Abstract
The liver is unique in its ability to regenerate in response to injury. A number of evolutionary safeguards have allowed the liver to continue to perform its complex functions despite significant injury. Increased understanding of the regenerative process has significant benefit in the treatment of liver failure. Furthermore, understanding of liver regeneration may shed light on the development of cancer within the cirrhotic liver. This review will provide an overview of the models of study currently utilized in liver regeneration, the molecular basis of liver regeneration, and the role of liver progenitor cells in regeneration of the liver. Specific focus will be placed on clinical applications of current knowledge in liver regeneration including small for size liver transplant. Furthermore, cutting edge topics in liver regeneration including in vivo animal models for xenogeneic human hepatocyte expansion and the use of decellularized liver matrices as a three dimensional scaffold for liver repopulation will be proposed. Unfortunately, despite 50 years of intense study, many gaps remain in the scientific understanding of liver regeneration.
Regeneration of the liver can be more correctly defined as compensatory hyperplasia where in the remaining liver tissue expands to meet the metabolic needs of the organism. Unlike anatomic trueregeneration, the expanding liver does not regain its original gross anatomical structure1. It is also important to note the origin of cells utilized to replace the missing hepatocytes. Contrary to true regeneration, in the case of partial hepatectomy and some chemical liver injuries the liver mass is replaced by replication of existing hepatocytes without activation of progenitor cells. In other cases of chemical liver injury including galactosamine toxicity, activation and replication of progenitor cells does occur2.
Timing of Regeneration
Certain aspects of liver regeneration vary according to circadian rhythms. Matsuo and colleagues demonstrated that following partial hepatectomy in mice, the transition from G2 to mitosis occurred at the same time of day despite variability in the time of day the partial hepatectomy was performed3. DNA synthesis, however, peaked at 36 hours after surgical intervention, irrespective of the light dark cycle employed. This data strongly supports that the transition from G2 to mitosis is controlled, at least in part, by circadian-dependent cell cycle-relate genes. Specifically, these genes modulate the expression of cyclin B1-Cdc2 kinase, an important regulator of mitosis. Matsuo further presentedwee1 as a candidate for the circadian regulator of hepatocyte division. At high levels, WEE1 phosphorylates Cdc2 kinase, disrupting the activity of the cyclin B1-Cdc2 kinase complex4. Therefore, the progression of hepatocytes into mitosis is postponed until levels of WEE1 are low.
In contrast to the circadian rhythm regulated hepatocyte mitosis, DNA replication is independent of circadian rhythm but appears to be an intrinsic property of hepatocytes. There is species variation in peak DNA synthesis following partial hepatectomy with rat DNA synthesis peaking 12-16 hours earlier compared to mice. Weglarz and Sandgren demonstrated the timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous5. They transplanted rat hepatocytes into the livers of mice after partial hepatectomy and found that the rat hepatocytes replicated earlier than mouse hepatocytes in the chimeric liver. This results defined DNA synthesis as cell autonomous and suggests that cytokines or growth factors may have a permissive but not an instructive role in hepatocyte progression to S phase.
Models for Liver Regeneration
A number of models have been proposed for the study of liver regeneration. The most completely studied model is that of liver regeneration following partial hepatectomy. A rodent model of two-thirds hepatectomy was first proposed by Higgins et al in 19316. The rodent liver is multilobar allowing for the removal of 3 of 5 liver lobes (⅔ of the liver mass). Within 5 - 7 days of surgical removal the remaining liver has regenerated to a size equivalent to the original mass. This model has remained a popular model of study as there is no injury to the residual liver. The resultant sequence of events can be clearly delineated without histologic evidence of damage to the residual liver tissue.
Zebrafish have been recognized as an exceedingly important model of developmental biology due to their prolific production of offspring and transparent embryos offering constant visualization and experimental manipulation. Furthermore, organogenesis occurs rapidly with presence of nearly all major organ systems by 2 days post fertilization; a mature liver is visualized under standard light microscope by 5 days post fertilization7. Forward genetic screening, the technique of targeting embryonic mutants defective in a particular process, has allowed researchers to identify essential genes for various processes of hepatogenesis within this vertebrate model8.
Chemical mediated hepatotoxic injury, including carbon tetrachloride, has also served as a common model of liver injury. The challenge of CCL4 mediated injury is that it triggers necrosis of lobular zones of the liver leading to acute inflammatory response. The inflammatory response is dominated by polymorphonuclear leukocytes and macrophages infiltrating the liver to remove necrotic hepatocytes. The intense inflammatory response is thought to affect both the onset and duration of liver regenerative response 9.
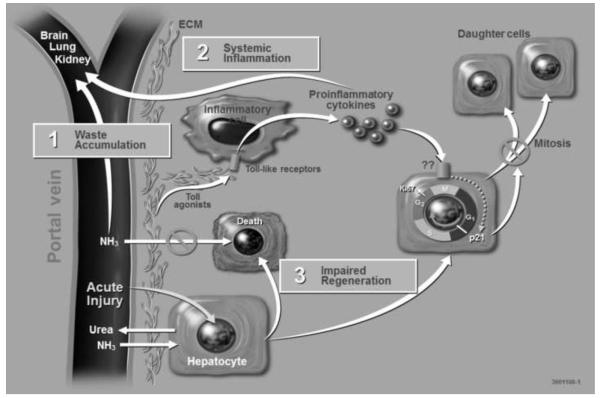
D-galactosamine is known to cause acute liver damage in animal models. The mechanism of D-galactosamine hepatotoxicity is not fully understood but D-galactosamine is believed to cause an intracellular deficiency of uridine metabolites leading to acute liver failure 10. As illustrated in Figure 1, acute liver injury by D-galactosamine is associated with waste accumulation, systemic inflammation and impaired regeneration. These three problems are also seen in humans and often contribute to death after drug induced acute liver injury which makes the porcine model of D-galactosamine acute liver failure an appropriate large animal model for testing extracorporeal liver assist devices.
Figure 1. Multi-hit hypothesis of Drug-Induced Acute Liver Injury.
Acute injury to hepatocytes by hepatotoxic drugs such as D-galactosamine and acetaminophen reduces their functional capacity leading to an accumulation of waste products such as ammonia in the blood.
Acute liver injury is also associated with the release of proinflammatory cytokines from the liver which have local adverse effects on hepatocytes leading to their impaired mitosis and extrahepatic effects such as systemic inflammation. The combination of waste molecules and proinflammatory cytokines systemically are believed to impair function of kidneys and lungs, and lead to edema formation in the brain. These systemic manifestations of acute liver failure are frequent causes of death in humans.
