Abstract
Survival median is commonly used to compare treatment groups in cancer-related research. The current literature focuses on developing tests for independent survival data. However, researchers often encounter dependent survival data such as matched pair data or clustered data. We propose a pseudo-value approach to test the equality of survival medians for both independent and dependent survival data. The Type I error and power of the proposed method are examined by a simulation study, in which we examine independent and dependent data. The simulation study shows that the proposed method performs equivalently to the existing methods for independent survival data and performs better for dependent survival data. The proposed method is illustrated by a study comparing survival median times for bone marrow transplants.
Keywords: pseudo-value approach, survival median, clustered data, censored data
1. Introduction
Survival analysis is widely used in cancer-related research, for example, bone marrow transplant studies [1]. Survival median is often provided to summarize survival data and to compare treatment groups of interest. Brookmeyer and Crowley [2] proposed a nonparametric test to compare the equality of survival medians (We refer to this method as the Brookmeyer-Crowley test in this paper). They considered a pooled-weighted Kaplan-Meier estimate based on the sample size of each group. Then, the pooled-sample median is calculated by the weighted Kaplan-Meier estimate and linear interpolation. If the survival medians of all groups are the same, the estimated survival rate of each group at the pooled-sample median should be close to 0.5. Thus, a chi-squared test was developed based on the difference between 0.5 and the estimated survival rate of each group at the pooled-sample median. Another chi-squared test was proposed by Rahbar et al. [3] with fewer assumptions than the Brookmeyer-Crowley test. In particular, they considered that the family of the underlying survival distributions or censoring distributions of some groups is different from those of the other groups in the simulation study. Their simulation study indicated that the performance of the Brookmeyer-Crowley test appears to be poor when the survival distribution family differs among groups. However, these two tests are limited to independent survival data.
Another limitation of [3] is that the test requires the existence of survival median for each group because the test statistic is based on the difference between the survival median of each group and the pooled survival median. On the other hand, the Brookmeyer-Crowley test only requires the existence of the pooled survival median. Thus, if the pooled survival median exists, the method is applicable even when the survival medians of some groups do not exist. See Section 4 for a concrete example.
The pseudo-value technique was proposed by Andersen et al. [4]. To explain the details of the pseudo-value approach for clustered or dependent data, we introduce some notations. Let dj be the sample size of cluster j, j = 1, …, m and n = Σj dj, where m is the number of clusters and n is the total number of individuals. The survival probability S(t) indicates the pooled survival probability at time t based on the total sample and Zij represents the covariate vector for the ith individual in the jth cluster. Assuming censoring times are independent of failure times, a pseudo-value at time t of the ith individual in the jth cluster is defined by Yij(t) = nŜ(t) − (n − 1)Ŝ−ij(t), i = 1, …, dj, j = 1, …, m, where Ŝ(t) is the Kaplan-Meier estimate of S(t) based on the pooled data and Ŝ−ij(t) is the Kaplan-Meier estimate of survival obtained by omitting the ith individual in the jth cluster. Logan et al. [5] studied marginal cumulative incidence and survival models for clustered data using the pseudo-value approach. Assuming the censoring times are a random sample from a common distribution, they showed that i) Yij(t) is approximately independent of Ykℓ for j ≠ ℓ as n → ∞; and ii) limn→∞ E(Yij(t)|Zij) = S(t|Zij). The pseudo-values can be used as a response variable in a generalized estimating equation (GEE) setting as described in [4], [5], and [6]. Since a single fixed time point is only considered in this paper, we illustrate the use of the GEE at a fixed time point τ. To obtain the marginal survival function at time τ given covariate Z, we consider g(S(τ|Z)) = β′Z, where β is a q × 1 parameter vector. Let μ = S(τ|Z) = g−1(β′Z), Yj = (Y1j(τ), …, Ydj,j (τ)), and μj = (μ1j(τ), …, μdj,j (τ)), j = 1, …, m. Then, the GEE is defined as follows:
| (1) |
where Vj is a dj × dj working covariance matrix for cluster j. The working covariance matrix Vj can be expressed by , where R(α) is a working correlation matrix and Aj is a diagonal matrix with elements var(Yij) = v(μij) for some known variance function v(·). Then, it was shown that converges in distribution to N(0, Σ) for some covariance matrix Σ, see [5], [7], and [8]. Thus, a Wald chi-square statistic mβ̂′Σ̂−1β̂ converges in distribution to the chi-squared distribution with degrees of freedom q under the assumption that β = 0 for an appropriate estimator Σ̂. To estimate Σ, the sandwich estimator is used:
where
Therefore, the dependent survival data are readily handled by considering within-cluster correlation between individuals.
