Abstract
Genome-wide association studies provide an unbiased approach in the identification of genes that increase the risk for osteochondritis dissecans (OCD). OCD is a disorder of the bone and cartilage that affects humans, horses, pigs, dogs, and other mammals. Recent genome-wide association studies in humans, horses, and pigs are reviewed and genes identified. The identified genes tended to cluster with respect to function and biological processes such as the protein secretion pathway, extracellular matrix molecules, and growth plate maturation. Genome-wide association studies in humans are a critical next step in the effort to provide a better understanding of the causes of OCD, which will, in turn, allow preventive strategies for treatment of adolescent and young adults who are at risk for the development of degenerative joint disease due to the effects of OCD.
Keywords: Osteochondritis dissecans, Osteochondrosis, Genome-wide association study, Osteoarthritis, Single Nucleotide Polymorphism, Human, Equine, Swine
INTRODUCTION
Osteochondritis dissecans (OCD) is a focal idiopathic alteration of subchondral bone with risk for instability and disruption of adjacent articular cartilage that may result in premature osteoarthritis (1). This condition is commonly found in horses, pigs, dogs and humans (2). König first used the term OCD to describe a condition that causes the formation of loose bodies in the joints of young individuals without arthritis or trauma (3). Although over 100 years have passed since König described OCD, little knowledge exists of the specific etiology and pathogenesis of this disease. Many etiological theories have been proposed for the onset and progression of OCD, including: trauma, diet, rapid growth, anatomic characteristics, lack of blood supply, necrosis of subchondral bone and heredity (recently reviewed in 4–8). Recent experiments have provided additional information regarding the underlying genetic traits that may predispose an individual to OCD.
König’s original description included inflammation as a contributor to the formation of OCD lesions and ‘joint mice’. However, recent research has suggested that inflammation is not involved in the etiology of OCD, and thus should be referred to as osteochondrosis (4, 8, 9). In the clinical and research settings, osteochondrosis represents a larger range of disorders than the original condition described by König (8). While the term OCD is used in human disease, much of the animal literature uses the term osteochondrosis to describe what may actually be the same condition.
In humans, OCD can affect both juveniles and adults (10), although onset is typically in skeletally immature patients. If untreated, OCD can lead to osteoarthritis (OA) as the age of the individual progresses (11, 12, 13). While both surgical and nonsurgical treatments are available for patients with OCD (14), a better understanding of the pathogenesis of OCD and the underlying genetics may provide opportunities for better diagnosis and additional treatment options (14).
Osteochondrosis has been investigated primarily in horses and pigs. Osteochondrosis is the leading cause of lameness and decreased performance in young athletic horses (15). Osteochondrosis is also considered the leading cause of leg weakness in pigs (16, 17). The most extensive analysis of genetic association has been completed for the disease in horses and pigs, to date.
Much of the previous research on the pathogenesis of OCD in humans has been through histological analysis of bone samples. A recent review by Shea et al. (4) found an overall lack of histological OCD research, with varying methodologies and findings among the studies reviewed. One issue inherent in histological analysis of tissue samples is that only a limited number of different proteins or markers can be tested and samples may at best represent end-stage or advanced stages of the disease. Tissue samples are only available during certain surgical procedures, and thus it can be difficult to obtain large or multiple sample sizes. Furthermore, biopsy specimens are unlikely to represent early stages of OCD.
Previous studies have analyzed candidate genes and their association with the onset of OCD (18, 19, 20) however, these efforts have been limited. With improved techniques and decreasing costs for genetic studies, the opportunity for genome-wide association studies (GWAS) and single-nucleotide polymorphism (SNP) studies has become available. The purpose of this review is two-fold: first, to review the most recent literature on GWAS and SNP studies on OCD and osteochondrosis, and second, to identify patterns within the genes of interest to serve as a guide for further investigation into the etiology of OCD.
In a GWAS study subjects are evaluated based on physical traits and then genotyped. The traits are compared to the gene sequences by a computer program, which calculates the correlation. Markers are designed and used to split the DNA at intervals that span the chromosomes. The markers are designed to identify single nucleotide polymorphisms (SNPs) where the genetic code tends to vary by a single base pair across individuals. Based on the locations of SNPs with high statistical significance (call rate), nearby genes can be identified as possible genes of interest for the selected trait. The gene regions with possible linkage to the trait are referenced as quantitative trait loci (QTL).
METHODS
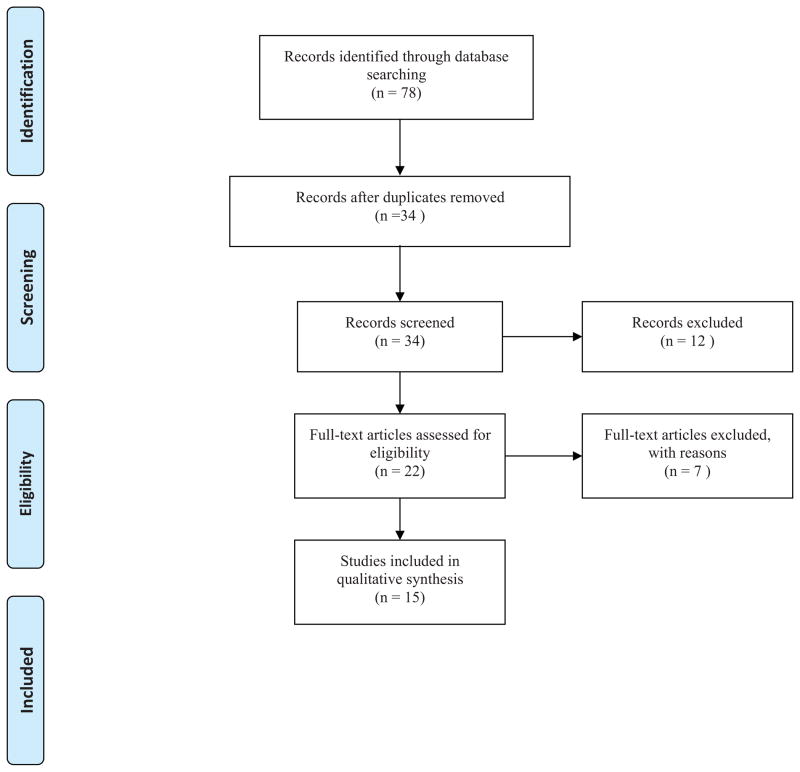
A Medline search was performed in March of 2013 to identify relevant genetic studies of OCD in all animals and humans (Figure 1). Searches were performed for both ‘osteochondritis dissecans’ and ‘osteochondrosis’ due to the historical interchange of terms with the following key words: ‘osteochondritis dissecans AND single nucleotide polymorphism’, ‘osteochondrosis AND single nucleotide polymorphism’, ‘osteochondritis dissecans AND genome-wide association study’, ‘osteochondrosis AND genome-wide association study’, ‘osteochondritis dissecans AND genome’, ‘osteochondrosis AND genome’, ‘osteochondritis dissecans AND cell signaling’, ‘osteochondrosis AND cell signaling’, ‘osteochondritis dissecans AND quantitative trait loci’, and ‘osteochondrosis AND quantitative trait loci’. All searches were completed with an English language filter.
Figure 1. PRISMA Flow Diagram.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines were used to select journal articles for inclusion and review (103).