(ECM, Extracellular Matrix)
Acetaminophen intoxication is a common clinical cause of acute liver failure. Following overdose of acetaminophen the liver cannot perform the necessary breakdown steps of glucuronidation and sulphationand the P450 system takes over. Subsequently a toxic accumulation of N-acetyl-benzoquinoneimine occurs leading to the formation of radicals and Kupffer cell activation11. The systemic manifestation of acetaminophen hepatotoxicity are believed to be mediated by proinflammatory cytokines and the innate immune system (see Figure 1). For example, mice with mutant Tol-like receptor 4 (TLR4) had significantly improved survival after acetaminophen overdose compared to normal wild-type mice. Furthermore, survival of wild-type mice was improved significantly both by depletion of Kupffer cells or pretreatment with a TLR 4 antagonist. Kupffer cells express high levels of TLR412. These studies show that reduction of TLR4 activity through clinical treatment is associated with mitigation of systemic inflammation and improved survival in a mouse model of acetaminophen-induced liver failure. They also show that the TLR4 activity of Kupffer cells is a main contributor to the systemic inflammatory response of acute liver failure, and that modulation of the TLR4 pathway by depletion of Kupffer cells or direct antagonism of TLR4 receptor leads to improved survival following acetaminophen-induced acute liver failure. Future studies should address whether improved survival is also the result of enhanced liver regeneration.
Genetically modified animals with inborn errors of metabolism have also been proposed to serve as models of liver regeneration. Most impressive may be theimmunodeficient, fumarylacetoacetate hydrolase (FAH)-deficient mouse model developed by Grompe et al. The livers of these triple knock-out mouse are capable of engraftment and significant repopulation with mature human hepatocytes following xenogeneic transplantation13. Nyberg et al has produced (FAH)-deficient swine to upscale production of high quality human hepatocytes14. This in vivo environment allows primary hepatocytes to expand when incorporated into the three-dimensional liver architecture and exposed to the complex signaling pathways required for liver regeneration which cannot be achieved in an in vitro setting. Bissig and colleagues achieved up to a 95% repopulation of immunodeficient, FAH-deficient mouse liver with human hepatocytes following intrasplenic injection15.
Molecular Basis of Liver Regeneration
The majority of evidence defining the molecular mechanisms associated with liver regeneration is derived from rodent models following partial hepatectomy. The numerous signaling pathways involved in liver regeneration are complex and interconnected. Genetic modifications resulting in defects in a single signaling pathway often result in delayed liver regeneration but do not completely prevent the regenerative process from occurring. Delays resulting from a single pathway disruption imply that the complex, often redundant, network of pathways is essential for liver regeneration to proceed in an optimal manner resulting in adequate hepatic mass9. The pathways involved in liver regeneration include cytokines, growth factors, and metabolic networks16.
Cytokine Signaling
Immediately following partial hepatectomy, greater than 100 immediate early genes are activated by transcription factors that are latent in the quiescent liver17, 18. IL-6 is responsible for activating approximately 40% of these genes19. Activation of the immediate early genes results in a series of events including DNA synthesis, cell replication, and increase in cell size over several days. These immediate early genes also allow the liver maintain its essential metabolic functions during the process of liver regeneration9.This process occurs in hepatocytes as well as the non-parenchymal liver cells, with hepatocytes replication occurring earlier than other cell types. Among hepatocytes there is an organized fashion by which DNA synthesis progresses, starting with the hepatocytes near the portal vein and proceeding to the cells adjacent to the central vein20.
The initiation of liver regeneration is driven by the innate immune system and cytokine release. TNF, NFΚB, and IL-6 are important mediators that result in activation of STAT3 in hepatocytes. Following partial hepatectomy, TNF binds to the TNF receptor 1 on non-parenchymal cells, primarily Kupffer cells. This leads to activation of NFΚB and production of IL-621. IL-6 acts on hepatocytes via the IL-6 receptor, activating the signal transducer and activator of transcription 3 (STAT3) and extracellular signal-related kinase 1 and 2 (ERK1/2) pathways22,23. Numerous studies have provided evidence relating this pathway to the initiation of liver regeneration. The additional of anti-TNF antibodies following partial hepatectomyinhibits IL-6 production and DNA replication in a rat model24. The IL-6 knockout mouse leads to delays in liver regeneration25. Hepatocyte proliferation and gene express can be corrected in this model with a single preoperative injection of IL-6. Interestingly, elevations in serum TNF following partial hepatectomy is not seen universally among all species studied. Tnfknockout mice proceed through a normal course of liver regeneration while Tnfr1 KO mice have multiple defects following partial hepatectomy suggesting that other ligands may bind to TNFr126,27.
The innate immune system plays a role in the initiation of the cytokinecascade. Lipopolysaccharide (LPS), C3a, and C5a, all components of the innate immune system, bind to their respective receptors on Kupffer cells, triggering liver regeneration. Rats with restricted production of LPS have a delay in regeneration28. Campbell and colleagues investigated the immune mediated signaling pathways involved in the initiation of liver regeneration29. They evaluated mice lacking Tlr2 and Tlr2, the LPS co-receptor Cd14, and Myd88. MyD88 is an adaptor protein for the TLR family of proteins. They determined that Myd88 knockout mice had decreased levels of IL-6 and tnf mRNA following partial hepatectomy. Activation of STAT3 and STAT3-responsive genes were blocked as well. Interestingly, none of the knockout mice showed a delay in DNA replication. They concluded that the LPS receptor (TLR4), TLR2, and CD14 do not play a role in regulating cytokine production or DNA replication but that Myd88 –dependent pathways are involved in TNF and IL-6 production. The specific MyD88 associated receptors involved in the process have yet to be identified. A recent study by Vaquero and colleagues showed that TLR4 signaling contributes to IL-6 activation, but the Tlr4-independent component is sufficient for intact signaling downstream of IL-630. They demonstrated an attenuated increase in IL-6 after partial hepatectomy in mice with TLR4 signaling defects, supporting a role for LPS in triggering IL-6 activation. Regarding the role of complement in the initiation of liver regeneration, mice deficient in C3a and C5a have significant defects in regeneration following partial hepatectomy31. There is decreased activation of the cytokine pathway, diminishedelevation in TNF,and IL-6 levels and impaired NFΚB and STAT3 activity. This phenotype can be reversed by reconstitution of C3a and C5a.
IL-6 has multiple functions during liver regeneration including its role in the acute phase response, hepatoprotection, and mitogenesis23. Many knockout mice studies have established that IL-6 is required for normal liver regeneration. However, as previously discussed, IL-6 is not the lone cytokine involved in the initiation of liver regeneration as the process is only delayed in the absence of IL-6. Following binding of IL-6 to its receptor on hepatocytes, the gp130 subunit is activated resulting in tyrosine kinase activity. This leads to activation and dimerization of STAT3 allowing for translocation to the nucleus where it activates transcription of target genes32. Stem cell factor (SCM) and oncostatin M (OCM) modulate and enhance the effects of IL-6 by activating STAT333,34. Studies in liver-specific stat3-null mice demonstrate a significant contribution of the IL-6 induced STAT3 pathway to immediate-early gene expression35. This observed decline in immediate-early gene expression in the stat3-null mice was similar but not identical to the IL-6 knockout mice gene expression. This was the first study to provide evidence that STAT3 promotes cell cycle progression and proliferation in vivo, blurring the lines between growth factor and cytokine regulated pathways. STAT3 also functions as the main IL-6-mediated effector of hepatoprotection. STAT3 blocks apoptosis by increasing anti-caspase regulators and decreasing oxidative injury by increasing levels of the antioxidant REF136. IL-6 provides hepatoprotectionfrom Fas-mediated injury and apoptosis. IL-6−/− mice demonstrate alterations in the apoptotic pathways with reduction in anti-apoptotic factors37.The IL-6 complex activation of gp130 also leads to activation of the MAPK signaling cascade. MAPK signaling is critical for cell proliferation. The stat3-null mice had normal activation of the MAPK pathway supporting the theory that not all effects of IL-6 on hepatocyte proliferation are mediated by STAT3.
