Abstract
WRKY transcription factor genes play critical roles in plant growth and development, as well as stress responses. WRKY genes have been examined in various higher plants, but they have not been characterized in Lotus japonicus. The recent release of the L. japonicus whole genome sequence provides an opportunity for a genome wide analysis of WRKY genes in this species. In this study, we identified 61 WRKY genes in the L. japonicus genome. Based on the WRKY protein structure, L. japonicus WRKY (LjWRKY) genes can be classified into three groups (I–III). Investigations of gene copy number and gene clusters indicate that only one gene duplication event occurred on chromosome 4 and no clustered genes were detected on chromosomes 3 or 6. Researchers previously believed that group II and III WRKY domains were derived from the C-terminal WRKY domain of group I. Our results suggest that some WRKY genes in group II originated from the N-terminal domain of group I WRKY genes. Additional evidence to support this hypothesis was obtained by Medicago truncatula WRKY (MtWRKY) protein motif analysis. We found that LjWRKY and MtWRKY group III genes are under purifying selection, suggesting that WRKY genes will become increasingly structured and functionally conserved.
1. Introduction
Transcription factors are crucial in regulating gene expression. Transcription factors present sequence-specific DNA binding sites and are able to modulate the transcription rate of downstream target genes [1]. WRKY genes have primarily been located in plants, where they are one of the most important transcription factor families [2]. WRKY genes are defined by having a unique WRKY domain of approximately 60 amino acid residues [2]. The WRKY domain contains a highly conserved amino acid sequence WRKYGQK at the N-terminal and a metal chelating zinc finger motif (C–X4-5–C–X22-23–H–X–H, (C2H2) or C–X5–8–C–X25–28–H–X1-2–C, (C2HXC)) at the C-terminal end [2–5]. In some WRKY genes, the WRKY domain can be characterized as WRRY, WSKY, WKRY, WVKY, or WKKY [6]. WRKY transcription factors interact with the W-box (TTGAC[T/C]) sequence in promoter regions to modulate gene expression [4, 7, 8]. In addition, WRKY transcription factors bind SURE, a novel cis-element in higher plants, to regulate sugar response [9].
Members of the WRKY family can be classified into three groups according to the number of WRKY domains and the pattern of the zinc finger motif [2]. Generally, group I WRKY transcription factors contain two WRKY domains with distinct functions. Previous studies have demonstrated that the C-terminal WRKY domain mediates sequence-specific binding to the target DNA [8, 10, 11]. It has been proposed that the N-terminal WRKY domain increases the affinity or specificity of these proteins to the target sites [2]. Group II and III WRKY transcription factors contain one WRKY domain with a C2H2 zinc finger motif and C2HXC zinc finger motif [2]. Based on a phylogenetic analysis of the WRKY family, the members of group II can be divided into five subgroups: IIa, IIb, IIc, IId, and IIe [2].
Since the cloning a WRKY gene cDNA from Ipomoea batatas [11], a large number of WRKY protein genes have been cloned from different plant species [3, 4, 12–24]. So far, only two WRKY homologues have been identified from nonplant species, Giardia lamblia [5] and Dictyostelium discoideum [25].
Plant WRKY genes regulate plant growth and development under normal and stressful conditions [26, 27]. Early studies found that WRKY genes play an important role in gene expression responses to sucrose [11]. Studies in Arabidopsis [17, 28, 29], rice [30], tobacco [31, 32], and parsley [33] have indicated that WRKY proteins play key roles in plant responses to pathogens [27, 34]. In addition, previous studies revealed the involvement of WRKY proteins in abiotic stress responses [27], for example, to high temperatures [35], low temperatures [13], salt and drought [24], H2O2 [36], and UV radiation [37]. WRKY proteins have also been reported to upregulate in response to herbivory [38], nematode damage [39], and wounding [40]. Moreover, WRKY genes may be involved in seed development [9, 41], dormancy and germination [42–44], plant senescence [45, 46] and regulation of metabolic pathways [9], trichome morphogenesis [47], and plant growth [48].
Lotus japonicus, an important forage crop in the legume family, is planted in many parts of the world. It has been used extensively in plant research as a model legume, due to its short life cycle (2-3 months), self-fertility, and relatively simple diploid genome [49]. Since the release of the whole genome sequence of L. japonicus, it is now possible to compare transcription factors in this plant with other plants. In this study, we analyzed 61 putative WRKY genes from the L. japonicus genome. We conducted a phylogenetic analysis to evaluate gene duplications, chromosomal localization, motif analysis, gene structure, and selection pressure analysis of group III WRKY genes to provide information about WRKY gene family evolution in L. japonicus.
2. Materials and Methods
2.1. Sequence Database Search
The L. japonicus genome sequence (build 2.5) was downloaded from http://www.kazusa.or.jp/lotus/. The complete set of WRKY gene sequences was identified using a deliberative process. First, a Hidden Markov Model (HMM) profile of the WRKY domain (PF03106) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/) [50]. We employed the WRKY domain as a query to identify all possible WRKY gene sequences in the L. japonicus genome database using the BLASTp program (P value = 0.001). Subsequently, a search on the Pfam database was used to confirm and classify each putative WRKY sequence. We located overlapping genes by aligning all of the candidate WRKY gene sequences using Clustal W [51]. Only the nonoverlapping WRKY sequences were used for further analysis.
2.2. Gene Duplication and Chromosomal Locations of WRKY Genes
To detect potential gene duplications, we aligned and calculated all of the relevant genes identified in L. japonicus genomes. We defined gene duplication between any two loci such that [52]: (1) the alignable nucleotide sequence covered >70% of the longer sequence; (2) the amino acid identity between the sequences was >70% identical.
In order to determine the physical locations of WRKY genes in chromosomes, we blasted each WRKY gene as a query against the L. japonicus genome (http://www.kazusa.or.jp/lotus/) to determine the initiation site of each gene. MapInspect software was used to draw the location images of the WRKY genes (http://www.plantbreeding.wur.nl/uk/software_mapinspect.html).
2.3. Multiple Sequence Alignments, Phylogenetic, and Gene Structure Analysis
To examine the domain organization of WRKY proteins in detail, multiple sequence alignments of WRKY domain sequences were performed using Clustal W [51]. Phylogenetic and molecular evolution analyses of WRKY proteins were conducted in MEGA 4.0 [53]. Phylogenetic trees were estimated with the Neighbor-Joining method. Bootstrap analyses of 1,000 repetitions were obtained for each tree to analyze statistical support for nodes. The complete amino acid sequences of group III WRKY genes were analyzed for evidence of selection pressure [54]. Gene structure display server (GSDS) software [55] was used to illustrate exon-intron organization for individual WRKY genes by comparing the cDNA sequence with the corresponding genomic DNA sequence.
2.4. Identification of Conserved Motifs
The program MEME 4.9 [56] was used to predict motifs in the WRKY domain with the following parameters: (1) any number of repetitions, (2) an optimum motif width between 6 and 200 residues, (3) and a maximum of 20 motifs. Structural motif annotation was performed using the Pfam database.