Duplicate articles were removed and abstracts were screened according to the following inclusion criteria: experimental research performing GWAS, studies of SNP, or refinements to previously completed articles. Articles that did not meet inclusion criteria were heritability reports, cell culture or tissue studies of genes or mRNA expression. Studies investigating SNPs within selected genes were also discarded due to the high degree of specificity of the SNP location.
Twenty-two articles were identified after screening. Of those, seven were excluded, which resulted in the inclusion of 15 in the current review. Of those 15 papers, one studied human OCD with a genome-wide linkage analysis, 12 studies were in equine, and two were genetic studies in swine. All articles were published between the years 2000 and 2013. A description of OCD or osteochondrosis, experimental methods, and results are found below organized by the species.
HUMAN
OCD in humans is primarily found in the knee, elbow, or ankle of adolescents. The subchondral bone is affected leading to disruption of articular cartilage and possible formation of osteochondral fragments. Familial cases of OCD were recently characterized by osteochondritic lesions in different joints, short statue, and early osteoarthritis (OA) due to mutations in the aggrecan gene (19).
Stattin et al. (19) studied 53 members of five generations of a Swedish family with inherited OCD, 15 of whom were diagnosed with OCD by MRI or X-ray. Blood samples were taken from the 53 subjects and genomic DNA was extracted from white blood cells. Genotyping was completed on 38 of the individuals using 400 microsatellite markers, which spanned all autosomes with an average spacing of 10 cM. Linkage was then analyzed using MCLINK from Myriad Genetics. A significant polymorphic marker was found on chromosome 15, revealing aggrecan (ACAN) to be a gene of interest. Further work was completed to test the mutated ACAN gene in vivo.
EQUINE
Osteochondrosis normally affects young horses at 5–8 months of age (21). Osteochondrosis in horses is defined as a disturbance of endochondral ossification at the epiphyseal growth plate. Disturbance during differentiation, development, and vascular invasion of the growth plate leads to alterations of the joint, formation of cartilage flaps, or osseous fragments (22). Osteochondrosis is primarily diagnosed via radiography, and is usually not identified until the formation of the loose bony or osteochondral fragments. Much of the recent literature identifies a disease stage where bony fragments are present, referred to as osteochondrosis dissecans (8). Teyssedre et al. (23) referred to osteochondrosis as a generic term for OCD and bone cysts. While osteochondrosis is present in many different joints in the horse, most research has focused on the tarsocrural (hock) joint due to the high incidence and defined diagnosis methods (24). Evidence for heritability has been found at frequencies between 0.2 and 0.64, depending on the location of the osteochondrosis lesion and the breed of the horse (22). Osteochondrosis is economically important and affects animal welfare in a significant manner (25).
After review, 12 articles were deemed relevant (2, 21, 23–32). Four of the included articles (28–31) were refinements to the work done in Dierks et al. (27). Two refinement studies were also completed by Wittwer and associates (25, 32) on the work of Wittwer et al. (21). In total, six studies completed GWAS to identify candidate genes responsible for osteochondrosis, and six studies were conducted on specific gene regions for fine mapping of previous work. In all studies, horses were diagnosed with osteochondrosis via radiography and scored for severity.
Corbin et al. (26) had 162 case horses with osteochondrosis and 168 control subjects. Dierks studies (27–32) used 76 horses with osteochondrosis and 28 unaffected horses for control. Orr et al. (24) used 90 osteochondrotic horses with 111 controls. In the research performed by Lykkjen et al. (2) 80 horses with osteochondrosis were compared to 82 control horses. The largest sample size was used by Teyssedre et al. (23) with 262 affected horses and 263 healthy horses for control. The initial work done by Wittwer et al. (21) had 96 osteochondrotic horses and 18 controls. Thirty-two affected and 64 healthy horses were included in studies by Wittwer et al. (32), while 96 horses were used in Wittwer et al. (25). Corbin et al. (26) tested Thoroughbred horses in their study. Hanoverian Warmblood horses were used in five of the studies (27–31). South German Coldblood horses were used in three studies (21, 25, 32). Dutch Warmblood horses, French trotter horses, and Norwegian Standardbred trotters were studied (2, 23, 24).
Genotyping was accomplished in four of the articles by using the Illumina Equine SNP50 BeadChip with 54,602 SNPs (2, 23, 24, 26). Each of the articles applied quality control measures, decreasing the amount of tested SNPs to 40,180 (26), 45,586 with an average call rate of 99.8% (24), 41,170 with a minimum call rate of 95% (2), and 41,249 with a minimum call rate of 98% (23). Initial genotyping was completed with 172 highly polymorphic microsatellites by Dierks et al. (27) with 88 additional microsatellites added for putative QTLs established in the first scan for a total of 260 microsatellite markers. Refinement studies added 49, 34, 11, and 37 new microsatellites on chromosomes 5, 16, 18, and 2 respectively (29–31).
Statistical analysis was completed using MERLIN (multipoint engine for rapid likelihood inference) software by Dierks (27–30) and Wittwer (21, 25, 32). Lykkjen et al. (2) used the whole-genome association analysis toolset PLINK for statistical calculations. Orr et al. (24) used GENABEL and HAPLOSTATS for their analysis work. REMPLF90 software was utilized by Teyssedre et al. (23) for statistical analysis calculations.
Corbin et al. (26) identified UDP-glucose dehydrogenase (UGDH) as a possible related gene to osteochondrosis. Dierks et al. (27) identified connections between SNPs found in their research and human genes. The genes identified were matrilin 1 (MATN1), laminin (LAMB1), solute carrier family 35 (SLC35D1), and parathyroid hormone receptor (PTH1R). Collagen type XXIV alpha 1 (COL24A1) was identified as a functional candidate gene in Lampe et al. (29). The hyaluronoglucosaminidase gene family members were identified as candidates in Lampe et al., (30). Refinement done by Lampe et al. (31) found parathyroid hormone 2 receptor (PTH2R) to be associated with osteochondrosis. The gene neurochondrin (NCDN) was concluded to be the functional candidate gene on ECA2 by Dierks et al. (28). Lykkjen et al. (2) found LOC100073151, chloride channel calcium activated family member 4 (CLCA4), COL24A1, F-box protein 25 (FBXO25), and TBC1 domain family member 22A (TBC1D22A) to be possible genes contributing to osteochondrosis in the horse. Lectin galactoside-binding soluble 3 (LGALS3), frizzled-related protein (FRZB), collagen type III alpha 1 (COL3A1), collagen type V alpha 2 (COL5A2), collagen type V alpha 1 (COL5A2), collagen type XXVII alpha 1 (COL27A1), calcitonin gene-related peptide-receptor component protein (RCP9), calneuron 1 (CALN1), and interleukin 6 (IL6) were also identified as candidate genes (21). Wittwer et al. (32) identified AOAH and PTH1R as other possible genes. Xin actin-binding repeat containing 2 (XIRP2) was shown to be a strong candidate gene.
SWINE
Osteochondrosis in swine is considered to be a generalized skeletal disease characterized by disturbed bone formation, cartilage retention, or necrosis of cartilage canal in articular cartilage (33, 34). Osteochondrosis is the primary cause of leg weakness in swine, impacting the fitness and longevity of animals (17). Previous studies have analyzed the relationship between growth rate, weight, and diet with osteochondrosis (8). Leg weakness is a general term that integrates many different diseases and conditions and may not be limited specifically to osteochondrosis. Heritability of osteochondrosis varies by breed but has been found to range from 0.06 to 0.5 (17).