Growth Factor-Mediated Pathways
Hepatocytes progress through the cell cycle in response to a collection of mitogenic growth factors. These growth factors override the G1 restriction point allowing hepatocytes to pass into the S phase. This passage involves Rb phosphorylation, increased expression of p107 and cyclins D, E, and A38,39. In addition, cdk4/cyclinD and cdk2/cyclinE complexes are formed. The gp130 receptor subunit is involved with downstream production of the cyclins required for progression through the cell cycle.
The epidermal growth factor (EGF) receptor ligand family and hepatocyte growth factor (HGF) are important growth factors during liver regeneration40. HGF is produced by stellate cells and act in a paracrine and endocrine fashion on hepatocytes. Pro-HGF is activated in the extracellular matrix by uPA41. HGF and c-met, the gene for the HGF receptor, are essential for liver regeneration42. HGF/c-met signaling results in activation of ERK1/243. ERK1/2 has been shown to lead to hepatocyte proliferation in vitro and DNA replication in vivo. Other studies have suggested that the HGF/c-met pathway plays an important role in hepatoprotection by up regulating kinases involved in cell survival, specifically PI3K and AKT 44. The EGF receptor ligand family includes EGF, TGFα, heparin-binding EGF-like growth factor (HBEGF), and amphiregulin (AR). These various ligands have different but often overlapping functions. EGF is produced by Brunner’s gland in the duodenum 45. TGFαis produced by hepatocytes in response to cell proliferation and functions in an autocrine fashion46. Increased levels ofTGFαresult in constitutive hepatocyte proliferation47. TGFα knockout models reveal normal liver regeneration following partial hepatectomy highlighting the overlapping roles of the multiple EGF receptor ligands48. HBEGF is expressed early in liver regeneration49. A HBEGFknockout model leads to delayed liver regeneration with earlier expression of TGFα as a compensatory mechanism50. Beyond the compensatory mechanisms within the EGF receptor ligand family, there is some evidence to suggest the EGF receptor and HGF/c-met pathways may compensate for one another.
Auxiliary mitogens include TNF, IL-6, norepinephrine, Notch and jagged, VEGF, insulin, bile acids, serotonin, complement, leptin, estrogens, and FGF1 and 251. Knockout models involving these mitogens will delay but not eliminate liver regeneration. Platelet-derived serotonin has been shown to mediate liver regeneration. The expression of 5-HT2A and 2B serotonin receptors increases in the liver following partial hepatectomy. In a series of experiments by Lesurtel et al, thrombocytopenia or impaired platelet activity results in failure to initiate hepatocyte proliferation in a mouse model52. Administration of a serotonin agonist in thrombocytopenic mice resulted in normal liver proliferation while administration of serotonin receptor antagonist led to inhibition of regeneration.
Overlap exists between the growth factor and cytokine mediated pathways. IL-6, HGF, and some EGF receptor ligands have been identified as promoting expression of ERK1/2. ERK1/2 activation results in DNA replication in vivo and proliferation in vitro 53,54. Additionally, the gene encoding for insulin-like growth-factor-binding protein (IGFBP) may be activated both IL-6 and HGF. IGFBP is a mitogenic and hepato-protective proteinup regulated during regeneration 55,56. IGFBP knockout models display impaired liver regeneration accompanied by delayed DNA synthesis, necrosis, and reduced expression of cyclins important for the S phase of the cell cycle. IGFBP is hepato-protective against Fas-mediated injury and regulates apoptotic proteins MMP9 and TGFβ57.
Metabolic Pathways
Following partial hepatectomy, the metabolic demands on the liver during regeneration are immense. The liver must continue to support the organism during the regenerative process by providing an adequate systemic energy requirement while attempting to meet the energy demands needed for DNA replication and cell division. Amino acids regulate hepatocyte proliferation through modulation of cyclin D1 expression 58. Studies in rats indicate that administration of amino acids leads to hepatocyte replication, while protein restriction impairs regeneration59,60. Translation is the control point that integrates nutrient levels with mitogenic signals; most proteins involved are downstream of mTOR (mammalian target of rapamycin)61. The mTOR complex may regulate regeneration by modulating cell size and proliferation based on energy demands62. Administration of rapamycin, an inhibitor of mTOR, inhibits DNA replication following partial hepatectomy63.
MicroRNAs
MicroRNAs (miRNAs) represent a relatively new class of gene expression regulators known to control cell proliferation in cancer. Work by Song, et al, has demonstrated the importance of miRNA in regulating hepatocyte proliferation during liver regeneration. Wild-type mice with inactivation of the DiGeorge syndrome critical region gene 8 (DGCR8), a critical region in the processing of miRNA,were found to exhibit delay in cell cycle progression from G1 to S phase. Post-mortem examination of livers from these mice demonstrated inhibition of miR-21 which is essential for DNA synthesis in hepatocytes following 2/3 hepatectomy; furthermore, miRNA-378 was repressed. miRNA-378 is responsible for inhibition of ornithine decarboxylase (Odc1) which in turn promotes DNA synthesis64. Additional research has demonstrated the influence of miRNA in embryonic development in zebrafish65. Current efforts are aimed at identifying potential therapeutic applications of miRNA in various hepatic disease states 66.
Termination of Liver Regeneration
Following partial hepatectomy, the liver rapidly regenerates to a size meeting the functional needs of the organism. The vast majority of research surrounding liver regeneration has focused on cytokine and growth factor mediated pathways involved in initiation and progression through the cell cycle. Yet, the mechanisms involved in termination of liver regeneration require critical review as they remain poorly understood. The termination process involves TGFβand feedback inhibition from the cytokine and growth factor pathways.
Suppressors of cytokine signaling (SOCS) are negative regulators of the cytokine signaling cascade. IL-6 signaling results in rapid up regulation of SOCS3. SOCS3 prevents phosphorylation of STAT3, leading to its down regulation67. This results in a blockade of the IL-6 signal. This negative feedback loop explains why overexpression of IL-6 can lead to increased liver injury and impaired cell growth following partial hepatectomy68. Without SOCS3, hepatocytes are hyperproliferative in response to growth factors in culture suggesting this protein is important in controlling the normal proliferative response in hepatocytes69.