We propose a pseudo-value-based method to test the equality of survival medians for independent and dependent data in this paper. In Section 2, we describe the proposed method and its asymptotic distribution. In Section 3, we compare the proposed method to the Brookmeyer-Crowley test and Rahbar et al. [3] in a simulation study. A bone marrow transplant example is illustrated in Section 4. Finally, we have a brief conclusion in Section 5.
2. Method
In this section, we lay out the proposed method. We assume that the censoring times are independent of the failure times and the censoring times are a random sample from a common distribution. The median survival time is given by
The Kaplan-Meier estimator Ŝ(t) is consistent even for dependent data under some mild conditions. For the detailed conditions, see [9]. Thus, ζ̂ defined by inf{t : Ŝ(t) ≤ 0.5} is also consistent for dependent data using Lemma 21.2 of [10]. Assume that we have ν groups to compare. Under the null hypothesis, we have ζ1 = · · · = ζν ≡ ζ0, where ζi is the survival median of group i, i = 1, …, ν. This is equivalent to testing H0 : S1(ζ0) = · · · = Sν(ζ0)(= 0.5), where Si(ζ0) is the survival rate of group i at time ζ0. Let ζ̂0 be an estimate of ζ0 based on the pooled data.
To compare survival probabilities at ζ0, we use the pseudo-value approach at a fixed time point ζ0. Let Ik be an indicator variable for group k such that for k = 1, …, ν,
Let β = (β1, …, βν)′ be the coefficient vector of the indicator variables in the GEE. To avoid an identifiability issue, without loss of generality we fix βν at 0 and estimate β−ν = (β1, …, βν−1)′. Next, we define pseudo-values Yij(ζ̂0) = nŜ(ζ̂0) − (n − 1)Ŝ−ij(ζ̂0), where Yij is a pseudo-value for the ith individual belonging to the jth cluster. Let Yj = {Yij(ζ̂0); i = 1, …, cj} and μj = {μij(ζ̂0); i = 1, …, cj}, which means that Yj includes all and only individuals belonging to the jth cluster and μj is the corresponding mean vector. Then, the GEE is defined as in (1). An identity link function or a logit link function can be used in practice. Assuming Zij = Ik, we have limn→∞ E(Yij(ζ0)|Zij) = S(ζ0|Ik) = Sk(ζ0) by [5]. Therefore, the null hypothesis S1(ζ0) = · · · = Sν(ζ0) is equivalent to testing β−ν = 0 given ζ0. Thanks to the consistency of ζ̂0, the test statistic is given by
where β̂−ν is found by solving the GEE numerically and Σ̂β−ν is the corresponding sandwich estimate of the covariance matrix of β̂−ν. By considering the sandwich estimate for Σβ−ν, we account for the dependency of survival data. In practice, the independence working correlation matrix was recommended for the pseudo-value approach [6]. Under the null hypothesis, X2 converges in distribution to a chi-squared distribution with ν − 1 degrees of freedom by the consistency of ζ̂0 and [5]. Note that like the Brookmeyer-Crowley test, the proposed pseudo-value approach only requires the existence of the pooled survival median, which is an advantage over [3].
3. Simulation Study
In the simulation study we compared four groups to examine the empirical Type I error rates and rejection rates of the proposed pseudo-value method, the Brookmeyer-Crowley test, and [3] at the significance level α = 0.05. To estimate the density functions of survival distributions for [3], we used bootstrap with 1000 replicates. Survival and censoring times were assumed to have exponential, log-normal, uniform, or Weibull distributions. Three censoring rates were considered: c = 0, 25, and 50 percent. The sample size for each group was fixed at ni = 100, or 200 for i = 1, …, 4, where ni is the sample size of group i. All experiments were replicated 5000 times. The logit link was considered for the pseudo-value approach. The independence working correlation matrix was used following the recommendation of [6]. We also examined the exchangeable working correlation matrix and the unstructured working correlation matrix for a portion of the simulation, but there was little difference from the result using the independence working correlation matrix.
Let ζ0 and c be the survival median and the censoring rate, respectively. The survival times were generated from i) the exponential distribution with mean ζ0/log(2); ii) the uniform distribution on (ζ0 − 3, ζ0 + 3); iii) the log-normal distribution with mean ζ0 exp(0.5) and variance ; and iv) the Weibull distribution with shape parameter 1.2 and scale parameter . For c > 0, the censoring times were generated from i) the exponential distribution with mean (1 − c)ζ0/c/log(2) for the exponential survival times; ii) the uniform distribution on (ζ0 − 3, ζ0 + 3 + 3(1 − 2c)/c) for the uniform survival times; iii) exp(log(ζ0) + U) where U was generated from the uniform distribution on (−2, 2 + (2 − 4c)/c) for the log-normal survival times; and iv) the Weibull distribution with shape parameter 1.2 and scale parameter {c log 2/ζ0/(1 − c)}−1/1.2 for the Weibull survival times.