Laenoi et al. (17) established a genetic link to leg weakness and osteochondrosis. They identified SNPs related to bone mineral density in addition to leg weakness and osteochondrosis, and performed genotyping with 79 microsatellites and three biallelic markers (a higher tolerance marker type) for an average marker interval of 31.57 cM. Additional SNPs in the COL10A1 and MMP3 loci were included, due to the relevance to cartilage quality. Andersson-Ekland (34) focused on phenotypic attributes and osteochondrosis. They used 236 markers across all autosomes. Leg weakness and osteochondrosis were also assessed from histological evaluation.
QTLs related to osteochondrosis were found on chromosomes 5 and 13 at 51 cM and 64 cM respectively by Andersson-Ekland et al. (34). The SNP at chromosome 5 was located between interferon-γ (IFNG) and insulin-like growth factor-1 (IGF1) with homology to human chromosome 12q14-q24 and the gene for cartilage homeoprotein 1 (CART1). Homology also exists in the region of the QTL on chromosome 13 for pituitary-specific positive transcription factor 1 (PIT1) and parathyroid hormone receptor (PTH1R) genes. Laenoi et al. (17) found QTLs on chromosomes 2, 3, 6, 10, and 14. The QTLs were located at 14 cM on chromosome 2, 13 cM on chromosome 3, 61 cM on chromosome 6, 70 cM on chromosome 10, and 0 cM for chromosome 14. Genes located at these identified polymorphisms are yet to be determined.
DISCUSSION
OCD in humans affects the subchondral bone, primarily in the knee, elbow, or ankle leading to disruption of articular cartilage and possible formation of osteochondral fragments (14). Etiology in humans may be related to several other factors, including trauma, ischemia, and disrupted endochondral ossification (4, 12, 35–37). In equine osteochondrosis, endochondral ossification of the epiphyseal growth plate is disrupted, leading to the formation of the loose bony osteochondral fragments. Different joints are affected with the highest incidence in the tarsocrural joint. Evidence supports inherited genetic traits underlying osteochondrosis in equine. Osteochondrosis in swine is characterized by disturbed bone formation, cartilage retention, or necrosis of cartilage canals in calcified cartilage (33, 34). Heritability of osteochondrosis in swine varies by breed; however, the argument for the existence of genetic factors is strong.
Candidate genes cluster into distinct groups
Candidate genes were identified in GWAS and through additional refinement of these studies. Analysis of these genes indicates a high prevalence for those encoding secreted extracellular matrix proteins, and those playing a role in the secretory pathway. Additionally, a cluster of genes regulating the differentiation and maturation of the growth plate was also observed. Notably, extracellular matrix molecules and parathyroid hormone signaling were identified in multiple studies and by multiple candidate genes.
Extracellular matrix proteins
COL24A1 was identified as a candidate gene in the refinement study completed by Lampe et al. (29) and by the GWAS analyzed by Lykkjen et al. (2). Wittwer et al. (21) found COL3A1, COL5A1, COL5A2, COL10A1, and COL27A1 to be significantly associated with incidence of osteochondrosis in horses. Additional extracellular matrix molecules included matrilin 1 (MATN1), laminin (LAMB1), and aggrecan (ACAN). Genes involved in extracellular matrix turnover include hyaluronoglucosaminidase (HYAL) and matrix metalloproteinase 3 (MMP3).
The hyaluronidases comprise a family of lysosomal β-endoglucosidases that are responsible for the turnover of hyaluronan, a ubiquitous ECM glycosaminoglycan, by cleaving internal β-1,4 linkages. Six hyaluronidase genes have been identified in humans (38, 39), clustered at chromosomal locations 3p21.3 (HYAL-1, HYAL-2, and HYAL-3) and at chr 7q31.3 (HYAL-4, HYALP1, and SPAM-1). Cartilage has been shown to express mRNA encoding HYAL-1, HYAL-2 AND HYAL-3 (40). TCF/Lef binding sites have been identified within the promoter regions, indicating a link to the Wnt signaling pathway (41). Tissue remodeling and growth during skeletal development relies on HYAL family member function.
Matrillins (MATNs) are a family of four oligomeric extracellular matrix proteins. Each family member contains a von Willebrand factor A domain, epidermal growth factor-like domains, and coiled–coil domains (42). Mutations in MATNs have been associated with osteochondrodysplasias including multiple epiphyseal dysplasia (43–47) and osteoarthritis (48–51).
Over 13,000 articles are indexed in Medline relating to cartilage and collagen, 44 of which address the connection between collagen and osteochondrosis. An additional 2,500+ studies report a connection between collagen and osteoarthritis. COL24A1 is described as a fibril diameter regulator with specific functions at specific sites for fibrillogenesis, especially for COL1A1 (52). Skagen et al. (53) studied biopsies of osteochondrotic lesions and determined that abnormal collagen fibrils are associated with OCD, with evidence supporting endoplasmic reticulum (ER) stress and an unfolded protein response (UPR) as identified by the extended ER by transmission electron microscopy. Additionally, collagen fibrils in the extracellular matrix appeared abnormal, and evidence of internal cellular accumulation of collagens was evident (53). A mutation in the gene COL24A1 represents a strong candidate for the effect seen on collagen fibril diameter in the cartilage extracellular matrix, since COL24A1 is known to regulate fibril diameter during fibrillogenesis, is expressed by osteoblasts (54) and can drive bone development through the TGF-β signaling pathway (55).
Secreted proteins associated with skeletal dysplasia and/or OCD
Familial cases of OCD have been reported, and in some forms of skeletal dysplasia, conditions resembling OCD had been identified in the patients. This suggests an underlying genetic factor in at least some cases of OCD. Mutations in genes encoding some secreted proteins have been shown to contribute to endoplasmic reticulum stress. Endoplasmic reticulum stress that induces an unfolded protein response in the cells of the growth plate may contribute to OCD. Cartilage ECM protein Cartilage Oligomeric Matrix Protein (COMP) causes pseudoachondroplasia (PSACH) (56) and matrilin 3 (MATN3) causes multiple epiphyseal dysplasia (MED) (57), and an unfolded protein response is observed in both cases. Short stature and early onset osteoarthritis was associated with familial OCD caused by mutations in ACAN (19).
Other secreted proteins
Additional secreted proteins identified from GWAS include acyloxyacyl hydrolase (AOAH), ficolin 3 (FCN3), and chloride channel, calcium activated, family member 4 (CLCA4). Ficolin 3 (FCN3) located on human chromosome 1.p36.11, encodes a secreted protein comprising a collagen-like domain and a fibrinogen-like domain. FCN3 has affinity for carbohydrate moieties, is able to activate the complement pathway, and is therefore associated with innate immunity. While no function for FCN3 in the growth plate has been reported, identification of FCN3 within the serum proteome associated with degenerative scoliosis may indicate a skeletal role (58). Chloride channel, calcium activated, family member 4 (CLCA4), located on human chromosome 1p22.3, encodes a calcium sensitive chloride conductance protein. Initially synthesized as a 125 kDa protein which is restricted to the endoplasmic reticulum, the protein undergoes specific proteolytic processing to yield a 90 kDa and 40 kDa fragments that are secreted and also found associated with the plasma membrane (59). Acyloxyacyl hydrolase (AOAH) located on chromosome 7p14.2, encodes a secreted enzyme capable of hydrolyzing acyloxyacyl-linked fatty acyl chains. A role for AOAH in OCD is currently not known.