2.5. Selection Pressure in Group III WRKY Proteins
The amino acid sequences of group III Medicago truncatula (MtWRKY) and L. japonicus WRKY (LjWRKY) proteins were used to estimate phylogenetic trees and then the trees were used to detect evidence of selection. The PAL2NAL program [57] was used for conversion of a protein sequence into the corresponding nucleotide sequence. We used PAML 4.7 [58] to analyze codon substitution patterns in a maximum likelihood framework, implementing a site-specific model. The program CODEML was employed to calculate the d N/d S ratio (or ω), the ratio of nonsynonymous/synonymous distances. Generally, ω = 1, >1, and <1 indicate neutral, positive, and purifying selection, respectively. We detected variation in ω among sites by employing a likelihood ratio test (LRT) between M0 versus M3 and M7 versus M8 [54]. The nodes were considered to have undergone positive selection, if they satisfied the following criteria [59]: (1) an estimate of ω > 1 under M8, (2) sites identified to be under positive selection by Bayes Empirical Bayes (BEB) analysis, (3) and a statistically significant LRT.
3. Results
3.1. Identification and Classification of WRKY Genes
In this study, a total of 71 WRKY genes in the L. japonicus genome (build 2.5) were identified (Table 1). Among these sequences, 10 WRKY genes were excluded from this study due to the divergent structures of the putative proteins in these genes, a lack of specific domains or motifs, and the short length of the WRKY domain, generally 2/3 of the normal WRKY domain length. Although the LjWRKY51 protein, including two WRKY domains, lacked a complete WRKY domain at the C-terminal region, this protein was retained for subsequent analyses.
Table 1.
WRKY family genes in Lotus japonicus genome.
| Annotation IDa | Gene name | Group | Chr | Location | Annotation IDa | Gene name | Group | Chr | Location |
|---|---|---|---|---|---|---|---|---|---|
| CM0012.480.r2.a | LjWRKY1 | I | 1 | 58,226,335 | CM0432.2010.r2.d | LjWRKY37 | IIx | 4 | 7,529,554 |
| CM0032.140.r2.d | LjWRKY2 | IIc | 1 | 17,061,228 | CM0536.110.r2.d | LjWRKY38 | IId | 4 | 13,585,366 |
| CM0032.170.r2.d | LjWRKY3 | IIc | 1 | 17,093,440 | CM1622.200.r2.a | LjWRKY39 | IIc | 4 | 37,600,439 |
| CM0105.740.r2.a | LjWRKY4 | III | 1 | 65,847,050 | CM0040.550.r2.d | LjWRKY40 | IIa | 5 | 5,190,496 |
| CM0122.1300.r2.m | LjWRKY5 | IIb | 1 | 62,472,860 | CM0040.670.r2.d | LjWRKY41 | IIa | 5 | 5,268,779 |
| CM0122.2160.r2.d | LjWRKY6 | IIe | 1 | 63,001,936 | CM0239.70.r2.d | LjWRKY42 | IIc | 5 | 25,979,555 |
| CM0123.240.r2.m | LjWRKY7 | IIb | 1 | 6,329,403 | CM0852.250.r2.d | LjWRKY43 | IId | 5 | 2,801,268 |
| CM0133.780.r2.m | LjWRKY8 | IIc | 1 | 23,799,861 | CM1574.880.r2.m | LjWRKY44 | IId | 5 | 18,726,557 |
| CM0147.250.r2.m | LjWRKY9 | IIc | 1 | 42,532,993 | CM0037.660.r2.m | LjWRKY45 | I | 6 | 14,396,956 |
| CM0315.160.r2.m | LjWRKY10 | IIb | 1 | 35,572,836 | CM1034.270.r2.d | LjWRKY46 | IIe | 6 | 19,990,091 |
| CM0320.110.r2.d | LjWRKY11 | III | 1 | 15,934,171 | LjT11E14.30.r2.m | LjWRKY47 | I | 6 | 9,459,959 |
| CM0433.90.r2.d | LjWRKY12 | IIa | 1 | 24,711,963 | LjT46F23.30.r2.d | LjWRKY48 | IId | 6 | 17,672,968 |
| CM1413.480.r2.d | LjWRKY13 | I | 1 | 34,039,015 | LjSGA_012799.2 | LjWRKY49 | IIc | nd | nd |
| CM1413.70.r2.d | LjWRKY14 | IIe | 1 | 33,660,455 | LjSGA_014003.1 | LjWRKY50 | I | nd | nd |
| CM0058.50.r2.m | LjWRKY15 | I | 2 | 34,643,392 | LjSGA_015261.1 | LjWRKY51 | I | nd | nd |
| CM0060.250.r2.a | LjWRKY16 | III | 2 | 31,851,611 | LjSGA_020468.1 | LjWRKY52 | I | nd | nd |
| CM0120.570.r2.d | LjWRKY17 | IIc | 2 | 20,356,545 | LjSGA_021425.1 | LjWRKY53 | III | nd | nd |
| CM0191.300.r2.a | LjWRKY18 | I | 2 | 43,680,266 | LjSGA_025444.2 | LjWRKY54 | IIe | nd | nd |
| CM0608.50.r2.m | LjWRKY19 | IIx | 2 | 20,505,843 | LjSGA_038153.1 | LjWRKY55 | nd | nd | nd |
| CM0608.70.r2.m | LjWRKY20 | IIc | 2 | 20,514,487 | LjSGA_038411.1 | LjWRKY56 | nd | nd | nd |
| CM1895.20.r2.m | LjWRKY21 | III | 2 | 34,148,852 | LjSGA_046833.1 | LjWRKY57 | IIa | nd | nd |
| LjB06N21.120.r2.a | LjWRKY22 | IIb | 2 | 42,171,450 | LjSGA_050544.1 | LjWRKY58 | nd | nd | nd |
| CM0243.470.r2.m | LjWRKY23 | IIb | 3 | 39,562,863 | LjSGA_055568.1 | LjWRKY59 | IIe | nd | nd |
| CM0936.60.r2.d | LjWRKY24 | I | 3 | 18,082,486 | LjSGA_058050.1 | LjWRKY60 | IIe | nd | nd |
| LjT41F16.60.r2.d | LjWRKY25 | III | 3 | 33,152,607 | LjSGA_058052.1 | LjWRKY61 | nd | nd | nd |
| LjT45M09.130.r2.d | LjWRKY26 | IIa | 3 | 474,491 | LjSGA_066036.1 | LjWRKY62 | nd | nd | nd |
| CM0004.1290.r2.m | LjWRKY27 | III | 4 | 40,133,355 | LjSGA_069579.1 | LjWRKY63 | IIc | nd | nd |
| CM0004.880.r2.d | LjWRKY28 | IIe | 4 | 39,837,881 | LjSGA_074792.1 | LjWRKY64 | IIe | nd | nd |
| CM0007.1120.r2.a | LjWRKY29 | IIc | 4 | 3,358,814 | LjSGA_084767.1 | LjWRKY65 | nd | nd | nd |
| CM0046.1360.r2.d | LjWRKY30 | IIe | 4 | 36,250,655 | LjSGA_086809.1 | LjWRKY66 | nd | nd | nd |
| CM0179.130.r2.m | LjWRKY31 | I | 4 | 28,951,362 | LjSGA_091481.1 | LjWRKY67 | nd | nd | nd |
| CM0227.580.r2.m | LjWRKY32 | I | 4 | 10,519,027 | LjSGA_107504.1 | LjWRKY68 | nd | nd | nd |
| CM0244.860.r2.m | LjWRKY33 | IId | 4 | 30,871,468 | LjSGA_116672.1 | LjWRKY69 | nd | nd | nd |
| CM0333.340.r2.d | LjWRKY34 | IIc | 4 | 32,856,509 | LjT10P12.70.r2.d | LjWRKY70 | IIb | nd | nd |
| CM0333.510.r2.a | LjWRKY35 | IIc | 4 | 32,985,615 | LjT45D24.80.r2.d | LjWRKY71 | IIb | nd | nd |
| CM0337.220.r2.d | LjWRKY36 | IIb | 4 | 3,852,684 |
aAnnotation IDs are from miyakogusa website http://www.kazusa.or.jp/lotus/.