Each cluster was assumed to have 8 individuals. The 8 individuals were divided into the 4 groups by 2. Thus, the ith group with ni = 100 and ni = 200 consisted of 50 and 100 clusters, respectively. Normal copulas were used to generate correlated survival times of each cluster. The 8 × 8 exchangeable correlation matrix
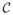 with correlation ρ = 0, 0.25 and 0.5 was used for the normal copulas, i.e.,
with correlation ρ = 0, 0.25 and 0.5 was used for the normal copulas, i.e.,
Note that ρ = 0 means that the survival times of the four groups are independent. After generating 8-dimensional random vectors on the unit cube [0, 1]8 from normal copulas given ρ, the survival times were generated corresponding to their marginal survival distributions. Independent of the survival times, the censoring times were randomly generated using normal copulas with ρ = 0. For the detailed use of copulas, see [11].
Table 1 shows the empirical Type I error rates when the survival time distributions of the four groups were the same. The true survival median was fixed at ζ0 = 6. ‘PV’, ‘BC’, and ‘Rahbar’ represent the pseudo-value approach, the Brookmeyer-Crowley test, and [3], respectively. ‘Exp’, ‘LN’, ‘Unif’, and ‘WB’ indicate the exponential distribution, log-normal distribution, uniform distribution, and Weibull distribution for survival time distributions. The censoring time distributions were chosen accordingly as discussed previously. Distributions in parentheses are the survival time distributions for four groups. For example, (Exp,Exp,Exp,Exp) indicates that all four groups have the exponential survival distributions. For the independent survival data (ρ = 0), the empirical Type I error rates of all three methods are close to the nominal rate 0.05. For the dependent data (ρ = 0.25 or 0.5), in contrast to the two methods, the pseudo-value approach controls Type I error rates very well. The Brookmeyer-Crowley test and [3] show much less Type I error rates than 0.05 in general. It appears that as the dependency of data increases, the Type I error rates of the Brookmeyer-Crowley test and [3] decreases towards 0.
Table 1.
Simulation 1: Empirical Type I error rates with α = 0.05 when the survival distributions of the four groups are the same. ‘PV’, ‘BC’, and ‘Rahbar’ indicate the pseudo-value approach, the Brookmeyer-Crowley test, and [3], respectively.
| ρ | ni | c | (Exp,Exp,Exp,Exp) | (LN,LN,LN,LN) | (WB,WB,WB,WB) | (Unif,Unif,Unif,Unif) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | |||
| 0 | 100 | 0 | 0.050 | 0.056 | 0.063 | 0.050 | 0.055 | 0.057 | 0.071 | 0.054 | 0.063 | 0.071 | 0.055 | 0.066 |
| 0.25 | 0.044 | 0.056 | 0.056 | 0.050 | 0.060 | 0.051 | 0.065 | 0.057 | 0.050 | 0.062 | 0.059 | 0.068 | ||
| 0.5 | 0.044 | 0.061 | 0.034 | 0.040 | 0.056 | 0.037 | 0.066 | 0.062 | 0.033 | 0.061 | 0.057 | 0.059 | ||
| 200 | 0 | 0.052 | 0.053 | 0.054 | 0.052 | 0.053 | 0.052 | 0.063 | 0.053 | 0.055 | 0.059 | 0.052 | 0.059 | |
| 0.25 | 0.044 | 0.050 | 0.057 | 0.050 | 0.057 | 0.053 | 0.062 | 0.055 | 0.059 | 0.066 | 0.064 | 0.067 | ||
| 0.5 | 0.046 | 0.055 | 0.049 | 0.051 | 0.058 | 0.048 | 0.060 | 0.061 | 0.049 | 0.058 | 0.055 | 0.061 | ||
|
| ||||||||||||||
| 0.25 | 100 | 0 | 0.070 | 0.032 | 0.033 | 0.072 | 0.032 | 0.028 | 0.070 | 0.032 | 0.033 | 0.070 | 0.032 | 0.036 |