TGFβ is an antiproliferative factor produced by stellate cells that is upregulated during liver regeneration in response to signaling from HGF and EGF70,71. This increase in TGFβ is countered by a decrease in TGFβ receptors on hepatocytes during the first 48 hours following partial hepatectomy allowing for rapid proliferation 72. In addition, norepinephrine may block the antiproliferative effects of TGFβearly during the regeneration process73. Much controversy remains surrounding the specific mechanisms by which TGFβ modulates the regenerative process and overall data to support TGFβ as the primary stimulus for termination is lacking. ATGFβreceptor knockout model also revealed normal regulation unless activin, a member of theTGFβfamily, was also eliminated74.
Other theories surrounding the termination of liver regeneration focus on re-establishing the pre-partial hepatectomy extracellular matrix. TGFβhas a role in the assembly of the extracellular matrix and sinusoidal networks at the end of the regenerative process. As the extracellular matrix is re-established, it binds HGF, preventing activation and returning hepatocytes to their quiescent state75. Additionally, TGFβ is increasingly bound to decorin in the extracellular matrix leading to a extracellular milieu similar to the pre-partial hepatectomy state. This return of HGF and TGFβ to baseline may lead to complete termination of liver regeneration51.
Recent work completed by Wuestefeld et.al. identified the kinase MKK4 as a master regulator of liver regeneration76. Silencing MKK4 resulted in an increased regenerative capacity of hepatocytes in mouse models of acute and chronic liver disease. MKK4 knockdown resulted in increased liver regeneration through faster hepatocyte entry into and progression through the cell cycle.
Liver Progenitor Cells
Liver progenitor cells are thought of as the second line of defense against liver injury, becoming active when mature hepatocytes are prevented from proliferating. Ongoing work in animal and human models of disease has helped to delineate the role of liver progenitor cells in the physiologic as well as the regenerating liver.
The first liver progenitor cells to be identified, termed oval cells, were described in 1956 by Farber et al77. Oval cells, named for the appearance of their nucleus, are small bipotent cells with a high nuclear-cytoplasmic ratio which are capable of differentiation into both choangiocytes and hepatocytes78. These cells have been shown to activate in animal studies in which native hepatocytes have been chemically blocked from proliferation in the setting of liver injury stimulating regeneration 79. Oval cells proliferate within the peri-portal region dependant on growth factors produced by stellate cells including HGF, FGF1, FGF2, and VEGF 80. The oval cell, capable of production of albumin and alpha-fetoprotein become basophilic hepatocytes within four to five days of activation. Eventually these cells can become mature hepatocytes 81.
Origin of Liver Progenitor Cells
The origin of liver progenitor cells has important clinical implications. If early progenitor cells can be identified and stimulated to proliferate within the injured liver, more rapid regeneration may occur. Initial work in bone marrow transplantation suggested liver progenitor cells were a continuous population with bone marrow stem cells. This hypothesis originated from studies demonstrating that female recipients of bone marrow transplants from male donors were found to have XY hepatocytes 82. Subsequent studies utilizing a murine model of hereditary tyrosinemia demonstrated that the liver could be completely regenerated with bone marrow stem cells 83. Additional studies, however, have indicated that fusion was mechanism behind the regeneration of the liver following a bone marrow transplant 84.
Liver progenitor cells have been noted to have both epithelial and mesenchymalmarkers 85. Furthermore, mesenchymal stem cells have been observed to convert to hepatocyte-like cells under appropriate conditions, thus researchers postulated that perhaps progenitor cells were derived from hepatocyte stellate cells. Subsequently Yang et al utilized the technique of fate mapping to demonstrate that stellate cells can go on to become oval cells following liver injury 86. This theory suggested that stellate cells contribute to both fibrosis and regeneration by transition from epithelial to mesenchymal cell lines with subsequent reversion. Additional fate mapping by other researchers, however, has failed to demonstrate this phenomenon and this theory remains controversial 87, 88.
Further study is needed to delineate the origin of progenitor cells. Current evidence is conflicting, however, many experts believe most liver progenitor cells are derived from in situ cells that are direct descendants of the fetal ductal plate 85. Current efforts are directed at stimulating proliferation of stem cells or transplanting additional progenitor cells into the affected liver.
Liver Progenitor Cells Role in the Physiologic Liver
The role of progenitor cells in normal liver physiology is not completely understood, however, it is thought that progenitor cells have little involvement in day to day liver remodeling89. Work by Suzuki et al demonstrates that patients with higher MELD scores demonstrate higher levels of progenitor cell activation over the normal liver 90. Regardless of etiology of disease (i.e. chronic alcoholism, viral hepatitis, or primary biliary cirrhosis), when native hepatocytes are blocked from proliferation, levels of progenitor cells increase 89, 91. Furthermore, work by Libberechtet al in patients with viral hepatitis has demonstrated that progenitor cells are surrounded by other liver progenitor cells of various differentiation suggesting ongoing maturation and repopulation from the progenitor cell lineage92.
Acute liver failure patients also demonstrate an increased degree of progenitor cell proliferation over native liver. It has been postulated that at 50% loss of hepatocytes with decreased proliferation of mature hepatocytes triggers proliferation of the progenitor cell population90. This further de-emphasizes the role of liver progenitor cells in the physiologic liver.
Clinical Implications
Among the transplant community, the increasing demand for high-quality transplantable livers far outweighs the donated organ supply. In an attempt to overcome this disparity, partial liver transplantation from a living donor is being performed with increasing frequency. The major limitation to this technique is providing the recipient with an adequately sized, functioning graft while maintaining a high safety profile for the donor operation. The success of partial liver transplant requires some degree of liver regeneration.Small-for-size-syndrome (SFSS) results from an inability of a small graft to regenerate and is the main limiting factor in expanding the role of partial liver transplant93. Small-for-size-syndrome is characterized clinically by prolonged cholestasis, intractable ascites, coagulopathy, and encephalopathy which manifest only 3 to 5 days following segmental liver transplant as the small partial graft is unable to meet the metabolic needs of the recipient94. In the most severe cases, SFSS progresses to acidosis, hypoglycemia, septic shock, renal and pulmonary failure, and death without retransplantation. SFSS results from a failure of liver regeneration due to a combination of inadequate parenchymal volume, portal hyperperfusion, arterial hypoperfusion, and venous pathology95. Significant parenchymal injury is not required for liver failure to develop in small grafts. A deficiency in cell cycle progression, via a p21 dependent block, causes liver failure in mice. This can be overcome by inhibition of p2196.Earlier data suggested that a graft-to-recipient weight ratio of <0.8% or a liver volume <30% of standard estimated liver volume were risk factors for SFSS97. Recent data suggests that the exposure of small grafts to high portal blood flow impairs liver regeneration as sinusoidal congestion and hemorrhage have been identified in partial liver grafts in pigs 98. Portal hyperperfusion results from a smaller liver volume compared to the native liver along with pre-existing portal hypertension93. As portal venous flow is relatively unregulated in the liver compared to arterial flow, the portal hyperperfusion leads to a compensatory decrease in arterial blood flow. This “arterial buffering response” further contributes to impaired liver regeneration99. Successful partial liver transplantation relies on balancing the portal venous and hepatic artery flow and ensuring adequate hepatic venous drainage.