Italics indicated pseudogenes.
Chr: chromosome.
nd: nondetermined.
LjWRKY, WRKY genes of Lotus japonicas.
Among these 61 WRKY genes, there were 12 group I WRKY genes, 42 group II WRKY genes, and 7 group III WRKY genes, based on the number of WRKY domains and the type of zinc finger motifs (Tables 1 and 2). To obtain a better classification within group II WRKY proteins, we constructed a phylogenetic tree with 42 group II LjWRKY using the Arabidopsis WRKY sequence as a reference [2] (Figure 1). It was found that LjWRKY genes in group II could be divided into six subgroups, including 5 members in subgroup IIa, 8 in subgroup IIb, 13 in subgroup IIc, 5 in subgroup IId, 9 in subgroup IIe, and 2 in subgroup IIx (Tables 1 and 2 and Figure 1). Among them, LjWRLY19 and LjWRKY37 genes did not cluster with Arabidopsis, and we temporarily named this group IIx (Figure 1).
Table 2.
The number of WRKY gene in eight plants.
| Group I | Group IIa | Group IIb | Group IIc | Group IId | Group IIe | Group IIx | Group III | Total | Genome size (Mb) | |
|---|---|---|---|---|---|---|---|---|---|---|
| LjWRKY | 12 | 5 | 8 | 13 | 5 | 9 | 2 | 7 | 61 | 472 |
| MtWRKY | 19 | 6 | 8 | 19 | 8 | 8 | 0 | 13 | 81 | 375 |
| AtWRKYa | 13 | 4 | 7 | 18 | 7 | 9 | 0 | 14 | 72 | 125 |
| OsWRKYa | 15 | 4 | 8 | 15 | 7 | 11 | 0 | 36 | 96 | 480 |
| CsWRKYa | 10 | 4 | 4 | 16 | 8 | 7 | 0 | 6 | 55 | 367 |
| VvWRKYa | 12 | 4 | 7 | 14 | 6 | 7 | 0 | 5 | 55 | 487 |
| BdWRKYb | 17 | 3 | 6 | 21 | 6 | 10 | 0 | 23 | 86 | 272 |
| PtWRKYc | 50 | 5 | 9 | 13 | 13 | 4 | 0 | 10 | 104 | 485 |
Figure 1.
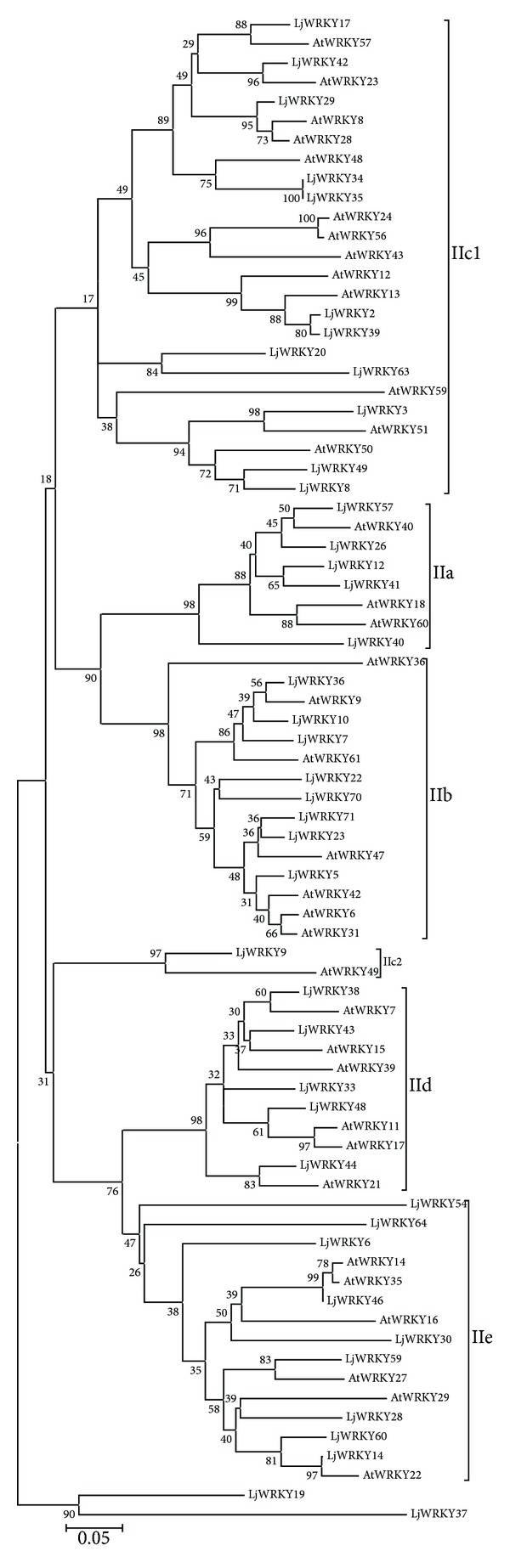
Phylogenetic tree of Arabidopsis WRLY and Lotus japonicus WRKY domain. The phylogenetic tree was constructed using MEGA 4.0 by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. The percentage bootstrap scores higher than 50% are indicated on the nodes. The AtWRKY domains from Eulgem et al. (2000).
We found 9 potential pseudogenes among 61 WRKY genes, with either a premature stop codon or a frame shift mutation. Among these pseudogenes, 3 genes were found in group I, 2 in group IIa, 2 in group IIc, and 2 in groups IIx and IIe (Table 1).
3.2. Gene Duplication and Chromosomal Locations of WRKY Genes
Gene duplication, including tandem and segmental duplication events, plays a crucial role in genomic expansions. Two or more duplicated genes located in the same chromosome are defined as tandem duplication, while other types of gene duplication are defined as segmental duplication events. We detected only one tandem duplication (LjWRKY34 and LjWRKY35) on chromosome 4 (Figure 2), suggesting that WRKY genes in L. japonicus are not recently generated by gene duplication.
Figure 2.
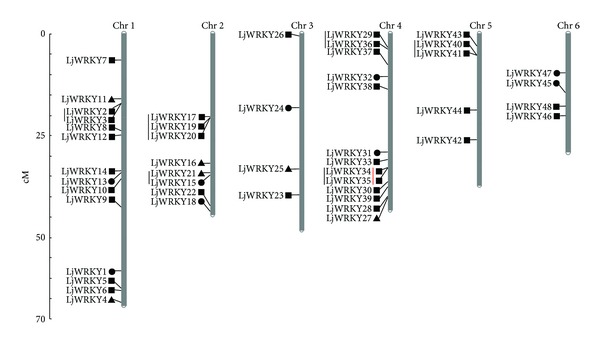
Chromosomal locations of Lotus japonicus WRKY genes. The chromosome numbers were shown at the top of each chromosome (chromosome; gray bars). The names on the left side of each chromosome correspond to the approximate location of each WRKY gene. The markers next to the gene names represented the groups to which each WRKY gene belongs (●: group I; ■: group II; ▲: group III). The black lines on the left side of the names of the WRKY genes indicated the clusters of gene on each chromosome. The red line on the left side of the markers indicated the duplicated genes. Unmapped WRKY genes were not shown.