| 0.25 | 0.064 | 0.034 | 0.036 | 0.062 | 0.033 | 0.029 | 0.071 | 0.035 | 0.032 | 0.066 | 0.033 | 0.042 | ||
| 0.5 | 0.062 | 0.042 | 0.032 | 0.062 | 0.037 | 0.025 | 0.063 | 0.042 | 0.034 | 0.058 | 0.035 | 0.040 | ||
| 200 | 0 | 0.053 | 0.024 | 0.032 | 0.056 | 0.023 | 0.031 | 0.053 | 0.023 | 0.033 | 0.056 | 0.023 | 0.037 | |
| 0.25 | 0.058 | 0.031 | 0.037 | 0.060 | 0.034 | 0.034 | 0.057 | 0.034 | 0.039 | 0.060 | 0.037 | 0.043 | ||
| 0.5 | 0.058 | 0.035 | 0.036 | 0.057 | 0.036 | 0.037 | 0.057 | 0.035 | 0.037 | 0.057 | 0.035 | 0.042 | ||
|
| ||||||||||||||
| 0.5 | 100 | 0 | 0.068 | 0.010 | 0.013 | 0.068 | 0.010 | 0.012 | 0.068 | 0.010 | 0.013 | 0.068 | 0.010 | 0.014 |
| 0.25 | 0.069 | 0.012 | 0.014 | 0.064 | 0.015 | 0.018 | 0.067 | 0.013 | 0.015 | 0.062 | 0.014 | 0.020 | ||
| 0.5 | 0.061 | 0.021 | 0.012 | 0.060 | 0.024 | 0.016 | 0.058 | 0.021 | 0.016 | 0.063 | 0.023 | 0.023 | ||
| 200 | 0 | 0.054 | 0.008 | 0.013 | 0.056 | 0.007 | 0.011 | 0.054 | 0.007 | 0.013 | 0.056 | 0.008 | 0.012 | |
| 0.25 | 0.062 | 0.012 | 0.020 | 0.060 | 0.015 | 0.018 | 0.061 | 0.014 | 0.017 | 0.065 | 0.014 | 0.022 | ||
| 0.5 | 0.057 | 0.021 | 0.022 | 0.059 | 0.025 | 0.020 | 0.054 | 0.020 | 0.021 | 0.055 | 0.021 | 0.025 | ||
Table 2 shows the simulation results of the empirical rejection rates when the survival time distributions of the four groups were the same. Four true survival medians were assumed to be 8, 8, 6, and 6 for (Exp,Exp,Exp,Exp) and (Unif,Unif,Unif,Unif). For (WB,WB,WB,WB) and (LN,LN,LN,LN), the true survival medians were 7.5, 7.5, 6, and 6. For ρ = 0, the pseudo-value approach appears to have higher power than [3] and comparable power to the Brookmeyer-Crowley test. For ρ = 0.25 and 0.5, the pseudo-value approach has greater power than [3] and the Brookmeyer-Crowley test.
Table 2.
Simulation 2: Empirical rejection rates with α = 0.05 when the survival distributions of the four groups are the same.
| ρ | ni | c | (Exp,Exp,Exp,Exp) | (LN,LN,LN,LN) | (WB,WB,WB,WB) | (Unif,Unif,Unif,Unif) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | |||
| 0 | 100 | 0 | 0.379 | 0.365 | 0.346 | 0.325 | 0.309 | 0.276 | 0.343 | 0.323 | 0.307 | 0.575 | 0.567 | 0.533 |
| 0.25 | 0.348 | 0.338 | 0.291 | 0.271 | 0.264 | 0.212 | 0.298 | 0.281 | 0.260 | 0.485 | 0.494 | 0.472 | ||
| 0.5 | 0.264 | 0.269 | 0.175 | 0.215 | 0.227 | 0.134 | 0.221 | 0.232 | 0.161 | 0.390 | 0.413 | 0.357 | ||
| 200 | 0 | 0.653 | 0.644 | 0.603 | 0.552 | 0.551 | 0.524 | 0.587 | 0.584 | 0.546 | 0.867 | 0.865 | 0.848 | |
| 0.25 | 0.595 | 0.600 | 0.536 | 0.473 | 0.476 | 0.432 | 0.530 | 0.531 | 0.503 | 0.793 | 0.804 | 0.774 | ||
| 0.5 | 0.473 | 0.482 | 0.411 | 0.362 | 0.378 | 0.306 | 0.418 | 0.428 | 0.539 | 0.683 | 0.697 | 0.650 | ||
|
| ||||||||||||||
| 0.25 | 100 | 0 | 0.452 | 0.329 | 0.312 | 0.377 | 0.265 | 0.236 | 0.395 | 0.278 | 0.276 | 0.653 | 0.544 | 0.514 |
| 0.25 | 0.388 | 0.304 | 0.258 | 0.312 | 0.230 | 0.184 | 0.344 | 0.258 | 0.235 | 0.543 | 0.462 | 0.447 | ||
| 0.5 | 0.294 | 0.245 | 0.148 | 0.233 | 0.197 | 0.121 | 0.246 | 0.202 | 0.141 | 0.434 | 0.390 | 0.343 | ||
| 200 | 0 | 0.732 | 0.637 | 0.603 | 0.643 | 0.518 | 0.496 | 0.661 | 0.558 | 0.539 | 0.918 | 0.876 | 0.856 | |
| 0.25 | 0.665 | 0.580 | 0.547 | 0.523 | 0.437 | 0.401 | 0.594 | 0.504 | 0.487 | 0.850 | 0.801 | 0.778 | ||