Secretory pathway proteins
Not only must the extracellular matrix molecules be synthesized by the cell, but they must also be assembled, modified, and secreted properly in order for the extracellular matrix to function properly. Genes that play a role in this process include Solute carrier family 35 member D1 (SLC35D1) and UDP-glucose dehydrogenase (UGDH). Solute carrier family 35 member D1 (SLC35D1, located at human chromosome 1p31.3, is responsible for the glycosylation of proteins within the endoplasmic reticulum and Golgi compartments of the protein secretion pathway. SLC35D1 transports UDP-glucuronic acid and UDP-N-acetylgalactosamine from the cytoplasm to the lumen of the endoplasmic reticulum. Chondroitin sulfate (CS) is a critical component of cartilage extracellular matrix, and SLC35D1 may participate in CS biosynthesis (60, 61).
Cell signaling pathways and growth plate maturation
Signaling pathways implicated by GWAS include PTH, PTHrP, Interleukin 6 (IL6), Wnt signaling through association with Frizzled-related protein (FRZB), Neurochondrin (NCDN), TBC1 domain family member 22A (TBC1D22A), and Xin actin binding repeat containing protein 2 (XIRP2).
Type 1 and 2 PTH/PTHrP receptor (PTH1R and PTH2R) belong to a family of G-protein-coupled receptors with seven membrane-spanning helices (62–64). PTH1R is expressed in bone and prehypertrophic chondrocytes and mediates mineral homeostasis (62, 65, 66). Parathyroid hormone and parathyroid hormone related peptide signaling were previously implicated in osteoarthritis and skeletal homeostasis (67). PTHrP regulates endochondral ossification during bone development and mice null for PTHrP expression show accelerated differentiation of chondrocytes in growing long bones (68). PTH stimulates proliferation of prechondrocytes and inhibits collagen and matrix synthesis, regulating cartilage growth and chondrocyte apoptosis (69). PTHrP acts as a local factor to regulate chondrogenesis and attenuating chondrocyte hypertrophy (64, 70). PTHrP is a central factor in the regulation of bone growth. Although the role of PTH1R and PTH2R in OCD is not currently known, a decrease in expression levels corresponding to an increase in OA severity has been reported in human subchondral osteoblasts (71). Osteoblasts may be responsible for the initiation of OCD and OA, indicated by in vitro studies utilizing a co-culture system comprising osteoarthritic osteoblasts and normal cartilage explants, in which cartilage degradation was initiated by enzymes produced by the osteoblasts (72). While PTH1R levels decrease, the levels of PTHrP have been shown to increase with increase OA severity (73). PTH1R levels in chondrocytes have been evaluated in a rabbit model system over time (74).
PTH1R was identified as a candidate gene for both equine and swine osteochondrosis (27, 34) while the related PTH2R was identified by Lampe et al. (31). PTH1R is activated by PTH and PTHrP, both of which have been shown to regulate bone metabolism and differentiation (70, 75–76). PTHrP interacts with Indian hedgehog (IHH) and transforming growth factor-beta (TGF-beta) to create a feedback loop that regulates the terminal differentiation of chondrocytes to hypertrophic cells (8). Without PTHrP, chondrocytes prematurely differentiate into hypertrophic cells. PTH1R is vital for the appropriate signaling by PTHrP in the correct cells for differentiation of chondrocytes (70). Previous experimentation by Semevolos et al. (76) found that PTHrP was upregulated in equine osteochondrosis by in situ hybridization and immunohistochemistry; however, PCR testing was inconclusive. A mutation associated with PTH1R can change the binding affinity of PTH and PTHrP, in turn, affecting the PTHrP, IHH, and TGF-β feedback loop essential for chondrocyte differentiation.
Neurochondrin (NCDN) located on human chromosome 1.p34.3, encodes a leucine-rich cytoplasmic protein that functions as a negative regulator of calcium/calmodulin-dependent protein kinase II phosphorylation (CaMKII) (77). NCDN is expressed by chondrocytes, osteoblasts and osteocytes within the skeleton, and evidence suggests that NCDN may be involved in bone metabolism (78). Both the Wnt and PTHrP signaling pathways regulate CaMKII activity to support normal growth plate development, regulating the proliferative potential of chondrocytes (79).
Cartilage (CART1; ALX1), located on human chromosome 12q21.31, encodes a homeobox transcription factor that plays a role in chondrocyte differentiation. (80, 81). Gene variants of FRZB have been previously associated with osteoarthritis (82). FRZB encodes the secreted frizzled-related protein 3, which is a soluble inhibitor of the Wnt signaling pathway. Wnt signaling activity is increased in patients with osteoarthritis (83–85),
TBC1 domain family member 22A (TBC1D22A), located on human chromosome 22q13.31, encodes a GTPase-activating protein. Xin actin-binding repeat containing 2 (XIRP2), located on chromosome 2q24.3, encodes a cytosolic protein that interacts with actin stress fibers and protects actin filaments from depolymerization (86) in the cells of the myotendinous junction of skeletal muscle, among other locations. Inorganic phosphate transporter 1 (PIT1;SLC20A1), located on human chromosome 2q13, encodes a sodium-phosphate symporter that plays a role in phosphate transport in osteoblasts (87) and cartilage calcification (88).
Galectin-3 (lectin, galactoside-binding soluble, 3 (LGALS3)), located on human chromosome 14q22.3, encodes an extracellular matrix protein which binds specifically to β-galactosides. Galectin-3 can also be found located in the nucleus and the cytoplasm of mature and early hypertrophic chondrocytes. Galectin plays an important role in skeletal development and in extracellular matrix assembly. Galectin-3 plays a role in the coordination between chondrocyte differentiation, hypertrophy, and subsequent vascular invasion essential for bone formation. In the absence of galectin-3, vascular invasion of hypertrophic cartilage is reduced (89).
Calcitonin gene-related peptide-receptor component protein (RCP9; CRCP), located on human chromosome 7q11.21, encodes a membrane protein that functions as part of a receptor complex for a small neuropeptide that increases intracellular cAMP levels (90).
Apart from COL24A1 and PTH1R, no other specific genes were identified by more than one study; however, the collagen family was implicated in multiple studies. Many other genes have been identified inconsistently. One possible reason for the large amount of QTLs identified may be the variation in breeds tested. Heritability varies by breed; the variability may be explained by different polymorphisms found in different breeds. PTH1R is a strong candidate gene for OCD, identified in both horse and pigs. Research on OCD and osteochondrosis has been completed in other animals including dog and rat, however; genetic studies have not yet been completed.
Mitochondrial trans-2-enoyl CoA reductase (MECR), located on human chromosome 1p35.3, encodes a mitochondrial enzyme. Although it is not clear what role MECR might play in the onset and progression of OCD, metabolic factors play a role in osteochondrosis (91). One significant marker region was shared by Orr et al. (24) and Teyssedre et al. (23) on chromosome 3 at 105 cM, however neither article suggested genes of interest in this region. A BLAST search was completed for genes in the region, revealing leucine aminopeptidase 3 (LAP3) as a possible gene of interest in that region. While no evidence currently supports a role for LAP3 in OCD, future studies may provide this information.