A total of 48 WRKY genes could be mapped to chromosomes 1–6, and the others could not be conclusively mapped to any chromosomes, because some WRKY genes have no precise location information. Fourteen WRKY genes including two group I, 10 group II, and two group III genes were located on chromosome 1. Thirteen genes were mapped to chromosome 4. Four genes were mapped to chromosome 3 (one group I, two group II, and one group III genes) and chromosome 6 (two group I and two group II genes), respectively. Eight (two group I, four group II, and two group III genes) and five WRKY genes (five group II genes) were found on chromosome 2 and chromosome 5, respectively (Figure 2).
A gene cluster is defined as a chromosome region with two or more genes located within 200 kb sequence [60]. Using this criterion, we found 13 WRKY genes forming six gene clusters. Chromosomes 2 and 4 each contain two gene clusters, while only one gene cluster was found on chromosomes 1 and 5, respectively (Figure 2). No clusters were found on chromosomes 3 and 6. Through chromosomal location and gene cluster analysis, we found that the number of genes on chromosomes is disproportionate to the number of gene clusters. For example, 14 WRKY genes were found on chromosome 1, which contains only one gene cluster.
3.3. Phylogenetic Analysis and Gene Structure
Amino acid residues of WRKYGQK are the distinguishing regions of the WRKY transcription factor [2, 27]. Multiple alignment analysis of LjWRKY domains found that mutations occurred at R, Y, and Q in the conserved WRKYGQK sequence (Figure 3). Further study showed that the variation arose from amino acid substitutions of R to K or L or from K to C and from Q to Y, E, L, or K (Figure 3). On the other hand, we identified a CX4CX22HXH zinc finger motif in subgroup In and IIx genes, a CX4CX23HXH motif in subgroup Ic and IIe genes, and a CX5CX23HXH motif in subgroup IIa, IIc, IId, and IIe genes (Figure 3). We found that the WRKY domain was replaced by WKKY in subgroup In and IIx genes, suggesting that subgroup IIx genes originated from N-terminal WRKY domain of group I genes. The subgroup IIa and IIe genes seem to have formed after the group I genes lost alternative WRKY domains.
Figure 3.
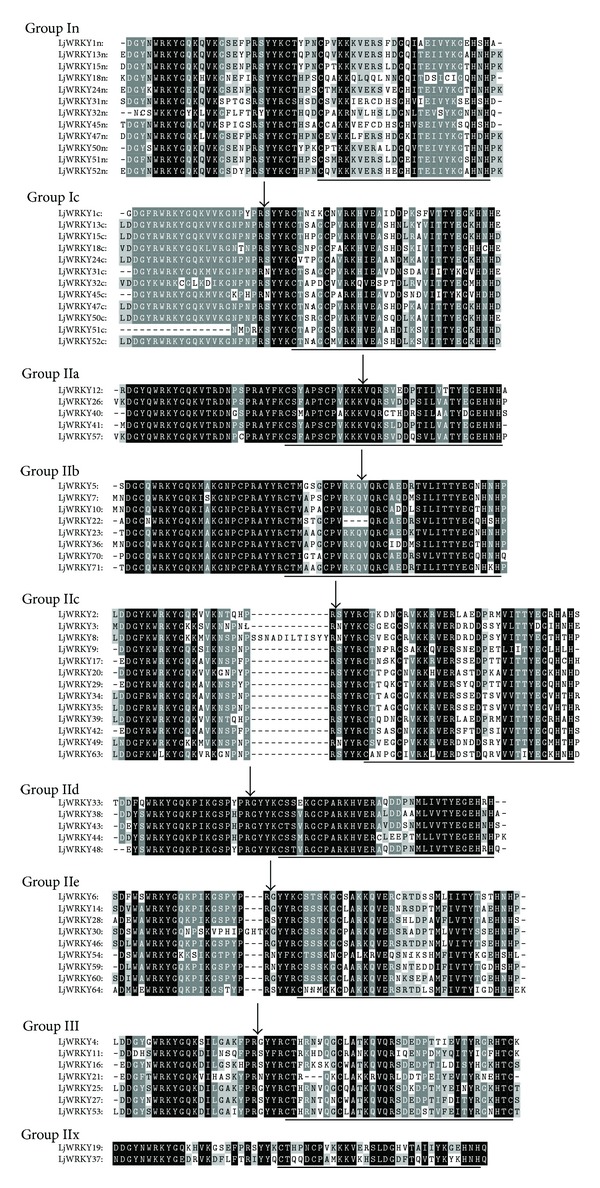
Amino acid residue alignment of Lotus japonicus WRKY domain. Alignment was performed using the Clustal W program and is displayed with the software Gendoc. Residues that were highly conserved within each of the major groups are in black. The position of a conserved intron was indicated by an arrowhead. The black lines indicated the conserved zinc finger motifs.
The WRKY domain phylogenetic tree can be subdivided into eight clades: In, Ic, IIa, IIb, IIc, IId, IIe, and III (IIx nested in In; Figure 4(a)). The group I proteins contain two WRKY domains located at the N-terminal domain (In) or the C-terminal domain (Ic). Although In and Ic belong to group I, members of In and Ic were clustered in different clades, representing the sequence divergence of WRKY domain in In and Ic.
Figure 4.
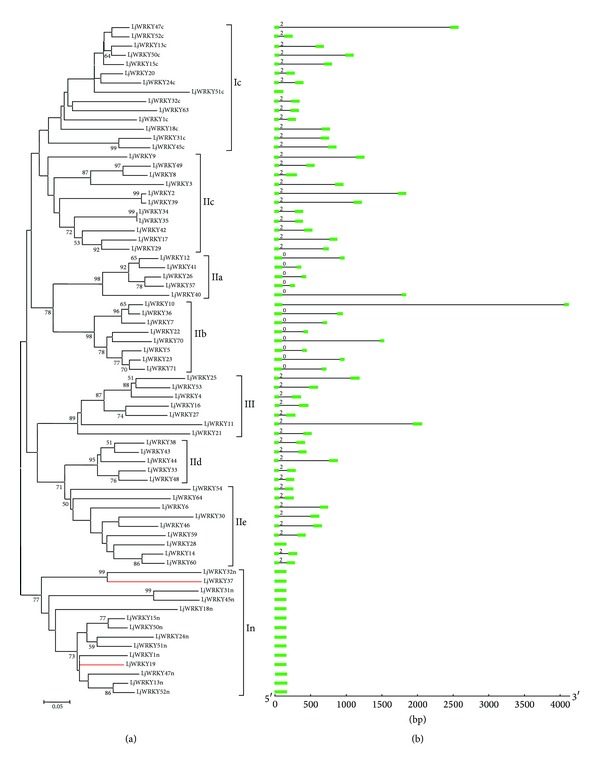
Phylogenetic relationships and gene structure of Lotus japonicus WRKY domains. (a) Multiple alignments of WRKY domain amino acids executed by Clustal W and the phylogenetic tree constructed using MEGA 4.0 by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. The red line indicated group IIx member. The percentage bootstrap scores higher than 50% were indicated on the nodes. (b) Exon-intron structures of WRKY domain genes from Lotus japonicus. Exons and introns were represented by green boxes and black lines, respectively. The number indicated introns phases. The sizes of exons and introns can be estimated using the scale at the bottom.