| 0.5 | 0.517 | 0.467 | 0.386 | 0.398 | 0.344 | 0.291 | 0.453 | 0.394 | 0.347 | 0.728 | 0.688 | 0.653 | ||
|
| ||||||||||||||
| 0.5 | 100 | 0 | 0.523 | 0.283 | 0.276 | 0.458 | 0.215 | 0.192 | 0.468 | 0.236 | 0.239 | 0.758 | 0.518 | 0.502 |
| 0.25 | 0.471 | 0.264 | 0.213 | 0.348 | 0.187 | 0.158 | 0.401 | 0.206 | 0.201 | 0.626 | 0.437 | 0.425 | ||
| 0.5 | 0.337 | 0.222 | 0.127 | 0.256 | 0.150 | 0.101 | 0.278 | 0.167 | 0.125 | 0.483 | 0.357 | 0.324 | ||
| 200 | 0 | 0.833 | 0.645 | 0.588 | 0.746 | 0.498 | 0.465 | 0.770 | 0.534 | 0.510 | 0.970 | 0.884 | 0.873 | |
| 0.25 | 0.760 | 0.571 | 0.532 | 0.597 | 0.407 | 0.362 | 0.688 | 0.477 | 0.464 | 0.915 | 0.803 | 0.784 | ||
| 0.5 | 0.595 | 0.448 | 0.376 | 0.435 | 0.315 | 0.251 | 0.506 | 0.366 | 0.314 | 0.781 | 0.676 | 0.640 | ||
Although the asymptotics of the proposed method works for a common censoring distribution, the performance is also of interest when some survival distributions or censoring distributions of the groups are different from the others [3]. Table 3 shows the empirical Type I error rates when the two survival time distributions of the four groups were different from the other two survival distributions. For example, (Exp,Exp,LN,LN) is two exponential survival distributions and two log-normal survival distributions. The true survival median was fixed at ζ0 = 6 as previous. For ρ = 0, the empirical Type I error rates of the three methods are close to 0.05 in general. For ρ = 0.25 and 0.5, in contrast to the other two, the pseudo-value approach controls Type I error rates very well. As in Table 1, the Brookmeyer-Crowley test and [3] show much less Type I error rates than 0.05 in general. Like Table 1, the Type I error rates of the Brookmeyer-Crowley test and [3] decreases to 0 as the dependency of the survival distributions increases.
Table 3.
Simulation 3: Empirical Type I error rates with α = 0.05 when the survival distributions of the two groups are different from those of the other two groups.
| ρ | ni | c | (Exp,Exp,LN,LN) | (Unif,Unif,LN,LN) | (Exp,Exp,WB,WB) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | ||||||
| 0 | 100 | 0 | 0.051 | 0.057 | 0.060 | 0.052 | 0.056 | 0.063 | 0.072 | 0.057 | 0.065 | |||
| 0.25 | 0.042 | 0.053 | 0.056 | 0.043 | 0.059 | 0.055 | 0.069 | 0.054 | 0.056 | |||||
| 0.5 | 0.041 | 0.054 | 0.034 | 0.042 | 0.063 | 0.047 | 0.062 | 0.060 | 0.043 | |||||
| 200 | 0 | 0.052 | 0.054 | 0.053 | 0.055 | 0.055 | 0.057 | 0.062 | 0.052 | 0.056 | ||||
| 0.25 | 0.047 | 0.051 | 0.052 | 0.055 | 0.060 | 0.054 | 0.054 | 0.053 | 0.057 | |||||
| 0.5 | 0.046 | 0.053 | 0.042 | 0.063 | 0.063 | 0.053 | 0.056 | 0.055 | 0.050 | |||||
|
| ||||||||||||||
| 0.25 | 100 | 0 | 0.074 | 0.032 | 0.030 | 0.074 | 0.034 | 0.034 | 0.074 | 0.032 | 0.032 | |||
| 0.25 | 0.061 | 0.030 | 0.030 | 0.062 | 0.034 | 0.034 | 0.067 | 0.030 | 0.032 | |||||
| 0.5 | 0.060 | 0.039 | 0.026 | 0.058 | 0.039 | 0.035 | 0.060 | 0.041 | 0.028 | |||||
| 200 | 0 | 0.055 | 0.022 | 0.032 | 0.057 | 0.024 | 0.034 | 0.057 | 0.022 | 0.034 | ||||
| 0.25 | 0.057 | 0.031 | 0.035 | 0.067 | 0.035 | 0.040 | 0.060 | 0.030 | 0.038 | |||||
| 0.5 | 0.059 | 0.038 | 0.032 | 0.064 | 0.041 | 0.038 | 0.054 | 0.035 | 0.036 | |||||
|
| ||||||||||||||
| 0.5 | 100 | 0 | 0.073 | 0.011 | 0.013 | 0.074 | 0.010 | 0.015 | 0.075 | 0.011 | 0.013 | |||