Various strategies are currently being investigated to overcome SFSS. Zhu and colleagues demonstrated improved serologic studies , decreased post-transplant hospital stay, and reduced infection related morbidity in patients receiving omega-3 fatty acid supplementation compared to patients receiving parenteral nutrition alone 100. Further evidence supporting the protective effect of fatty acid supplementation following partial hepatectomy is provided by Yan et.al101. Following 70% partial hepatectomy, rats administered polyunsaturated fatty acids showed enhanced expression of the LKB1-AMPK signaling pathway resulting in improved tight junction integrity and improved postoperative hepatic function. Another potential strategy for overcoming SFSS utilizes the secreted factors from mesenchymal stem cells to promote liver regeneration. Following partial hepatectomy, mice treated with mesenchymal stem cell conditioned culture media demonstrated improved regenerative capacity with upregulation of cytokines and growth factors involved in cell proliferation, angiogenesis, and anti-inflammatory responses102. Infusion of bone marrow mesenchymal stem cells also promotes proliferation of hepatocytes following extended hepatectomy103. Pentoxifylline (a TNFα inhibitor that enhances activation of the IL-6 signaling pathway) was evaluated in a recent study including 101 noncirrhotic patients undergoing major liver resection. Administration of the drug resulted in better volumetry in patients with small remnant livers104. Finally, surgical interventions including splenic artery ligation, splenectomy, and creation of a portocaval shunt have been described in clinical and animal studies to decrease the portal hyperperfusion associated with SFSS105.
As primary hepatocytes rapidly proliferate in vivo in response to the complex signaling pathways of liver regeneration and under the support of the liver’s three-dimensional architecture, efforts are underway to engineer liver scaffolds that can be transplanted and used for replacement of the liver function in patients with a failing liver. One such example is a recellularized liver graft using a decellularized liver matrix which has been successful using a rat model106. Upscaling this technique to a larger, porcine animal model may allow for the development of recellularized liver matrices functioning as an auxiliary graft for transplantation in humans.
Unlike the liver regeneration that occurs following living donor liver transplantation, which follows the same patternas seen in rodent models after partial hepatectomy, in humans liver regeneration occurs more frequently after injury from an ischemic or toxic insult107. With the majority of animal models utilizing partial hepatectomy as the inciting event for liver regeneration, it is difficult to correlate these results to the human liver’s regenerative strategy following damage by drug overdose, viral infection, or excessive alcohol consumption. More research will be required to better understand the pathways involved in liver regeneration following a toxic insult, where progenitor cells play a larger role. Additionally, cirrhosis, steatosis, and age can have detrimental effects on the liver’s ability to regenerate23. Little evidence exists to explain how altered liver pathology affects the process of liver regeneration.
Concluding Remarks
Significant advances in biologic understanding and clinical applications of the regeneration of the liver have occurred in the past several decades. The field of liver regeneration has provided an appropriate model for the study of signal transduction and cell cycle in vivo. From a clinical perspective better understanding of the role of liver regeneration has allowed for more aggressive liver resections in the setting of malignancy and treatment strategies for cirrhosis. Multiple gaps exist in our current knowledge, the understanding of which would further support clinical treatment strategies including small for size transplantation and availability of high quality transplantable organs. Current research efforts including the use of animal models as in vivo vectors for high quality human hepatocytes represents a unique and important frontier in the field of liver regeneration.
Acknowledgments
All authors report no potential financial or personal conflicts of interest. All authors have read the journal’s policy on disclosure of potential conflicts of interest. No editorial support has been utilized in preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farber JL, Gerson RJ. Mechanisms of cell injury with hepatotoxic chemicals. Pharmacological reviews. 1984;36(2 Suppl):71S–75S. [PubMed] [Google Scholar]
- 2.Lemire JM, Shiojiri N, Fausto N. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. The American Journal of Pathologyxs. 1991;139(3):535–52. [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuo T, Yamaguchi S, Mitsui S, et al. Control Mechanism of the Circadian Clock for Timing of Cell Division in Vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 4.Schibler U. Liver Regeneration Clocks On. Science. 2003;302(5643):234–235. doi: 10.1126/science.1090810. [DOI] [PubMed] [Google Scholar]
- 5.Weglarz TC, Sandgren EP. Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proceedings of the National Academy of Sciences. 2000;97(23):12595–12600. doi: 10.1073/pnas.220430497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GM H, RM A. Experimental pathology of the liver, I: Restoration of the liver of the white rat following partial surgical removal. Arch Pathology. 1931;12:186–202. [Google Scholar]
- 7.Chu J, Sadler KC. New school in liver development: lessons from zebrafish. Hepatology. 2009;50(5):1656–63. doi: 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadler KC, Krahn KN, Gaur NA, et al. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1570–5. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalopoulos GK. Liver regeneration. Journal of Cellular Physiology. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decker K, Keppler D. Galactosamine induced liver injury. Progress in liver diseases. 1972;4:183–99. [PubMed] [Google Scholar]
- 11.Rahman TM, Hodgson HJ. Animal models of acute hepatic failure. International journal of experimental pathology. 2000;81(2):145–57. doi: 10.1046/j.1365-2613.2000.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher JE, McKenzie TJ, Lillegard JB, et al. Role of Kupffer cells and toll-like receptor 4 in acetaminophen-induced acute liver failure. The Journal of surgical research. 2013;180(1):147–55. doi: 10.1016/j.jss.2012.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nature biotechnology. 2007;25(8):903–10. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickey RD, Lillegard JB, Fisher JE, et al. Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology. 2011;54(4):1351–9. doi: 10.1002/hep.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bissig K-D, Wieland SF, Tran P, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. The Journal of Clinical Investigation. 2010;120(3):924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(S1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 17.Arai M, Yokosuka O, Chiba T, et al. Gene Expression Profiling Reveals the Mechanism and Pathophysiology of Mouse Liver Regeneration. Journal of Biological Chemistry. 2003;278(32):29813–29818. doi: 10.1074/jbc.M212648200. [DOI] [PubMed] [Google Scholar]