Clade Ic contained 14 members in L. japonicus, including 12 members with WRKY domains at the C-terminal region and two group II members (LjWRKY20 and LjWRKY63) (Figure 4(a)). These two group II members were clustered with LjWRKY24c and LjWRKY32c, respectively, indicating a common origin of their domains. Moreover, two group II members (LjWRKY19 and LjWRKY37) were also found in the In clade.
Group II can be divided into five clades with high bootstrap values (Figure 4(a)). Further analysis revealed that IIa and IIb form a single clade and IId and IIe form a clade, while IIc contained 11 LjWRKY members form a clade (Figure 4(a)). This suggests that the domains had a recent gene ancestor or formed under similar selective pressures. Clade III contained seven members which are more similar to clade IId and clade IIe than any other members (Figure 4(a)), suggesting that they may have shared an ancestor before divergence of group II and group III.
During analysis of the cDNA and DNA sequences, we found that most of the LjWRKY genes contained two types of introns in their WRKY domains. The phase-2 intron is spliced exactly after the R position, similar to the splicing position observed in Arabidopsis [2]. We designate the phase-2 intron as an R-type intron. A phase-0 intron is located before the V position, at the sixth amino acid after the second C residue in the C2H2 zinc finger motif [22]. We designate the phase-0 intron as a V-type intron. Interestingly, in subgroups Ic, IIc, IId, IIe, and III, the R-type intron is located before the zinc finger motif region in the WRKY domain of genes, while in subgroups IIa and IIb, the V-type intron is found within the zinc finger motif region in the WRKY domain (Figure 4(b)). Furthermore, introns have been lost from LjWRKY28 (subgroup IIe), LjWRKY51c (subgroup Ic), and subgroups In and IIx (Figure 4(b)). Intron loss can be considered as the result of intron turnover, the result of homologous recombination between an intron-containing allele and a mature mRNA [22].
3.4. Conserved Motifs in LjWRKY Proteins
With the exception of the conserved 60 amino acid residues, no functional or structural homologies were previously known from the remainder of the WRKY protein sequences [2]. Analysis of the 20 motifs revealed that LjWRKY motif lengths ranged from 11 to 113 and the distribution of 20 motifs in each amino acid sequence varied greatly (Table 3 and Figure 5). In addition, the function of the majority of LjWRKY motifs could not be predicted. Unexpectedly, we observed a herpes virus glycoprotein motif (motif14) in LjWRKY34 and LjWRKY35, which has not been reported in previous studies of WRKY genes. It will be interesting to analyze the function of this motif in LjWRKY genes in the future.
Table 3.
Consensus sequences of WRKY motif in Lotus japonicus proteins.
| Motif | Width | Consensus sequencesa |
|---|---|---|
| Motif1 | 29 | ILDDGYRWRKYGQKVIKGNPYPRSYYRCT |
| Motif2 | 29 | PGCPVKKHVERSAEDPSMVITTYEGEHNH |
| Motif3 | 21 | WRKYGQKVVKGSPFPRSYYKC |
| Motif4 | 21 | RSLDGHVTEITYKGxHNCPKP |
| Motif5 | 15 | VREPRVAVQTKSEVD |
| Motif6 | 28 | LVEAMAAALTADPNFTAALAAAISSIIG |
| Motif7 | 39 | EELERLNAENKKLREMLDQMNENYNALQMHLVELMQKQK |
| Motif8 | 26 | PAAMAMASTTSAAASMLLSGSMPSAD |
| Motif9 | 11 | HPNCPAKKKVE |
| Motif10 | 113 | QNLQVQNFQNQNVQQAAPSFSTTPSFNAATPVPPISYRNAIIPLPFNVANPAPNTH FNTSSFGSFLQGINGVDFVQSSRFMANNNQGLLRNNGLLQDMFVPAHMEFGGGGGRR |
| Motif11 | 24 | CSASMATLSASAPFPTITLDLTQS |
| Motif12 | 42 | DITEIISAPTSVTNSPILDLDLLDNVELDSNFPFNTPELFS |
| Motif13 | 15 | KKRKMRVKRTVRVPA |
| Motif14 | 91 | MPPPPPSPPLPPLSPLPLPPFHQQTSSFGSFKDLLTIEDFDPALFD WNPTTTTNTAAAADVTSPPLFTTQISHPVPSPATSNILPEDSFDV |
| Motif15 | 27 | SGGGELDDDEPDAKRWKGEGENDGYSA |
| Motif16 | 15 | LLLNRTGHARFRRAP |
| Motif17 | 26 | MEHLHKWEQKALINELIQGMELARKL |
| Motif18 | 14 | MDDDWDLHAVVRGC |
| Motif19 | 19 | MSVMPEFLQQGLNSSPVNV |
| Motif20 | 39 | QCSGDIEGELTGEAMELGGEKAIESARALLSIGFEIKPC |
aConsensus amino acid sequence was derived from MEME program.
Figure 5.
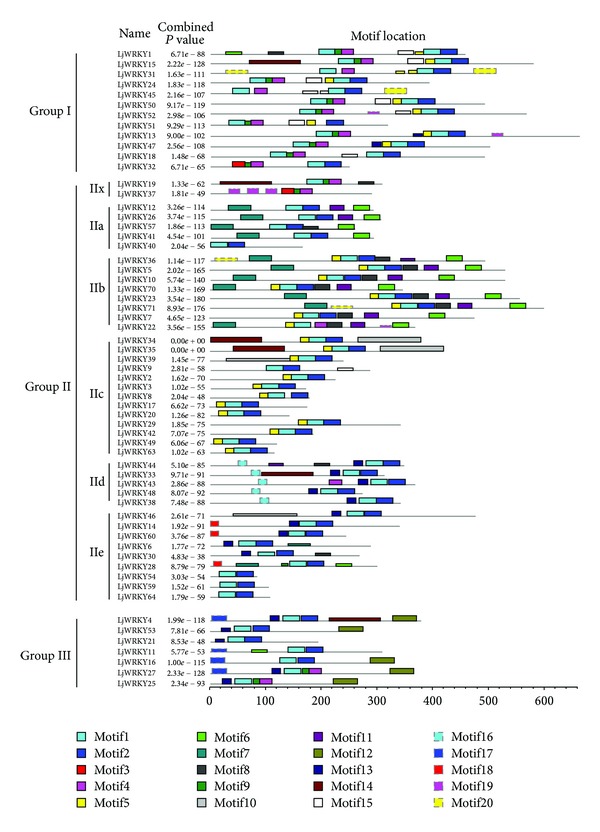
Distribution of 20 predicted conserved motifs in Lotus japonicus WRKY proteins. Motifs of Lotus japonicus WRKY proteins were identified by MEME program. The conserved amino acid sequences and length of each motif are shown in Table 3.