CONCLUSION
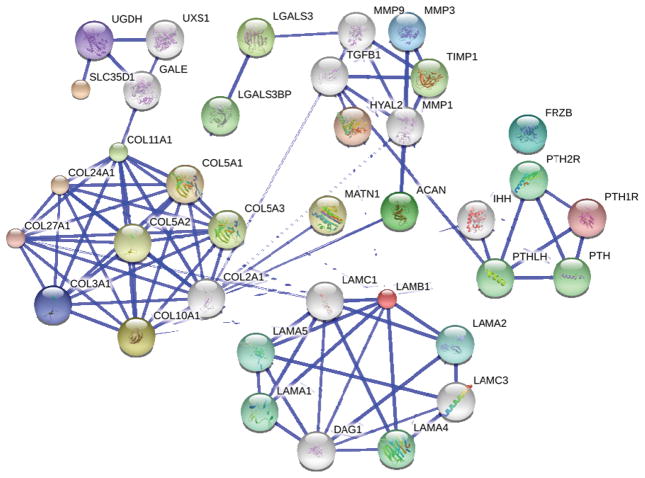
Although progress has been made, much about the genetic basis of OCD remains unknown. To understand OCD susceptibility genes, additional genetic analysis must be carried out in an unbiased fashion. GWAS provide this approach. While accuracy and density of genotyping information may represent a limitation in the effort to match genotype with phenotype for OCD, continuing technical advances coupled with decreasing costs of genome analysis hold promise for the utilization of whole genome DNA and RNA sequencing for patients. This review summarizes GWAS to date, and highlights the strength of genome-based approaches that utilize high throughput methods in the search for causes and potential therapeutic targets for the treatment of OCD. Results from GWAS tend to cluster into thematic groups (Figure 2).
Figure 2. STRING interaction network.
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations. STRING quantitatively integrates interaction data. The database currently covers 5,214,234 proteins from 1133 organisms (104). The interaction network was generated by seeding with genes identified through GWAS (see Table 3). Additional interacting proteins were identified through STRINGS and are included in the network as likely candidates for contributing roles in OCD. Blue lines connecting each node are weighted for confidence of interaction, based on peer-reviewed, published information. More data exists to support an interaction between those nodes connected by the heavier blue lines. Genes cluster into 1) a collagen cluster, 2) a laminin cluster, 3) a cell signaling cluster, 4) a matrix turnover cluster, 5) a posttranslational modification cluster. Each of these clusters is linked by multiple connections.
A common theme within the genes identified by GWAS reviewed here include secreted proteins of the extracellular matrix and the genes encoding proteins that mediate vesicular transport, posttranslational modification, and safe passage through the cellular secretory pathway. A second theme that emerges is that of cell signaling pathways known to regulate cellular differentiation for cells that play a critical role in growth plate development and maturation. Two pathways directly implicated are the PTH/PTHrP (by the identification of PTH1R and PTH2R) and Wnt (by the identification of FRZB) signaling pathways, both essential regulators of growth plate development. Established links exist between the gene clusters (Figure 2) since growth factors and cytokines regulate cellular responses that in turn lead to increases or decreases in the expression level and secretion of cell and tissue type-specific genes. The intersection of gene groups emerging from these studies may serve to guide future studies in OCD.
Much work has been done to understand OCD and osteochondrosis in humans and in animals, and an initial understanding of the etiology of this disease is forming. Genome-wide association studies provide an unbiased approach for the identification of contributing genes. Horses have been investigated in several genetic studies, which often have included larger subject populations than studies performed in pigs. Swine research did not focus solely on osteochondrosis but included other phenotypic trains relating to leg weakness. Additionally, many more SNPs were analyzed in horse studies compared to swine. Future studies are anticipated in the equine, swine and humans.
While similarities exist between human OCD and osteochondrosis in the equine and swine systems, comparison of the results of equine and swine studies to OCD in humans must be based on similarities between these conditions. Osteochondrosis in humans is different than OCD in humans. However, improved disease definitions, as well as classification and comparison criteria would advance the study of OCD irrespective of species. OCD is a multifactorial disease, alluding researchers due in part to low levels of statistical power and inconsistent results from study to study. Genome-wide association studies stand to improve both the statistical power and consistency from study to study.
Additional Genome-wide approaches for future studies
Differences in DNA methylation may be involved in the pathogenesis of OCD and must be analyzed in future studies. Subchondral bone homeostasis is perturbed in OCD, suggesting an interruption in the cyclic and sequential proliferation and differentiation of osteoclast and osteoblast precursor cells required for normal bone remodeling and growth. Methylation of cytosines in CpG-rich regions of DNA influence gene transcription and cell differentiation in the osteoblast lineage and osteoblast-osteocyte transition (92, 93) as shown by genome-wide methylation profiling of bone tissue in patients with osteoporosis and osteoarthritis (94). Future studies should include genome wide methylation profiling for OCD to identify regions of differential methylation and to understand the relationship between methylation and gene expression in OCD. This approach has been successful in the analysis of osteoporosis and for osteoarthritis, as DNA methylation represents an epigenetic level of regulation, methylation of CpG rich sequences of the promoter regions represses genes known to play roles in bone formation and bone resorption. Epigenetic mechanisms modulate the expression of proteases and other cartilage constituents, recently reviewed by Reynard (95) and Barter (96). However, little is known about the role of methylation in pathogenesis of osteochondral changes in OCD. Unlike the genome, epigenetic markers may be transmitted unmodified through cell division or, alternatively, change with the cell environment and stresses to the skeletal system. Age-related changes in DNA methylation patterns may provide information regarding the age of onset for OCD. Further studies are required to identify the mechanisms involved.
Small noncoding microRNAs (miRNAs) are important regulators of coordinated gene expression (97) and must be included in future studies. miRNAs act by forming base pairs with their complementary messenger RNA molecules, usually in the 3′-untranslated region of specific genes to silence gene expression (98). miRNAs have been shown to be necessary for normal skeletal development (99) and may also play a role during disease progression. For example, miR-140 was shown to be expressed only in cartilaginous tissues (100), and in its absence, miR-140(−/−) mice manifested a mild skeletal phenotype with a short stature. miR-140(−/−) mice experienced proteoglycan loss and fibrillation of articular cartilage (101).
Studies in humans will not only identify possible genes that cause OCD, but will also validate the work done in other species. McCoy and colleagues recently highlighted the similarities between osteochondrosis in animals and OCD in humans includinghistological appearance of end stage lesions, radiographic/MRI changes, clinical presentation, and predilection sites, strengthening the case for common pathogenesis (102). Additional studies will help to discover the role that secretion of extracellular matrix proteins such as COL24A1 and signaling pathways such as the ones mediated by PTH1R play in the onset and progression of OCD. Further use of genomic and molecular analysis techniques including GWAS, comprehensive genomic analysis of DNA methylation patterns, and analysis of miR expression to collect information from multiple species will improve our understanding of the pathogenesis of OCD, and identify molecular targets for the development of targeted preventions of joint damage and progression to OA. Future work should utilize the strengths of high efficiency methods that are becoming increasingly available to understand the molecular mechanisms underlying OCD and the link between the protein secretion pathway and cell signaling pathways that regulate chondrocyte and osteoblast differentiation during growth plate maturation.
Table 1.
Results from equine GWAS.