| 0.25 | 0.065 | 0.016 | 0.016 | 0.066 | 0.016 | 0.020 | 0.075 | 0.014 | 0.018 | |||||
| 0.5 | 0.064 | 0.026 | 0.014 | 0.068 | 0.027 | 0.020 | 0.069 | 0.028 | 0.019 | |||||
| 200 | 0 | 0.057 | 0.009 | 0.011 | 0.060 | 0.010 | 0.013 | 0.057 | 0.010 | 0.014 | ||||
| 0.25 | 0.055 | 0.011 | 0.015 | 0.059 | 0.016 | 0.020 | 0.063 | 0.015 | 0.020 | |||||
| 0.5 | 0.063 | 0.024 | 0.019 | 0.062 | 0.025 | 0.025 | 0.060 | 0.022 | 0.023 | |||||
Table 4 shows the simulation results of the empirical rejection rates when the two survival time distributions of the four groups were different from the other two survival distributions. For (Exp,Exp,LN,LN), survival medians are 8, 6, 8, and 6, respectively. The survival medians of (Exp,Exp,WB,WB) and (Unif,Unif,LN,LN) are 7.5, 6, 7.5, and 6, respectively. For ρ = 0, it appears that the three methods have comparable power. For ρ = 0.25 and 0.5, the pseudo-value approach has greater power than [3] and the Brookmeyer-Crowley test as expected.
Table 4.
Simulation 4: Empirical rejection rates with α = 0.05 when the survival distributions of the two groups are different from those of the other two groups
| ρ | ni | c | (Exp,Exp,LN,LN) | (Unif,Unif,LN,LN) | (Exp,Exp,WB,WB) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PV | BC | Rahbar | PV | BC | Rahbar | PV | BC | Rahbar | ||||||
| 0 | 100 | 0 | 0.447 | 0.431 | 0.390 | 0.466 | 0.442 | 0.415 | 0.255 | 0.277 | 0.264 | |||
| 0.25 | 0.365 | 0.360 | 0.312 | 0.383 | 0.384 | 0.343 | 0.223 | 0.249 | 0.237 | |||||
| 0.5 | 0.283 | 0.298 | 0.191 | 0.296 | 0.319 | 0.248 | 0.170 | 0.203 | 0.146 | |||||
| 200 | 0 | 0.653 | 0.754 | 0.694 | 0.749 | 0.741 | 0.731 | 0.426 | 0.504 | 0.475 | ||||
| 0.25 | 0.595 | 0.653 | 0.599 | 0.649 | 0.664 | 0.617 | 0.393 | 0.465 | 0.422 | |||||
| 0.5 | 0.473 | 0.522 | 0.444 | 0.528 | 0.556 | 0.498 | 0.301 | 0.366 | 0.305 | |||||
|
| ||||||||||||||
| 0.25 | 100 | 0 | 0.514 | 0.395 | 0.352 | 0.535 | 0.414 | 0.382 | 0.289 | 0.235 | 0.234 | |||
| 0.25 | 0.430 | 0.337 | 0.285 | 0.429 | 0.351 | 0.316 | 0.251 | 0.211 | 0.202 | |||||
| 0.5 | 0.310 | 0.268 | 0.177 | 0.309 | 0.286 | 0.226 | 0.191 | 0.172 | 0.123 | |||||
| 200 | 0 | 0.819 | 0.735 | 0.686 | 0.831 | 0.742 | 0.726 | 0.500 | 0.481 | 0.454 | ||||
| 0.25 | 0.720 | 0.638 | 0.595 | 0.720 | 0.653 | 0.627 | 0.439 | 0.425 | 0.395 | |||||
| 0.5 | 0.566 | 0.505 | 0.418 | 0.562 | 0.534 | 0.482 | 0.326 | 0.320 | 0.228 | |||||
|
| ||||||||||||||
| 0.5 | 100 | 0 | 0.614 | 0.360 | 0.316 | 0.638 | 0.379 | 0.358 | 0.355 | 0.187 | 0.195 | |||
| 0.25 | 0.495 | 0.292 | 0.250 | 0.501 | 0.322 | 0.297 | 0.299 | 0.177 | 0.161 | |||||
| 0.5 | 0.347 | 0.238 | 0.148 | 0.350 | 0.246 | 0.199 | 0.213 | 0.140 | 0.101 | |||||
| 200 | 0 | 0.900 | 0.735 | 0.682 | 0.907 | 0.746 | 0.729 | 0.607 | 0.446 | 0.436 | ||||
| 0.25 | 0.796 | 0.626 | 0.589 | 0.795 | 0.636 | 0.614 | 0.521 | 0.399 | 0.369 | |||||
| 0.5 | 0.630 | 0.498 | 0.410 | 0.627 | 0.512 | 0.462 | 0.447 | 0.307 | 0.252 | |||||
In summary, the proposed pseudo-value approach works properly for independent and dependent data. In particular, it controls Type I error a lot better and has higher power than the other two methods for dependent data. In addition, the proposed method appears to work very well even when some of the survival distributions or censoring distributions are different from the others.