Compared with motifs of the WRKY protein from Arabidopsis [2], Populus trichocarpa [61], and Oryza sativa [22], we detected three conserved motifs in LjWRKY genes, including Leu zipper, HARF, and NLS motifs. Subgroup IIa (LjWRKY12, LjWRKY26, LjWRKY41, and LjWRKY51), IIb (LjWRKY5, LjWRKY10, LjWRKY22, LjWRKY23, LjWRKY36, LjWRKY70, and LjWRKY71), and IIe (LjWRKY28) genes contained a Leu zipper motif (motif7). This motif is a hypothetical structure common to a new class of DNA binding proteins. A HARF motif (motif16) was distributed in subgroup IId (LjWRKY33, LjWRKY38, LjWRKY43, LjWRKY44, and LjWRKY48) genes and an NLS motif (motif13) was observed in group I, subgroup IId, subgroup IIe, and group III (LjWRKY13, LjWRKY47, LjWRKY33, LjWRKY38, LjWRKY43, LjWRKY44, LjWRKY48, LjWRKY6, LjWRKY14, LjWRKY30, LjWRKY46, LjWRKY60, LjWRKY4, LjWRKY21, LjWRKY25, LjWRKY27, and LjWRKY53) genes, but their functions are not clear. Previous studies have shown that WRKY proteins contain a coactivator motif (LXXLL or LXLXLX. L, leucine; X, any amino acid), suggesting the role of these motifs in plant immune responses [22, 61]. In this study, we found a probable coactivator motif in group III genes (LjWRKY4, LjWRKY16, LjWRKY25, LjWRKY27, and LjWRKY53), suggesting the involvement of group III genes in response to pathogens.
3.5. Evolutionary Analysis of Group III Genes in Plants to Determine Selection Pressure in L. japonicus and M. truncatula
In order to study the phylogenetic relationships of group III genes, a phylogenetic tree was estimated using the WRKY domain of seven species, including monocots and dicots. Group III LjWRKY genes did not form a clade; on the contrary, group III LjWRKY genes formed a clade with MtWRKY genes (Figure 6). This suggests that we did not find any paralogs of group III LjWRKY genes, but it suggests that group III LjWRKY genes are orthologous to MtWRKY genes. Paralogous relationships were observed among WRKY genes in other species (i.e., MtWRKY, AtWRKY, PtWRKY, OsWRKY, and BdWRKY). Gene duplication events are considered as the most likely process to result in paralogous copies of genes [59].
Figure 6.
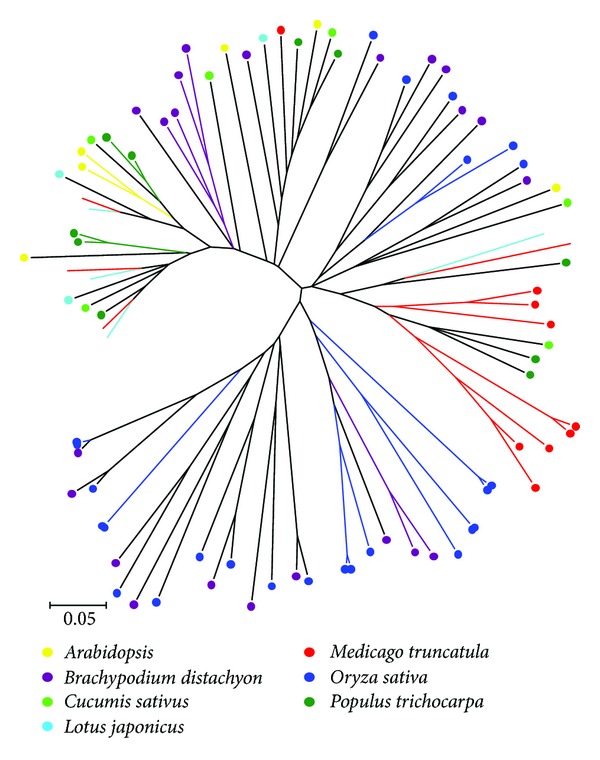
Phylogenetic tree of group III WRKY domains from Arabidopsis, Brachypodium distachyon, Cucumis sativus, Lotus japonicus, Medicago truncatula, Oryza sativa, and Populus trichocarpa. The phylogenetic tree was constructed using MEGA 4.0 by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. Each WRKY predicted orthologous gene was indicated in a specific color.
To detect whether selection pressure affected group III LjWRKY genes, ω (d N/d S) was calculated for phylogenetic nodes in PAML (Table 4 and Figure 7). In L. japonicus and M. truncatula, the ML estimations of ω values for all nodes under the model M0 were <1 (Table 4), suggesting that group III LjWRKY and MtWRKY genes have been under purifying selection during evolution. Nevertheless, the log likelihood ratio differences between models M3 and M0 were statistically significant for all nodes tested, except nodes 1 and 2 in MtWRKY (Table 4). This indicates that some genes may be under positive selection. Interestingly, we further analyzed the positive selection in group III genes with models M8 and M7. The ω values for all nodes were ≥1 under M8. However, only one node identified one positive selection site in group III MtWRKY genes under model M8 (Table 4). This result shows that group III WRKY genes in L. japonicus and M. truncatula have not undergone positive selection.
Table 4.
Likelihood ratio test results of group III Lotus japonicus WEKY and Medicago truncatula WRKY genes.
| Nodea | d N/d S under M0b | 2ΔlnL M3 versus M0 | 2ΔlnL M8 versus M7 | M8 estimatesc | Number of positive selection sited |
|---|---|---|---|---|---|
| Group III LjWRKY | |||||
| 1 | 0.29349 | 67.57** | 0.000074 | ω = 1 (P = 0.72, q = 1.2) | 0 |
| 2 | 0.29349 | 67.57** | 0.000074 | ω = 1 (P = 0.72, q = 1.22) | 0 |
| 3 | 0.25749 | 74.47** | 0.095776 | ω = 1.13 (P = 0.84, q = 2) | 0 |
| 4 | 0.23855 | 112.17** | 0.000198 | ω = 1 (P = 0.8, q = 1.8) | 0 |
|
| |||||
| Group III MtWRKY | |||||
| 1 | 0.31282 | 3.45 | 0.000016 | ω = 1 (P = 0.77, q = 1.44) | 0 |
| 2 | 0.28717 | 4.31 | 0.000042 | ω = 1 (P = 1.9, q = 4.18) | 0 |
| 3 | 0.27266 | 27.39** | 0.178716 | ω = 2.19 (P = 0.52, q = 0.93) | 1 |
| 4 | 0.26004 | 70.18** | 0.000078 | ω = 1 (P = 0.76, q = 1.51) | 0 |
| 5 | 0.24433 | 81.88** | 0.000204 | ω = 1 (P = 0.74, q = 1.48) | 0 |
| 6 | 0.00517 | 43.43** | 0.029352 | ω = 34.48 (P = 1.09, q = 61.42) | 0 |
| 7 | 0.20188 | 209.83** | 0.000302 | ω = 1 (P = 0.64, q = 1.68) | 0 |
| 8 | 0.18564 | 233.88** | 0.000154 | ω = 2.5 (P = 0.6, q = 1.8) | 0 |
*P < 0.05 and **P < 0.01 (χ 2 test).
aNode number from the phylogenetic tree.
b d N/d S is the average ratio over sites under a codon model with one ratio.
c ω was estimated under model M8; P and q are the parameters of the beta distribution.
dThe number of amino acid sites estimated to have undergone positive selection under M8.
Figure 7.