Identified genes and research methods in horses are outlined by paper.
| Author(s) | Year | Journal | Population size | Number of SNP markers | Map Length | Equine SNP location | Corresponding genes | Additional genes from refinement studies |
|---|---|---|---|---|---|---|---|---|
| Corbin et al. | 2012 | Mamm Genome | 330 | 40,180 | All autosomes | Chr 3 at 88.49 cM | - | - |
| Dierks et al. | 2007 | Mamm Genome | 104 | 260 | All autosomes | Chr 2 at 22.00–43.41 cM | Matrilin 1 (MATN1) | Neurochondrin (NCDN), Ficolin3 (FCN3), and Mitochondrial trans-2-enoyl CoA reductase (MECR) |
| Chr 4 at 7.70–24.30 cM | Laminin (LAMB1) | Hyaluronogluco saminidase (HYAL) family | ||||||
| Chr 5 at 79.30 cM | Solute carrier family 35 member D1 (SLC35D1) | Collage type XXIV α1 (COL24A1) | ||||||
| Chr 16 at 33.00–45.00 cM | Parathyroid hormone receptor (PTH1R) | - | ||||||
| Chr 18 at 74.94–82.25 cM* | - | Parathyroid hormone 2 receptor (PTH2R) | ||||||
| Lykkjen et al. | 2010 | Anim Genet | 162 | 41,170 | All autosomes | Chr 1 at 139 cM | - | - |
| Chr 3 at 113 cM | - | - | ||||||
| Chr 5 at 42, 77, and 79 cM | Chloride channel, calcium activated, family member 4 (CLCA4); Collagen type XXIV α1 (COL24A1) | - | ||||||
| Chr 10 at 80 cM | LOC10007351 | - | ||||||
| Chr 18 at 59 cM | - | - | ||||||
| Chr 28 at 43 cM | TBC1 domain family member 22A (TBC1D22A) | - | ||||||
| Orr et al. | 2012 | Anim Genet | 201 | 45,586 | All autosomes | Chr 3 at 105 cM | - | - |
| Chr 10 at 48 cM | - | - | ||||||
| Chr 16 at 47 cM | - | - | ||||||
| Chr 21 at 11 cM | - | - | ||||||
| Chr 31 at 22 cM | - | - | ||||||
| Teyssedre et al. | 2012 | J AnimSci | 525 | 41,249 | All autosomes | Chr 3 at 105 cM | - | - |
| Chr 13 at 9 cM | - | - | ||||||
| Chr 14 at 73 cM | - | - | ||||||
| Chr 15 at 87 cM | - | - | ||||||
| Wittwer et al. | 2007 | Anim Genet | 117 | 157 | All autosomes and X-chr | Chr 1 at 150.0–194.2 cM | lectin, galactoside-binding soluble, 3 (LGALS3) | - |
| Chr 4 at 7.8–38.0 cM | Calcitonin gene-related peptide-receptor component protein (RCP9); Calneuron 1 (CALN1) | - | ||||||
| Chr 4 at 70.0 cM | Interleukin 6 (IL6) | Acyloxyacyl hydrolase (AOAH) | ||||||
| Chr 18 at 45.9–87.6 cM | Collagen type III α1 (COL3A1); Collagen type V α2 (COL5A2); Frizzled-related protein (FRZB) | Xin actin-binding repeat containing 2 (XIRP2) | ||||||
| Chr 25 at 30.1 cM | Collagen type V α1 (COL5A1); Collagen XXVII α1 (COL27A1) | - |
Table 2.
Results of GWAS from swine
Identified genes and research methods in pigs are outlined by paper.
| Author(s) | Year | Journal | Population size | Number of SNP markers | Map length | Pig SNP location | Corresponding human genes |
|---|---|---|---|---|---|---|---|
| Andersson-Eklund et al. | 2000 | Genet. Res. | 195 | 236 | 2300 cM | Chr 5 at 51 cm | Cartilage homeoprotein 1 (CART1) |
| Chr 13 at 64 cm | PIT1, Parathyroid hormone receptor (PTHR) | ||||||
| Laenoi et al. | 2011 | Genet Sel Evol | 310 | 79 microsatellites, 3 biallelic markers | 2588.7 cM | Chr 2 at 14 cM | - |
| Chr 3 at 13 cM | - | ||||||
| Chr 6 at 61 cM | - | ||||||
| Chr 10 at 70 cM | - | ||||||
| Chr 14 at 0 cM | - |
Table 3.
Genes identified from GWAS used to seed STRING network (see Figure 2).
Genes identified and reviewed were input into the STRING network. All genes included in the STRING network and a description of the genes are included.
|
|
LAMB1 | laminin, beta 1; Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components (1786 aa) |
|
|
SLC35D1 | solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1; Transports both UDP-glucuronic acid (UDP-GlcA) and UDP-N-acetylgalactosamine (UDP-GalNAc) from the cytoplasm to into the endoplasmic reticulum lumen. Participates in glucuronidation and/or chondroitin sulfate biosynthesis (355 aa) |
|
|
COL10A1 | collagen, type X, alpha 1; Type X collagen is a product of hypertrophic chondrocytes and has been localized to presumptive mineralization zones of hyaline cartilage (680 aa) |
|
|
LGALS3 | lectin, galactoside-binding, soluble, 3; Mediates with the alpha-3, beta-1 integrin the stimulation by CSPG4 of endothelial cells migration. Together with DMBT1, required for terminal differentiation of columnar epithelial cells during early embryogenesis (250 aa) |
|
|
ACAN | aggrecan; This proteoglycan is a major component of extracellular matrix of cartilaginous tissues. A major function of this protein is to resist compression in cartilage. It binds avidly to hyaluronic acid via an N-terminal globular region (2416 aa) |
|
|
PTH2R | parathyroid hormone 2 receptor; This is a specific receptor for parathyroid hormone. The activity of this receptor is mediated by G proteins which activate adenylyl cyclase. PTH2R may be responsible for PTH effects in a number of physiological systems. (550 aa) |
|
|
FRZB | frizzled-related protein; Soluble frizzled-related proteins (sFRPS) function as modulators of Wnt signaling through direct interaction with Wnts. They have a role in regulating cell growth and differentiation in specific cell types. SFRP3/FRZB is involved in limb skeletogenesis. Antagonist of Wnt8 signaling. Regulates chondrocyte maturation and long bone development (325 aa) |
|
|
MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase); Can degrade fibronectin, laminin, gelatins of type I, III, IV, and V; collagens III, IV, X, and IX, and cartilage proteoglycans. Activates procollagenase (477 aa) |
|
|
COL3A1 | collagen, type III, alpha 1; Collagen type III occurs in most soft connective tissues along with type I collagen (1466 aa) |
|
|
UGDH | UDP-glucose dehydrogenase; Biosynthesis of glycosaminoglycans; hyaluronan, chondroitin sulfate, and heparan sulfate (494 aa) |
|
|
PTH1R | parathyroid hormone 1 receptor; This is a receptor for parathyroid hormone and for parathyroid hormone-related peptide. The activity of this receptor is mediated by G proteins which activate adenylyl cyclase and also a phosphatidylinositol-calcium second messenger system (593 aa) |
|
|
COL27A1 | collagen, type XXVII, alpha 1; Plays a role during the calcification of cartilage and the transition of cartilage to bone (1860 aa) |
|
|
HYAL2 | hyaluronoglucosaminidase 2; Hydrolyzes high molecular weight hyaluronic acid to produce an intermediate-sized product which is further hydrolyzed by sperm hyaluronidase to give small oligosaccharides. Associates with and negatively regulates MST1R (473 aa) |
|
|
COL24A1 | collagen, type XXIV, alpha 1; Participates in regulating type I collagen fibrillogenesis during fetal development (1714 aa) |
|
|
COL5A1 | collagen, type V, alpha 1; Type V collagen is a member of group I collagen (fibrillar forming collagen). It is a minor connective tissue component of nearly ubiquitous distribution. Type V collagen binds to heparan sulfate, thrombospondin, heparin, and insulin (1838 aa) |
|
|
MATN1 | matrillin 1, cartilage matrix protein; A major component of the extracellular matrix of cartilage. It binds to collagen (496 aa) |
|
|
COL5A2 | collagen, type V, alpha 2; Type V collagen is a member of group I collagen (fibrillar forming collagen). It is a minor connective tissue component of nearly ubiquitous distribution. Type V collagen binds to heparan sulfate, thrombospondin, heparin, and insulin. Type V collagen is a key determinant in the assembly of tissue-specific matrices (1499 aa) |
KEY POINTS.