4. Example
The data we considered were collected by the Center for International Blood and Marrow Transplant Research (CIBMTR) [12] and consisted of pediatric patients (< 18 years) with myelodysplastic syndrome undergoing a first allogeneic transplantation from 1993 to 2006. We limited the data to patients with advanced disease status in this example. The study population consisted of 1609 patients at 53 transplant centers. The center size was from 6 to 216 patients and the overall censoring rate was 50%. The data were correlated because a significant center effect (p-value = 0.007) on disease-free survival (DFS) rates was found using the random effect score test of [13]. To test the equality of survival medians, we examined two cases: donor groups and disease groups. Although we found that the choice of the working correlation matrix made little difference in the results under our simulation setting, three working correlation matrices were examined to choose a working correlation matrix based on the quasi-likelihood under the independence model criterion (QIC) [14]: the unstructured correlation matrix, the exchangeable correlation matrix, and the independence correlation matrix.
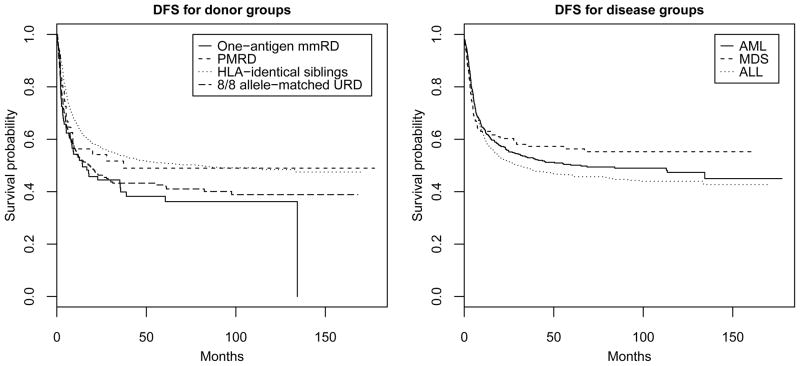
For the donor groups, we considered four donor groups: 97 patients with one-antigen mismatched related donors (mmRD), 51 patients with phenotypically matched nonsibling related donors (PMRD), 1197 patients with human leukocyte antigen (HLA) identical sibling donors, 264 patients with 8/8 allele-matched unrelated donors (URD). The censoring rates of mmRD, PMRD, HLA identical sibling donors, and URD were 42%, 51%, 52%, and 42%, respectively. The left plot of Figure 1 shows the Kaplan-Meier disease-free survival curves of the four donor groups. The DFS distributions of the four groups were different based on [15] (p-value = 0.0001). The estimated survival medians of mmRD, PMRD, HLA identical sibling donors, and URD were 14.2, 27.9, 78.9, and 17.6 months, respectively. It appears that the survival medians of some of the four groups are different. The p-values of the Brookmeyer-Crowley test and [3] are 0.107 and 0.513, respectively. For the pseudo-value approach, the independence working correlation matrix was chosen by QIC. The pseudo-value approach found a significant difference within the four donor groups with p-value 0.004.
Figure 1.
Disease-free survival rates for donor groups and disease groups
To compare different disease groups, we further limited the data to three disease groups: 553 patients with acute myeloid leukemia (AML), 756 patients with acute lymphoblastic leukemia (ALL), and 156 patients with myelodysplastic syndrome (MDS). The censoring rates of AML, ALL, and MDS were 52%, 57%, and 47%, respectively. The right plot of Figure 1 shows the Kaplan-Meier disease-free survival curves of the three disease groups. The estimated survival medians of AML and ALL were 60 and 29 months, respectively. The estimated survival median of MDS did not exist. Although the DFS distributions of the three groups were not different based on [15] (p-value = 0.120), the survival medians of the three disease groups appear to be different to each other. Because the estimated survival median of the MDS group did not exist, [3] was not applicable. The p-value of the Brookmeyer-Crowley test is 0.060. The exchangeable working correlation matrix was selected by QIC for the pseudo-value approach. The pseudo-value approach found a significant difference with p-value 0.031 at the significance level 0.05.