- 18.Haber BA, Mohn KL, Diamond RH, et al. Induction patterns of 70 genes during nine days after hepatectomy define the temporal course of liver regeneration. The Journal of Clinical Investigation. 1993;91(4):1319–1326. doi: 10.1172/JCI116332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Liang X, Leu JI, et al. Global changes in interleukin-6–dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology. 2001;33(6):1377–1386. doi: 10.1053/jhep.2001.24431. [DOI] [PubMed] [Google Scholar]
- 20.Grisham JW. A Morphologic Study of Deoxyribonucleic Acid Synthesis and Cell Proliferation in Regenerating Rat Liver; Autoradiography with Thymidine-H3. Cancer Research. 1962;22(7 Part 1):842–849. [PubMed] [Google Scholar]
- 21.FitzGerald M, Webber E, Donovan J, et al. Rapid DNA binding by nuclear factor kappa B in hepatocytes at the start of liver regeneration. Cell Growth Differ. 1995;6(4):417–427. [PubMed] [Google Scholar]
- 22.Cressman DE, Diamond RH, Taub R. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 1995;21(5):1443–1449. [PubMed] [Google Scholar]
- 23.Taub R. Liver regeneration: from myth to mechanism. Nature Reviews Molecular Cell Biology. 2004;5(10):836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 24.Akerman P, Cote P, Yang SQ, et al. Antibodies to tumor necrosis factor-alpha inhibit liver regeneration after partial hepatectomy. American Journal of Physiology - Gastrointestinal and Liver Physiology. 1992;263(4):G579–G585. doi: 10.1152/ajpgi.1992.263.4.G579. [DOI] [PubMed] [Google Scholar]
- 25.Cressman DE, Greenbaum LE, DeAngelis RA, et al. Liver Failure and Defective Hepatocyte Regeneration in Interleukin-6-Deficient Mice. Science. 1996;274(5291):1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Kirillova I, Peschon JJ, et al. Initiation of Liver Growth by Tumor Necrosis Factor: Deficient Liver Regeneration in Mice Lacking Type I Tumor Necrosis Factor Receptor. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(4):1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi H, Nagaki M, Imose M, et al. Normal liver regeneration and liver cell apoptosis after partial hepatectomy in tumor necrosis factor-α-deficient mice. Liver International. 2005;25(1):162–170. doi: 10.1111/j.1478-3231.2005.01029.x. [DOI] [PubMed] [Google Scholar]
- 28.Cornell RP, Liljequist BL, Bartizal KF. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology. 1990;11(6):916–922. doi: 10.1002/hep.1840110603. [DOI] [PubMed] [Google Scholar]
- 29.Campbell JS, Riehle KJ, Brooling JT, et al. Proinflammatory Cytokine Production in Liver Regeneration Is Myd88-Dependent, but Independent of Cd14, Tlr2, and Tlr4. The Journal of Immunology. 2006;176(4):2522–2528. doi: 10.4049/jimmunol.176.4.2522. [DOI] [PubMed] [Google Scholar]
- 30.Vaquero J, Campbell JS, Haque J, et al. Toll-like receptor 4 and myeloid differentiation factor 88 provide mechanistic insights into the cause and effects of interleukin-6 activation in mouse liver regeneration. Hepatology. 2011;54(2):597–608. doi: 10.1002/hep.24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strey CW, Markiewski M, Mastellos D, et al. The Proinflammatory Mediators C3a and C5a Are Essential for Liver Regeneration. The Journal of Experimental Medicine. 2003;198(6):913–923. doi: 10.1084/jem.20030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich PC, Behrmann I, Haan S, et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren X, Hogaboam C, Carpenter A, et al. Stem cell factor restores hepatocyte proliferation in IL-6 knockout mice following 70% hepatectomy. The Journal of Clinical Investigation. 2003;112(9):1407–1418. doi: 10.1172/JCI17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura K, Nonaka H, Saito H, et al. Hepatocyte proliferation and tissue remodeling is impaired after liver injury in oncostatin M receptor knockout mice. Hepatology. 2004;39(3):635–644. doi: 10.1002/hep.20086. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Liang X, Kellendonk C, et al. STAT3 Contributes to the Mitogenic Response of Hepatocytes during Liver Regeneration. Journal of Biological Chemistry. 2002;277(32):28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 36.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. The Journal of Clinical Investigation. 2003;112(7):978–980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovalovich K, Li W, DeAngelis R, et al. Interleukin-6 Protects against Fas-mediated Death by Establishing a Critical Level of Anti-apoptotic Hepatic Proteins FLIP, Bcl-2, and Bcl-xL. Journal of Biological Chemistry. 2001;276(28):26605–26613. doi: 10.1074/jbc.M100740200. [DOI] [PubMed] [Google Scholar]
- 38.M M, K I, M N. Regulation of G1 cyclin-dependent kinases in liver regeneration. J Gastroenterol Hepatol. 1998;13(Suppl):S100–5. [PubMed] [Google Scholar]
- 39.Albrecht JH, Hoffman JS, Kren BT, et al. Cyclin and cyclin-dependent kinase 1 mRNA expression in models of regenerating liver and human liver diseases. Am J Physiol. 1993;265(5 Pt 1):G857–64. doi: 10.1152/ajpgi.1993.265.5.G857. [DOI] [PubMed] [Google Scholar]
- 40.Michalopoulos GK, Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology. 2005;128(2):503–506. doi: 10.1053/j.gastro.2004.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu M, Hara A, Okuno M, et al. Mechanism of retarded liver regeneration in plasminogen activator-deficient mice: Impaired activation of hepatocyte growth factor after Fas-mediated massive hepatic apoptosis. Hepatology. 2001;33(3):569–576. doi: 10.1053/jhep.2001.22650. [DOI] [PubMed] [Google Scholar]
- 42.Huh C-G, Factor VM, Sánchez A, et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borowiak M, Garratt AN, Wüstefeld T, et al. Met provides essential signals for liver regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(29):10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozaki M, Haga S, Zhang HQ, et al. Inhibition of hypoxia/reoxygenation-induced oxidative stress in HGF-stimulated antiapoptotic signaling: role of PI3-K and Akt kinase upon rac1. Cell Death & Differentiation. 2003;10(5):508. doi: 10.1038/sj.cdd.4401172. [DOI] [PubMed] [Google Scholar]
- 45.Skov Olsen P, Boesby S, Kirkegaard P, et al. Influence of epidermal growth factor on liver regeneration after partial hepatectomy in rats. Hepatology. 1988;8(5):992–6. doi: 10.1002/hep.1840080503. [DOI] [PubMed] [Google Scholar]