PTH1R is a strong candidate gene for OCD, identified in both horse and pigs, indicating the potential for involvement of pathways that mediate transition from cartilage to bone during endochondral ossification and growth plate maturation.
Genes identified include secreted proteins of the extracellular matrix and the genes encoding proteins that mediate the cellular secretory pathway.
The identified genetic loci may also indicate a higher risk for osteoarthritis.
Acknowledgments
Authors acknowledge the technical assistance of Barbara Jibben. Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P20 RR016454 and P20 GM103408, the Boise State University Student Research Initiative Program and the Department of Biological Sciences (BIOL451).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edmonds E, Shea K. Osteochondritis Dissecans: Editorial Comment. Clin Orthop Relat Res. 2013;471:1105–6. doi: 10.1007/s11999-013-2837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lykkjen S, Dolvik N, McCue M, et al. Genome-Wide Association Analysis of Osteochondrosis of the Tibiotarsal Joint in Norwegian Standardbred Trotters. Anim Genet. 2010;41 (Suppl 2):111–20. doi: 10.1111/j.1365-2052.2010.02117.x. [DOI] [PubMed] [Google Scholar]
- 3.König F. The Classic: On Loose Bodies in the Joint 1888 reprinted. Clin Orthop Relat Res. 2013;471:1107–15. doi: 10.1007/s11999-013-2824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shea K, Jacobs J, Jr, Carey J, et al. Osteochondritis Dissecans Knee Histology Studies Have Variable Findings and Theories of Etiology. Clin Orthop Relat Res. 2013;471:1127–1136. doi: 10.1007/s11999-012-2619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edmonds E, Polousky J. A review of knowledge in osteochondritis dissecans:123 years of minimal evolution from König to the ROCK study group. Clin OrthopRelat Res. 2013;471:1118–26. doi: 10.1007/s11999-012-2290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thacker M, Dabney K, Mackenzie W. Osteochondritis dissecans of the talarhead: natural history and review of literature. J Pediatr Orthop B. 2012;21:373–6. doi: 10.1097/BPB.0b013e328346c076. [DOI] [PubMed] [Google Scholar]
- 7.Polousky J. Juvenile osteochondritis dissecans. Sports Med Arthrosc. 2011;19:56–63. doi: 10.1097/JSA.0b013e31820b94b9. [DOI] [PubMed] [Google Scholar]
- 8.Ytrehus B, Carlson C, Ekman S. Etiology and Pathogenesis of Osteochondrosis. Vet Pathol. 2007;44:429–48. doi: 10.1354/vp.44-4-429. [DOI] [PubMed] [Google Scholar]
- 9.Kocher M, Tucker R, Ganley T, et al. Management of osteochondritis dissecans of the knee: current concepts review. Am J Sports Med. 2006;34:1181–91. doi: 10.1177/0363546506290127. [DOI] [PubMed] [Google Scholar]
- 10.Cahill B. Osteochondritis Dissecans of the Knee: Treatment of Juvenile and Adult Forms. J Am Acad Orthop Surg. 1995;3:237–47. doi: 10.5435/00124635-199507000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Stattin E, Tegner Y, Domellof M, et al. Familial Osteochondritis Dissecans Associated with Early Osteoarthritis and Disproportionate Short Stature. Osteoarthritis Cartilage. 2008;16:890–6. doi: 10.1016/j.joca.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Lindén B, Telhag H. Osteochondritis dissecans. A histologic and autoradiographic study in man. Acta Orthop Scand. 1977;48:682–6. doi: 10.3109/17453677708994817. [DOI] [PubMed] [Google Scholar]
- 13.Twyman R, Desai K, Aichroth P. Osteochondritis dissecans of the knee. A long-term study. J Bone Joint Surg Br. 1991;73:461–4. doi: 10.1302/0301-620X.73B3.1670450. [DOI] [PubMed] [Google Scholar]
- 14.Chambers H, Shea K, Anderson A, et al. Diagnosis and Treatment of Osteochondritis Dissecans. J Am Acad Orthop Surg. 2011;19:297–306. doi: 10.5435/00124635-201105000-00007. [DOI] [PubMed] [Google Scholar]
- 15.McIlwraith C. Inferences from referred clinical cases of osteochondritis dissecans. Equine Veterinary Journal. 25:S16, 2042–3306. [Google Scholar]
- 16.Jorgensen B, Arnbjerg J, Aaslyng M. Pathological and Radiological Investigations on Osteochondrosis in Pigs, Associated with Leg Weakness. Zentralbl Veterinarmed. 1995;42:489–504. doi: 10.1111/j.1439-0442.1995.tb00404.x. [DOI] [PubMed] [Google Scholar]
- 17.Laenoi W, Uddin J, Cinar M, et al. Quantitative Trait Loci Analysis for Leg Weakness-Related Traits in a Duroc X Pietrain Crossbred Population. Genet Sel Evol. 2011;43:13. doi: 10.1186/1297-9686-43-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffcott L. Osteochondrosis in the Horse--Searching for the Key to Pathogenesis. Equine Vet J. 1991;23:331–8. doi: 10.1111/j.2042-3306.1991.tb03733.x. [DOI] [PubMed] [Google Scholar]
- 19.Stattin E, Wiklund F, Lindblom K, et al. A missense mutation in the aggrecan C-type lectin domain disrupts extracellular matrix interactions and causes dominant familial osteochondritis dissecans. Am J Hum Genet. 2010;86:126–37. doi: 10.1016/j.ajhg.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mubarak S, Carroll N. Familial osteochondritis dissecans of the knee. ClinOrthop Relat Res. 1979;140:131–6. [PubMed] [Google Scholar]
- 21.Wittwer C, Lohring K, Drogemuller C, et al. Mapping Quantitative Trait Loci for Osteochondrosis in Fetlock and Hock Joints and Palmar/Plantar Osseus Fragments in Fetlock Joints of South German Coldblood Horses. Anim Genet. 2007;38:350–7. doi: 10.1111/j.1365-2052.2007.01610.x. [DOI] [PubMed] [Google Scholar]
- 22.Wittwer C, Hamann H, Rosenberger E, et al. Genetic Parameters for the Prevalence of Osteochondrosis in the Limb Joints of South German Coldblood Horses. J Anim Breed Genet. 2007;124:302–7. doi: 10.1111/j.1439-0388.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- 23.Teyssedre S, Dupuis M, Guerin G, et al. Genome-Wide Association Studies for Osteochondrosis in French Trotter Horses. J Anim Sci. 2012;90:45–53. doi: 10.2527/jas.2011-4031. [DOI] [PubMed] [Google Scholar]
