Craig J, Mackey DA, Kalaydjieva L, Riazuddin S, Inglehearn CF. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84:664–71. doi: 10.1016/j.ajhg.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narooie-Nejad M, Paylakhi SH, Shojaee S, Fazlali Z, Rezaei Kanavi M, Nilforushan N, Yazdani S, Babrzadeh F, Suri F, Ronaghi M, Elahi E, Paisán-Ruiz C. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum Mol Genet. 2009;18:3969–77. doi: 10.1093/hmg/ddp338. [DOI] [PubMed] [Google Scholar]
- 5.Weber AJ. Ex Vivo and Whole-Mount Retinal Preparations. In: Levin LA, Di Polo A, editors. Ocular Neuroprotection. New York: Marcel Dekker Inc; 2003. p. 205–224. [Google Scholar]
- 6.Hughes A. Population magnitudes and distribution of the major modal classes of cat retinal ganglion cell as estimated from HRP filling and a systematic survey of the soma diameter spectra for classical neurons. J Comp Neurol. 1981;197:303–39. doi: 10.1002/cne.901970209. [DOI] [PubMed] [Google Scholar]
- 7.Hughes A, Wieniawa-Narkiewicz E. A newly identified population of presumptive microneurons in the cat retinal ganglion cell layer. Nature. 1980;284:468–70. doi: 10.1038/284468a0. [DOI] [PubMed] [Google Scholar]
- 8.Wӓssle H, Chun MH, Muller F. Amacrine cells in the ganglion cell layer of the cat retina. J Comp Neurol. 1987;265:391–408. doi: 10.1002/cne.902650308. [DOI] [PubMed] [Google Scholar]
- 9.Wong RO, Hughes A. The morphology, number, and distribution of a large population of confirmed displaced amacrine cells in the adult cat retina. J Comp Neurol. 1987;255:159–77. doi: 10.1002/cne.902550202. [DOI] [PubMed] [Google Scholar]
- 10.Boycott BB, Wassle H. The morphological types of ganglion cells of the domestic cat's retina. J Physiol. 1974;240:397–419. doi: 10.1113/jphysiol.1974.sp010616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone J. The number and distribution of ganglion cells in the cat's retina. J Comp Neurol. 1978;180:753–71. doi: 10.1002/cne.901800407. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan BC, Levatte TL, Garnier KL, Tremblay F, Pang I, Clark AF, Archibald ML. Semiquantitative optic nerve grading scheme for determining axonal loss in experimental optic neuropathy. Invest Ophthalmol Vis Sci. 2006;47:634–40. doi: 10.1167/iovs.05-1206. [DOI] [PubMed] [Google Scholar]
- 13.Martin KR, Quigley HA, Zack DJ, Levkovitch-Verbin H, Kielczewski J, Valenta D, Baumrind L, Pease ME, Klein RL, Hauswirth WW. Gene therapy with brain-derived neurotrophic factor as a protection: retinal ganglion cells in a rat glaucoma model. Invest Ophthalmol Vis Sci. 2003;44:4357–65. doi: 10.1167/iovs.02-1332. [DOI] [PubMed] [Google Scholar]
- 14.Marina N, Bull ND, Martin KR. A semiautomated targeted sampling method to assess optic nerve axonal loss in a rat model of glaucoma. Nat Protoc. 2010;5:1642–51. doi: 10.1038/nprot.2010.128. [DOI] [PubMed] [Google Scholar]
- 15.McLellan GJ, Kemmerling JP, Kiland JA. Validation of the TonoVet(®) rebound tonometer in normal and glaucomatous cats. Vet Ophthalmol. 2013;16:111–8. doi: 10.1111/j.1463-5224.2012.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. <http://www.R-project.org/>.
- 17.Bates D, Maechler M, Bolker B. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.0–4. <http://lme4.r-forge.r-project.org/ >
- 18.Donovan A. The nerve fibre composition of the cat optic nerve. J Anat. 1967;101:1–11. [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes A, Wassle H. The cat optic nerve: fibre total count and diameter spectrum. J Comp Neurol. 1976;169:171–84. doi: 10.1002/cne.901690204. [DOI] [PubMed] [Google Scholar]
- 20.Brooks DE, Komaromy AM, Kallberg ME. Comparative retinal ganglion cell and optic nerve morphology. Vet Ophthalmol. 1999;2:3–11. doi: 10.1046/j.1463-5224.1999.00047.x. [DOI] [PubMed] [Google Scholar]
- 21.Evans A, Jeffery G. The fascicular organization of the cat optic nerve. Exp Brain Res. 1992;91:79–84. doi: 10.1007/BF00230015. [DOI] [PubMed] [Google Scholar]
- 22.Williams RW, Chalupa LM. An analysis of axon caliber within the optic nerve of the cat: evidence of size groupings and regional organization. J Neurosci. 1983;3:1554–64. doi: 10.1523/JNEUROSCI.03-08-01554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison JC, Johnson EC, Cepurna W, Jia L. Understanding mechanisms of pressure-induced optic nerve damage. Prog Retin Eye Res. 2005;24:217–40. doi: 10.1016/j.preteyeres.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Anderson MG, Libby RT, Gould DB, Smith RS, John SW. High-dose radiation with bone marrow transfer prevents neurodegeneration in an inherited glaucoma. Proc Natl Acad Sci USA. 2005;102:4566–71. doi: 10.1073/pnas.0407357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levkovitch-Verbin H, Martin KR, Quigley HA, Baumrind LA, Pease ME, Valenta D. Measurement of amino acid levels in the vitreous humor of rats after chronic intraocular pressure elevation or optic nerve transsection. J Glaucoma. 2002;11:396–405. doi: 10.1097/00061198-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Pease ME, Zack DJ, Berlinicke C, Bloom K, Cone F, Wang Y, Klein RL, Hauswirth WW, Quigley HA. Effect of CNTF on retinal ganglion cell survival in experimental glaucoma. Invest Ophthalmol Vis Sci. 2009;50:2194–200. doi: 10.1167/iovs.08-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebneter A, Franzco RJC, Wood JPM, Chidlow G. Estimation of axon counts in a rat model of glaucoma: comparison of fixed-pattern sampling with targeted sampling. Clin Experiment Ophthalmol. 2012;40:626–33. doi: 10.1111/j.1442-9071.2011.02741.x. [DOI] [PubMed] [Google Scholar]
- 28.Reynaud J, Cull G, Wang L, Fortune B, Gardiner S, Burgoyne CF, Cioffi GA. Automated Quantification of Optic Nerve Axons in Primate Glaucomatous and Normal Eyes—Method and Comparison to Semi-Automated Manual Quantification. Invest Ophthalmol Vis Sci. 2012;53:2951–9. doi: 10.1167/iovs.11-9274. [DOI] [PMC free article] [PubMed] [Google Scholar]