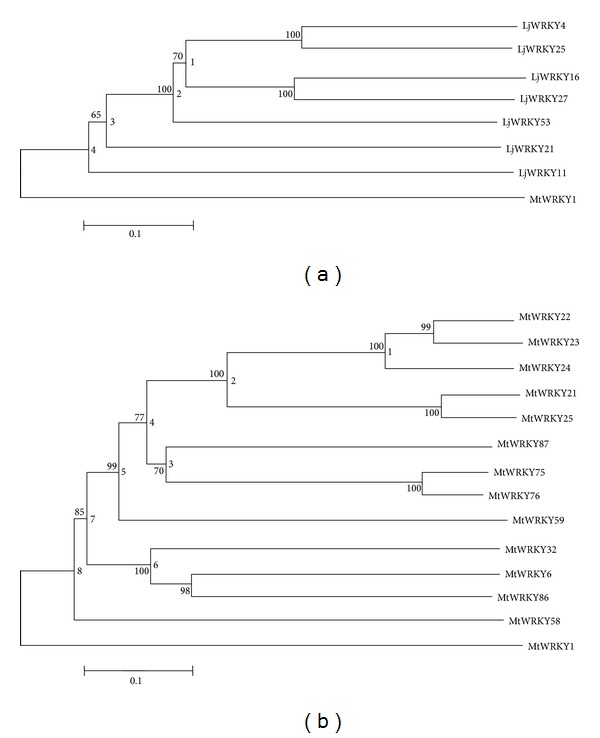
Phylogenetic tree of group III WRKY genes of Lotus japonicus and Medicago truncatula. The phylogenetic trees were constructed using MEGA 4.0 by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. Numbers on the left of each internal node represented bootstrap support values and numbers on the right of each node represented the nodes that were used for positive selection analysis. MtWRKY1 gene was used as outgroup. The tree represented phylogenetic relationships among LjWRKY genes (a) and MtWRKY genes (b).
4. Discussion
WRKY genes are commonly found in land plants and many WRKY genes have been identified and classified in Arabidopsis [2], Oryza sativa [6, 22, 62], Hordeum vulgare [63], Cucumis sativus [59], Brachypodium distachyon [64], and Populus trichocarpa [61]. However, little information has been reported on WRKY genes in leguminous forage crops. In 2008, approximately 67% of the L. japonicus genome (472 Mb) sequences were available on public databases, representing 91.3% coverage of the gene space [49]. In our current work, we conducted an analysis of 61 WRKY genes in the L. japonicus genome.
One hundred and four WRKY genes were identified in P. trichocarpa, while only 55 WRKY genes were discovered in C. sativus and Vitis vinifera genomes, respectively (Table 2). The relative number of WRKY genes was not associated with genome size. For example, the number of WRKY genes was about 2x greater in P. trichocarpa (104) than in C. sativus (55), while these two plants have an approximately equal genome size (458 Mb and 487 Mb; Table 2). By analyzing the number of WRKY gene groups or subgroups (except for subgroup IIx) in each species where they have been characterized, we discovered an uneven distribution of the number of WRKY genes in each group. In rice, the largest number of WRKY genes (36) was found in group III, but only four genes were found to belong to subgroup IIa (Table 2). In C. sativus, however, more WRKY genes (16) are classified as subgroup IIc and fewer WRKY genes (4) are categorized in subgroup IIa (Table 2).
Although gene duplication events seem to have led to the expansion of WRKY genes in the Arabidopsis [2] and Oryza [22] genomes, duplicated WRKY genes were not detected in C. sativus [59]. It is not yet clear whether gene duplication typically occurs during LjWRKY gene evolution. Among the 61 WRKY genes, we found that, in L. japonicus, only 2 of them were involved in duplication events. In contrast, 11 gene duplication events were identified in the model plant M. truncatula. In addition, there were 42 PtWRKY gene duplication events identified in the P. trichocarpa genome [61], with 29 out of 42 PtWRKY genes arising from segmental duplication. This comparison suggests that the expansion of LjWRKY genes is not necessarily dependent on gene duplication events.
We found few duplicated LjWRKY genes in the L. japonicus genome and there are at least three possible explanations: (1) most duplicated genes have been lost after segmental duplication events in LjWRKY genes [65]; (2) LjWRKY genes that are nonfunctional or duplicate the function of other copies are inclined to disappear to avoid fitness cost; and (3) the draft sequenced genome in L. japonicus may not yet contain all WRKY genes present in the genome.
The WRKY gene sequences contain two types of conserved introns (R-type and V-type) [22], and we found them in this study. In Arabidopsis [2] and C. sativus [59] subgroup IIa and IIb WRKY genes, R-type introns are inserted in domains located at the fourth amino acid (K residue) after the second C residue in the zinc finger region and there are no V-type introns. These results suggest that introns in subgroup IIa and IIb WRKY genes may have different origins.
The average length of the conserved introns (597 bp) in L. japonicus WRKY domains was longer than that in Arabidopsis (241 bp) but shorter than that in M. truncatula (705 bp). In nematodes and mammals, there is a dramatic decline in average intron size when there is increased gene expression [66]. However, the correlation between expression of WRKY genes in various plants and the intron length needs to be tested.
We found an interesting phenomenon which may provide context to interpret group I, II, and III WRKY gene origin and evolutionary relationships. Two conserved motifs (motif4 and/or motif9) were observed after motif1 (contains the conserved sequence WRKYGQK) in the N-terminal region, while, in the C-terminal region, other conserved motifs (motif5 or/and motif13) occur before motif1 (Figure 5). In addition, analysis of group II and III LjWRKY gene motifs revealed that most LjWRKY genes contain motif1, motif5, and/or motif13. However, motif1, motif4, and/or motif9 in the N-terminal region of group I genes are distributed in LjWRKY19, LjWRKY37, (group II) and LjWRKY25 and LjWRKY27 (group III), respectively (Figure 5). Previous research showed that group II and III WRKY genes evolved from group I through the loss of the N-terminal WRKY domain [2, 5]. However, our results indicate that some WRKY genes in groups II and III may have originated from the N-terminal region of group I. From the phylogenetic relationship, we found that LjWRK19 and LjWRKY37 (group II) clustered with group In with well-supported bootstrap values, suggesting that they have a common origin or ancestry. Moreover, we consider that some group III WRKY genes could have resulted from a mutation in the zinc finger motif in the N-terminal of group I genes. To confirm our inference, we analyzed the conserved motifs in MtWRKY genes. The same phenomenon was detected in MtWRKY genes based on analysis of location of motifs (data not shown) and 13 MtWRKY genes might have evolved from N-terminal of group I genes.
The natural selection pressure imposed by pathogens is expected to be diverse in different plants [67] and WRKY genes and NBS-LRR genes forming a fused gene may effectively resist a wide variety of pathogens. Fusion genes contain the C-terminal WRKY motif and a NBS-LRR (nucleotide-binding site-leucine-rich repeat) motif in the R gene was identified in AtWRKY [68, 69] and OsWRKY [22] genes. The R gene is mainly involved in a pathogen response pathway in plants [70]. Fusion gene, one gene included WRKY and NBS-LRR domains and/or motifs, was not detected in L. japonicus.
The majority of group III AtWRKY genes under positive selection are expressed in response to various abiotic stresses [59]. In contrast, the expression of group III CsWRKY genes shows that these genes are under purifying selection and are specialized to respond as single type of stress [59]. Ling et al. showed that positive selection may have resulted in the functional divergence of duplicated genes during the expansion of group III WRKY genes in Arabidopsis [59]. Similarly, our selection pressure study of group III LjWRKY and MtWRKY genes found that expansion of these genes may be under purifying selection, although gene duplication events occurred within these genes. Purifying selection may generate genes with conserved functions or pseudogenization [71] in duplicated group III MtWRKY genes. Therefore, we speculate that group III LjWRKY and MtWRKY genes may be more conservative in their response to stress.