- 46.Mead JE, Fausto N. Transforming growth factor alpha may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proceedings of the National Academy of Sciences. 1989;86(5):1558–1562. doi: 10.1073/pnas.86.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webber EM, Wu JC, Wang L, et al. Overexpression of transforming growth factor-alpha causes liver enlargement and increased hepatocyte proliferation in transgenic mice. Am J Pathol. 1994;145(2):398–408. [PMC free article] [PubMed] [Google Scholar]
- 48.Russell WE, Kaufmann WK, Sitaric S, et al. Liver regeneration and hepatocarcinogenesis in transforming growth factor-alpha-targeted mice. Mol Carcinog. 1996;15(3):183–9. doi: 10.1002/(SICI)1098-2744(199603)15:3<183::AID-MC4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 49.Kiso S, Kawata S, Tamura S, et al. Role of heparin-binding epidermal growth factor-like growth factor as a hepatotrophic factor in rat liver regeneration after partial hepatectomy. Hepatology. 1995;22(5):1584–90. [PubMed] [Google Scholar]
- 50.Mitchell C, Nivison M, Jackson LF, et al. Heparin-binding Epidermal Growth Factor-like Growth Factor Links Hepatocyte Priming with Cell Cycle Progression during Liver Regeneration. Journal of Biological Chemistry. 2005;280(4):2562–2568. doi: 10.1074/jbc.M412372200. [DOI] [PubMed] [Google Scholar]
- 51.Michalopoulos GK. Liver Regeneration after Partial Hepatectomy: Critical Analysis of Mechanistic Dilemmas. The American Journal of Pathology. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lesurtel M, Graf R, Aleil B, et al. Platelet-Derived Serotonin Mediates Liver Regeneration. Science. 2006;312(5770):104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 53.Talarmin H, Rescan C, Cariou S, et al. The Mitogen-Activated Protein Kinase Kinase/Extracellular Signal-Regulated Kinase Cascade Activation Is a Key Signalling Pathway Involved in the Regulation of G1 Phase Progression in Proliferating Hepatocytes. Molecular and Cellular Biology. 1999;19(9):6003–6011. doi: 10.1128/mcb.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coutant A, Rescan C, Gilot D, et al. PI3K-FRAP/mTOR pathway is critical for hepatocyte proliferation whereas MEK/ERK supports both proliferation and survival. Hepatology. 2002;36(5):1079–1088. doi: 10.1053/jhep.2002.36160. [DOI] [PubMed] [Google Scholar]
- 55.Leu JI, Crissey MAS, Leu JP, et al. Interleukin-6-Induced STAT3 and AP-1 Amplify Hepatocyte Nuclear Factor 1-Mediated Transactivation of Hepatic Genes, an Adaptive Response to Liver Injury. Molecular and Cellular Biology. 2001;21(2):414–424. doi: 10.1128/MCB.21.2.414-424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weir E, Chen Q, DeFrances MC, et al. Rapid induction of mRNAs for liver regeneration factor and insulin-like growth factor binding protein-1 in primary cultures of rat hepatocytes by hepatocyte growth factor and epidermal growth factor. Hepatology. 1994;20(4 Pt 1):955–60. doi: 10.1002/hep.1840200426. [DOI] [PubMed] [Google Scholar]
- 57.Leu JI, Crissey MAS, Taub R. Massive hepatic apoptosis associated with TGF-β1 activation after Fas ligand treatment of IGF binding protein-1–deficient mice. The Journal of Clinical Investigation. 2003;111(1):129–139. doi: 10.1172/JCI16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelsen CJ, Rickheim DG, Timchenko NA, et al. Transient Expression of Cyclin D1 Is Sufficient to Promote Hepatocyte Replication and Liver Growth in Vivo. Cancer Research. 2001;61(23):8564–8568. [PubMed] [Google Scholar]
- 59.McGowan J, Atryzek V, Fausto N. Effects of protein-deprivation on the regeneration of rat liver after partial hepatectomy. Biochem J. 1979;180(1):25–35. doi: 10.1042/bj1800025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mead JE, Braun L, Martin DA, et al. Induction of Replicative Competence (“Priming”) in Normal Liver. Cancer Research. 1990;50(21):7023–7030. [PubMed] [Google Scholar]
- 61.Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- 62.Kim DH, Sabatini DM. Raptor and mTOR: subunits of a nutrient-sensitive complex. Curr Top Microbiol Immunol. 2004;279:259–70. doi: 10.1007/978-3-642-18930-2_15. [DOI] [PubMed] [Google Scholar]
- 63.Jiang Y-P, Ballou LM, Lin RZ. Rapamycin-insensitive Regulation of 4E-BP1 in Regenerating Rat Liver. Journal of Biological Chemistry. 2001;276(14):10943–10951. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 64.Song G, Sharma AD, Roll GR, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51(5):1735–43. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hand NJ, Master ZR, Eauclaire SF, et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136(3):1081–90. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell JS, Prichard L, Schaper F, et al. Expression of suppressors of cytokine signaling during liver regeneration. The Journal of Clinical Investigation. 2001;107(10):1285–1292. doi: 10.1172/JCI11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wüstefeld T, Rakemann T, Kubicka S, et al. Hyperstimulation With Interleukin 6 Inhibits Cell Cycle Progression After Hepatectomy in Mice. Hepatology. 2000;32(3):514–522. doi: 10.1053/jhep.2000.16604. [DOI] [PubMed] [Google Scholar]
- 69.Riehle KJ, Campbell JS, McMahan RS, et al. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. The Journal of Experimental Medicine. 2008;205(1):91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 71.Michalopoulos GK, Bowen WC, Mulé K, et al. Histological Organization in Hepatocyte Organoid Cultures. The American Journal of Pathology. 2001;159(5):1877–1887. doi: 10.1016/S0002-9440(10)63034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chari RS, Price DT, Sue SR, et al. Down-regulation of transforming growth factor beta receptor type I, II, and III during liver regeneration. The American Journal of Surgery. 1995;169(1):126–132. doi: 10.1016/s0002-9610(99)80120-2. [DOI] [PubMed] [Google Scholar]
- 73.Houck KA, Cruise JL, Michalopoulos G. Norepinephrine modulates the growth-inhibitory effect of transforming growth factor-beta in primary rat hepatocyte cultures. J Cell Physiol. 1988;135(3):551–5. doi: 10.1002/jcp.1041350327. [DOI] [PubMed] [Google Scholar]
- 74.Oe S, Lemmer ER, Conner EA, et al. Intact signaling by transforming growth factor β is not required for termination of liver regeneration in mice. Hepatology. 2004;40(5):1098–1105. doi: 10.1002/hep.20426. [DOI] [PubMed] [Google Scholar]
- 75.Ichikawa T, Zhang YQ, Kogure K, et al. Transforming growth factor beta and activin tonically inhibit DNA synthesis in the rat liver. Hepatology. 2001;34(5):918–25. doi: 10.1053/jhep.2001.29132. [DOI] [PubMed] [Google Scholar]
- 76.Wuestefeld T, Pesic M, Rudalska R, et al. A Direct in Vivo RNAi Screen Identifies MKK4 as a Key Regulator of Liver Regeneration. Cell. 2013;153(2):389–401. doi: 10.1016/j.cell.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 77.Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3'-methyl-4-dimethylaminoazobenzene. Cancer research. 1956;16(2):142–8. [PubMed] [Google Scholar]
- 78.Lazaro CA, Rhim JA, Yamada Y, et al. Generation of hepatocytes from oval cell precursors in culture. Cancer research. 1998;58(23):5514–22. [PubMed] [Google Scholar]