5. Conclusion
We proposed the pseudo-value approach to test survival medians for independent and dependent data. The simulation study showed that the pseudo-value approach performed equivalently to the Brookmeyer-Crowley test and [3] for independent data. For dependent data, the existing methods ignoring dependency were found to be too conservative. However, the pseudo-value approach controlled Type I error satisfactorily and had higher power than the two existing methods for dependent data. Although this paper focuses on the inference of survival median, the proposed method can be readily applied to other survival quantiles. Choosing an appropriate working correlation matrix may improve the performance of the pseudo-value approach [6]. We selected a working correlation matrix by QIC in the example. It may be interesting to investigate whether the working correlation matrix selected by QIC or other criteria such as the correlation information criterion [16] improves the performance of the pseudo-value approach. Exploring a pseudo-value approach for testing the equality of quantiles of cumulative incidence rates for competing risks data may also be an interesting future research question.
Acknowledgments
This work was partially supported by the U.S. National Science Foundation (DMS-1021896). The authors would like to thank Ms. Elizabeth Cho for her proof reading and thank two anonymous reviewers for their helpful comments and suggestions.
References
- 1.Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. 2. Springer-Verlag; New York: 2003. [Google Scholar]
- 2.Brookmeyer R, Crowley J. A k-sample median test for censored data. Journal of American Statistics Association. 1982;77:433–440. [Google Scholar]
- 3.Rahbar MH, Chen Z, Jeon S, Gardinerd JC, Ninge J. A nonparametric test for equality of survival medians. Statistics in Medicine. 2012;31:844–854. doi: 10.1002/sim.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen PK, Klein JP, Rosthøj S. Generalised linear models for correlated pseudo-observations, with applications to multi-state models. Biometrika. 2003;90:15–27. [Google Scholar]
- 5.Logan B, Zhang MJ, Klein JP. Marginal models for clustered time to event data with competing risks using pseudovalues. Biometrics. 2011;67:1–7. doi: 10.1111/j.1541-0420.2010.01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein JP, Andersen PK. Regression modeling of competing risks data based on pseudo-values of the cumulative incidence function. Biometrics. 2005;61:223–229. doi: 10.1111/j.0006-341X.2005.031209.x. [DOI] [PubMed] [Google Scholar]
- 7.Graw F, Schumacher MTAG. On pseudo-values for regression analysis in competing risks models. Lifetime Data Analysis. 2009;15:241–255. doi: 10.1007/s10985-008-9107-z. [DOI] [PubMed] [Google Scholar]
- 8.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 9.Ying Z, Wei LJ. The Kaplan-Meier estimate for dependent failure time observations. Journal of Multivariate Analysis. 1994;50:17–29. [Google Scholar]
- 10.van der Vaart AW. Asymptotic Statistics. Cambridge University Press; New York: 1998. [Google Scholar]
- 11.Yan J. Enjoy the joy of copulas: with a package copula. Journal of Statistical Software. 2007;21:1–21. [Google Scholar]
- 12.Shaw PJ, Kan F, Ahn KW, Spellman SR, Aljurf M, Ayas M, Burke M, Cairo MS, Chen AR, Davies SMHF, et al. Outcomes of pediatric bone marrow transplantation for leukemia and myelodysplasia using matched sibling, mismatched related, or matched unrelated donors. Blood. 2010;116:4007–4015. doi: 10.1182/blood-2010-01-261958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Commenges D, Andersen PK. Score test of homogeneity for survival data. Lifetime Data Analysis. 1995;1:145–156. doi: 10.1007/BF00985764. [DOI] [PubMed] [Google Scholar]
- 14.Pan W. Akaike’s information criterion in generalized estimating equations. Biometrics. 2001;57:120–125. doi: 10.1111/j.0006-341x.2001.00120.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee EW, Wei LJ, Amato DA. Cox-Type Regression Analysis for Large Numbers of Small Groups of Correlated Failure Time Observations. In: Klein JP, Goel PK, editors. Survival Analysis: State of the Art. Kluwer Academic Publishers; Dordrecht, Netherlands: 1992. pp. 237–247. [Google Scholar]
- 16.Hin LY, Wang YG. Working-correlation-structure identification in generalized estimating equations. Statistics in Medicine. 2009;28:642–658. doi: 10.1002/sim.3489. [DOI] [PubMed] [Google Scholar]