Acknowledgments
This research was supported by grants from the National Basic Research Program of China (2014CB138702). The authors are grateful to the members of the State Key Laboratory of Grassland Agro-ecosystems for their assistance in this study. They thank associate editor Professor John Parkinson and an anonymous reviewer for taking extensive time and care for providing thoughtful comments on the manuscript.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Molecular Biology. 2002;50(6):925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 2.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5(5):199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 3.Liu J-J, Ekramoddoullah AKM. Identification and characterization of the WRKY transcription factor family in Pinus monticola . Genome. 2009;52(1):77–88. doi: 10.1139/G08-106. [DOI] [PubMed] [Google Scholar]
- 4.Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO Journal. 1996;15(20):5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology. 2005;5:p. 1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Z, Zhang Z-L, Zou X, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiology. 2005;137(1):176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Molecular Biology. 2008;68(1-2):81–92. doi: 10.1007/s11103-008-9353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO Journal. 1999;18(17):4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15(9):2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Pater S, Greco V, Pham K, Memelink J, Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis . Nucleic Acids Research. 1996;24(23):4624–4631. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiguro S, Nakamura K. Characterization of a cDNA encodine a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Molecular and General Genetics. 1994;244(6):563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 12.Goff SA, Ricke D, Lan T-H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296(5565):92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 13.Huang T, Duman JG. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara . Plant Molecular Biology. 2002;48(4):339–350. doi: 10.1023/a:1014062714786. [DOI] [PubMed] [Google Scholar]
- 14.Kato N, Dubouzet E, Kokabu Y, et al. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica . Plant and Cell Physiology. 2007;48(1):8–18. doi: 10.1093/pcp/pcl041. [DOI] [PubMed] [Google Scholar]
- 15.Kim D-J, Smith SM, Leaver CJ. A cDNA encoding a putative SPF1-type DNA-binding protein from cucumber. Gene. 1997;185(2):265–269. doi: 10.1016/s0378-1119(96)00665-8. [DOI] [PubMed] [Google Scholar]
- 16.Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytologist. 2009;184(1):48–70. doi: 10.1111/j.1469-8137.2009.02955.x. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16(2):319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantri NL, Ford R, Coram TE, Pang ECK. Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics. 2007;8, article 303 doi: 10.1186/1471-2164-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchive C, Mzid R, Deluc L, et al. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. Journal of Experimental Botany. 2007;58(8):1999–2010. doi: 10.1093/jxb/erm062. [DOI] [PubMed] [Google Scholar]
- 20.Pnueli L, Hallak-Herr E, Rozenberg M, et al. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam . Plant Journal. 2002;31(3):319–330. doi: 10.1046/j.1365-313x.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 21.Ülker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology. 2004;7(5):491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Wu K-L, Guo Z-J, Wang H-H, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research. 2005;12(1):9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- 23.Young ND, Debellé F, Oldroyd GED, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q-Y, Tian A-G, Zou H-F, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnology Journal. 2008;6(5):486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 25.Glöckner G, Eichinger L, Szafranski K, et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum . Nature. 2002;418(6893):79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 26.Ramamoorthy R, Jiang S-Y, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology. 2008;49(6):865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- 27.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Asai T, Tena G, Plotnikova J, et al. Map kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 29.Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13(7):1527–1539. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Z-J, Kan Y-C, Chen X-J, Li D-B, Wang D-W. Characterization of a rice WRKY gene whose expression is induced upon pathogen attack and mechanical wounding. Acta Botanica Sinica. 2004;46(8):955–964. [Google Scholar]
- 31.Chen C, Chen Z. Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Molecular Biology. 2000;42(2):387–396. doi: 10.1023/a:1006399311615. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Schiff M, Dinesh-Kumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant Journal. 2004;38(5):800–809. doi: 10.1111/j.1365-313X.2004.02085.x. [DOI] [PubMed] [Google Scholar]
- 33.Cormack RS, Eulgem T, Rushton PJ, Köchner P, Hahlbrock K, Somssich IE. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochimica et Biophysica Acta. 2002;1576(1-2):92–100. doi: 10.1016/s0167-4781(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 34.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology. 2007;10(4):366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Fu Q, Huang W, Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Reports. 2009;28(4):683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 36.Vandenabeele S, van der Kelen K, Dat J, et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izaguirre MM, Scopel AL, Baldwin IT, Ballaré CL. Convergent responses to stress. Solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora . Plant Physiology. 2003;132(4):1755–1767. doi: 10.1104/pp.103.024323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skibbe M, Qu N, Galis I, Baldwin IT. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell. 2008;20(7):1984–2000. doi: 10.1105/tpc.108.058594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grunewald W, Karimi M, Wieczorek K, et al. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiology. 2008;148(1):358–368. doi: 10.1104/pp.108.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yong HC, Chang H-S, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis . Plant Physiology. 2002;129(2):661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America. 2005;102(48):17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zentella R, Zhang Z-L, Park M, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis . Plant Cell. 2007;19(10):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z-L, Xie Z, Zou X, Casaretto J, Ho T-HD, Shen QJ. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiology. 2004;134(4):1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou X, Neuman D, Shen QJ. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiology. 2008;148(1):176–186. doi: 10.1104/pp.108.123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001;213(3):469–473. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- 46.Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes and Development. 2002;16(9):1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson CS, Kolevski B, Smyth DR. Transparent Testa Glabra2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14(6):1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiology. 2002;129(2):706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato S, Nakamura Y, Kaneko T, et al. Genome structure of the legume, Lotus japonicus . DNA Research. 2008;15(4):227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn RD, Mistry J, Schuster-Böckler B, et al. Pfam: clans, web tools and services. Nucleic acids research. 2006;34:247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou T, Wang Y, Chen J-Q, et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Molecular Genetics and Genomics. 2004;271(4):402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Gu S, Wang X, Li W, Tang Z, Xu C. Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. Journal of Molecular Evolution. 2008;67(3):266–277. doi: 10.1007/s00239-008-9143-z. [DOI] [PubMed] [Google Scholar]
- 55.Guo A-Y, Zhu Q-H, Chen X, Luo J-C. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–1026. [PubMed] [Google Scholar]
- 56.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proceedings of the 3rd International Conference on Intelligent Systems for Molecular Biology; 1995; pp. 21–29. [PubMed] [Google Scholar]
- 57.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 59.Ling J, Jiang W, Zhang Y, et al. Genome-wide analysis of WRKY gene family in Cucumis sativus . BMC Genomics. 2011;12, article 471 doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Houb EB. The arms race is ancient history in Arabidopsis, the wildflower. Nature Reviews Genetics. 2001;2(7):516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 61.He H, Dong Q, Shao Y, et al. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa . Plant Cell Reports. 2012;31(7):1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- 62.Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2007;49(6):827–842. [Google Scholar]
- 63.Mangelsen E, Kilian J, Berendzen KW, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics. 2008;9, article 194 doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi P, Rabara RC, Langum TJ, et al. The WRKY transcription factor family in Brachypodium distachyon . BMC Genomics. 2012;13, article 270 doi: 10.1186/1471-2164-13-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165(3):937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- 66.Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nature Genetics. 2002;31(4):415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- 67.Li J, Ding J, Zhang W, et al. Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Molecular Genetics and Genomics. 2010;283(5):427–438. doi: 10.1007/s00438-010-0527-6. [DOI] [PubMed] [Google Scholar]
- 68.Deslandes L, Olivier J, Peeters N, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deslandes L, Olivier J, Theulières F, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(4):2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J. Evolution by gene duplication: an update. Trends in Ecology and Evolution. 2003;18(6):292–298. [Google Scholar]