- 79.Dunsford HA, Karnasuta C, Hunt JM, et al. Different lineages of chemically induced hepatocellular carcinoma in rats defined by monoclonal antibodies. Cancer research. 1989;49(17):4894–900. [PubMed] [Google Scholar]
- 80.Evarts RP, Hu Z, Fujio K, et al. Activation of hepatic stem cell compartment in the rat: role of transforming growth factor alpha, hepatocyte growth factor, and acidic fibroblast growth factor in early proliferation. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1993;4(7):555–61. [PubMed] [Google Scholar]
- 81.Evarts RP, Nagy P, Nakatsukasa H, et al. in Vivo differentiation of rat liver oval cells into hepatocytes. Cancer research. 1989;49(6):1541–7. [PubMed] [Google Scholar]
- 82.Theise ND, Saxena R, Portmann BC, et al. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30(6):1425–33. doi: 10.1002/hep.510300614. [DOI] [PubMed] [Google Scholar]
- 83.Lagasse E, Connors H, Al-Dhalimy M, et al. Purified hematopoietic stem cells can differentiate into hepatocytes in Vivo. Nature medicine. 2000;6(11):1229–34. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 84.Willenbring H, Bailey AS, Foster M, et al. Myelomonocytic cells are sufficient for therapeutic cell fusion in liver. Nature medicine. 2004;10(7):744–8. doi: 10.1038/nm1062. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L, Theise N, Chua M, et al. The stem cell niche of human livers: symmetry between development and regeneration. Hepatology. 2008;48(5):1598–607. doi: 10.1002/hep.22516. [DOI] [PubMed] [Google Scholar]
- 86.Yang L, Jung Y, Omenetti A, et al. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem cells. 2008;26(8):2104–13. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scholten D, Osterreicher CH, Scholten A, et al. Genetic labeling does not detect epithelial-to-mesenchymal transition of cholangiocytes in liver fibrosis in mice. Gastroenterology. 2010;139(3):987–98. doi: 10.1053/j.gastro.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taura K, Miura K, Iwaisako K, et al. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology. 2010;51(3):1027–36. doi: 10.1002/hep.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lowes KN, Brennan BA, Yeoh GC, et al. Oval cell numbers in human chronic liver diseases are directly related to disease severity. The American journal of pathology. 1999;154(2):537–41. doi: 10.1016/S0002-9440(10)65299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katoonizadeh A, Nevens F, Verslype C, et al. Liver regeneration in acute severe liver impairment: a clinicopathological correlation study. Liver international : official journal of the International Association for the Study of the Liver. 2006;26(10):1225–33. doi: 10.1111/j.1478-3231.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 91.Libbrecht L, Roskams T. Hepatic progenitor cells in human liver diseases. Seminars in cell & developmental biology. 2002;13(6):389–96. doi: 10.1016/s1084952102001258. [DOI] [PubMed] [Google Scholar]
- 92.Libbrecht L, Desmet V, Van Damme B, et al. Deep intralobular extension of human hepatic 'progenitor cells' correlates with parenchymal inflammation in chronic viral hepatitis: can 'progenitor cells' migrate? The Journal of pathology. 2000;192(3):373–8. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH700>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 93.Dahm F, Georgiev P, Clavien P-A. Small-for-Size Syndrome After Partial Liver Transplantation: Definition, Mechanisms of Disease and Clinical Implications. American Journal of Transplantation. 2005;5(11):2605–2610. doi: 10.1111/j.1600-6143.2005.01081.x. [DOI] [PubMed] [Google Scholar]
- 94.Tucker ON, Heaton N. The ‘small for size’ liver syndrome. Current Opinion in Critical Care. 2005;11(2):150–155. doi: 10.1097/01.ccx.0000157080.11117.45. [DOI] [PubMed] [Google Scholar]
- 95.Gruttadauria S, Pagano D, Luca A, et al. Small-for-size syndrome in adult-to-adult living-related liver transplantation. World J Gastroenterol. 2010 Oct 28;16(40):5011–5. doi: 10.3748/wjg.v16.i40.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lehmann K, Tschuor C, Rickenbacher A, et al. Liver Failure After Extended Hepatectomy in Mice Is Mediated by a p21-Dependent Barrier to Liver Regeneration. Gastroenterology. 2012;143(6):1609–1619. doi: 10.1053/j.gastro.2012.08.043. e4. [DOI] [PubMed] [Google Scholar]
- 97.Kiuchi T, Kasahara M, Uryuhara K, et al. Impact of graft size mismatching on graft prognosis in liver transplantation from living donors. Transplantation. 1999;67(2):321–7. doi: 10.1097/00007890-199901270-00024. [DOI] [PubMed] [Google Scholar]
- 98.Kelly DM, Demetris AJ, Fung JJ, et al. Porcine partial liver transplantation: A novel model of the “small-for-size” liver graft. Liver Transplantation. 2004;10(2):253–263. doi: 10.1002/lt.20073. [DOI] [PubMed] [Google Scholar]
- 99.Ayuse T, Brienza N, O'Donnell CP, et al. Pressure-flow analysis of portal vein and hepatic artery interactions in porcine liver. American Journal of Physiology - Heart and Circulatory Physiology. 1994;267(4):H1233–H1242. doi: 10.1152/ajpheart.1994.267.4.H1233. [DOI] [PubMed] [Google Scholar]
- 100.Zhu XH, Wu YF, Qiu YD, et al. Liver-protecting effects of omega-3 fish oil lipid emulsion in liver transplantation. World J Gastroenterol. 2012;18(42):6141–7. doi: 10.3748/wjg.v18.i42.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan XP, Wang S, Yang Y, et al. Effects of n-3 Polyunsaturated Fatty Acids on Rat Livers after Partial Hepatectomy via LKB1-AMPK Signaling Pathway. Transplantation Proceedings. 2011;43(10):3604–3612. doi: 10.1016/j.transproceed.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 102.Fouraschen SM, Pan Q, de Ruiter PE, et al. Secreted factors of human liver-derived mesenchymal stem cells promote liver regeneration early after partial hepatectomy. Stem Cells Dev. 2012;21(13):2410–9. doi: 10.1089/scd.2011.0560. [DOI] [PubMed] [Google Scholar]
- 103.Yu J, Yin S, Zhang W, et al. Hypoxia preconditioned bone marrow mesenchymal stem cells promote liver regeneration in a rat massive hepatectomy model. Stem Cell Res Ther. 2013;4(4):83. doi: 10.1186/scrt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petrowsky H, Breitenstein S, Slankamenac K, et al. Effects of pentoxifylline on liver regeneration: a double-blinded, randomized, controlled trial in 101 patients undergoing major liver resection. Ann Surg. 2010;252(5):813–22. doi: 10.1097/SLA.0b013e3181fcbc5e. [DOI] [PubMed] [Google Scholar]
- 105.Yagi S, Uemoto S. Small-for-size syndrome in living donor liver transplantation. Hepatobiliary Pancreat Dis Int. 2012;11(6):570–6. doi: 10.1016/s1499-3872(12)60227-6. [DOI] [PubMed] [Google Scholar]
- 106.Uygun BE, Soto-Gutierrez A, Yagi H, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nature medicine. 2010;16(7):814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koniaris LG, McKillop IH, Schwartz SI, et al. Liver regeneration. Journal of the American College of Surgeons. 2003;197(4):634–659. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]