- 24.Orr N, Hill E, Gu J, et al. Genome-Wide Association Study of Osteochondrosis in the Tarsocrural Joint of Dutch Warmblood Horses Identifies Susceptibility Loci on Chromosomes 3 and 10. Anim Genet. 2012 Dec 25; doi: 10.1111/age.12016. [DOI] [PubMed] [Google Scholar]
- 25.Wittwer C, Hamann H, Distl O. The Candidate Gene Xirp2 at a Quantitative Gene Locus on Equine Chromosome 18 Associated with Osteochondrosis in Fetlock and Hock Joints of South German Coldblood Horses. J Hered. 2009;100:481–6. doi: 10.1093/jhered/esp006. [DOI] [PubMed] [Google Scholar]
- 26.Corbin L, Blott S, Swinburne J, et al. A genome-wide association study of osteochondritis dissecans in the Thoroughbred. Mamm Genome. 2012;23:294–303. doi: 10.1007/s00335-011-9363-1. [DOI] [PubMed] [Google Scholar]
- 27.Dierks C, Lohring K, Lampe V, et al. Genome-Wide Search for Markers Associated with Osteochondrosis in Hanoverian Warmblood Horses. Mamm Genome. 2007;18:739–47. doi: 10.1007/s00335-007-9058-9. [DOI] [PubMed] [Google Scholar]
- 28.Dierks C, Komm K, Lampe V, et al. Fine Mapping of a Quantitative Trait Locus for Osteochondrosis on Horse Chromosome 2. Anim Genet. 2010;41:87–90. doi: 10.1111/j.1365-2052.2010.02113.x. [DOI] [PubMed] [Google Scholar]
- 29.Lampe V, Dierks C, Distl O. Refinement of a Quantitative Trait Locus on Equine Chromosome 5 Responsible for Fetlock Osteochondrosis in Hanoverian Warmblood Horses. Anim Genet. 2009;40:553–5. doi: 10.1111/j.1365-2052.2009.01865.x. [DOI] [PubMed] [Google Scholar]
- 30.Lampe V, Dierks C, Komm K, et al. Identification of a new quantitative trait locus on equine chromosome 18 responsible for osteochondrosis in Hanoverian warmblood horses. J Anim Sci. 2009;87:3477–81. doi: 10.2527/jas.2009-1946. [DOI] [PubMed] [Google Scholar]
- 31.Lampe V, Dierks C, Distl O. Refinement of a quantitative gene locus on equine chromosome 16 responsible for osteochondrosis in Hanoverian warmblood horses. Animal. 2009;3:1224–31. doi: 10.1017/S1751731109004765. [DOI] [PubMed] [Google Scholar]
- 32.Wittwer C, Dierks C, Hamann H, et al. Associations between Candidate Gene Markers at a Quantitative Trait Locus on Equine Chromosome 4 Responsible for Osteochondrosis Dissecans in Fetlock Joints of South German Coldblood Horses. J Hered. 2008;99:125–9. doi: 10.1093/jhered/esm106. [DOI] [PubMed] [Google Scholar]
- 33.Laenoi W, Rangkasenee N, Uddin M, et al. Association and Expression Study of Mmp3, Tgf beta 1 and Col10a1 as Candidate Genes for Leg Weakness-Related Traits in Pigs. Mol Biol Rep. 2012;39:3893–901. doi: 10.1007/s11033-011-1168-5. [DOI] [PubMed] [Google Scholar]
- 34.Andersson-Eklund L, Uhlhorn H, Lundeheim N, et al. Mapping Quantitative Trait Loci for Principal Components of Bone Measurements and Osteochondrosis Scores in a Wild Boar X Large White Intercross. Genet Res. 2000;75:223–30. doi: 10.1017/s0016672399004371. [DOI] [PubMed] [Google Scholar]
- 35.Fairbank H. Osteochondritis dissecans. Br J Surg. 1933;21:67–82. [Google Scholar]
- 36.Laor T, Zbojniewicz A, Eismann E, et al. Juvenile osteochondritisdissecans: is it a growth disturbance of the secondary physis of the epiphysis? AJR Am J Roentgenol. 2012;199:1121–8. doi: 10.2214/AJR.11.8085. [DOI] [PubMed] [Google Scholar]
- 37.Chiroff R, Cooke C., 3rd Osteochondritis dissecans: a histologic and microradiographic analysis of surgically excised lesions. J Trauma. 1975;15:689–96. [PubMed] [Google Scholar]
- 38.Csóka A, Scherer S, Stern R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics. 1999;15:356–61. doi: 10.1006/geno.1999.5876. [DOI] [PubMed] [Google Scholar]
- 39.Csoka A, Frost G, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- 40.Flannery C, Little C, Hughes C, et al. Expression and activity of articular cartilage hyaluronidases. Biochem Biophys Res Commun. 1998;251:824–9. doi: 10.1006/bbrc.1998.9561. [DOI] [PubMed] [Google Scholar]
- 41.Chow G, Knudson W. Characterization of promoter elements of the human HYAL-2 gene. J Biol Chem. 2005;280:26904–12. doi: 10.1074/jbc.M413845200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deak F, Wagener R, Kiss I, et al. The matrilins: a novel family of oligomeric extracellular matrix proteins. Matrix Biology. 1999;18:55–64. doi: 10.1016/s0945-053x(98)00006-7. [DOI] [PubMed] [Google Scholar]
- 43.Chapman K, Mortier G, Chapman K, et al. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. NatureGenetics. 2001;28:393–6. doi: 10.1038/ng573. [DOI] [PubMed] [Google Scholar]
- 44.Mostert A, Dijkstra P, Jansen B, et al. Familial multiple epiphyseal dysplasia due to a matrilin-3 mutation: further delineation of the phenotype including 40 years follow-up. American Journal of Medical Genetics. 2003;120:490–7. doi: 10.1002/ajmg.a.20034. [DOI] [PubMed] [Google Scholar]
- 45.Makitie O, Mortier G, Czarny-Ratajczak M, et al. Clinical and radiographic findings in multiple epiphyseal dysplasia caused by MATN3 mutations: description of 12 patients. American Journal of Medical Genetics. 2004;125:278–284. doi: 10.1002/ajmg.a.20486. [DOI] [PubMed] [Google Scholar]
- 46.Mabuchi A, Haga N, Maeda K, et al. Novel and recurrentmutations clustered in the von Willebrand factor A domain of MATN3 in multiple epiphyseal dysplasia. Human Mutation. 2004;24:439–40. doi: 10.1002/humu.9286. [DOI] [PubMed] [Google Scholar]
- 47.Borochowitz Z, Scheffer D, Adir V, et al. Spondylo-epi-metaphyseal dysplasia (SEMD) matrilin 3 type: homozygote matrilin 3mutation in a novel form of SEMD. Journal of Medical Genetics. 2004;41:366–72. doi: 10.1136/jmg.2003.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefansson S, Jonsson H, Ingvarsson T, et al. Genomewide scan for hand osteoarthritis: a novel mutationin matrilin-3. American Journal of Human Genetics. 2003;72:1448–1459. doi: 10.1086/375556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pullig O, Weseloh G, Klatt A, et al. Matrilin-3 in human articular cartilage: increased expression in osteoarthritis. Osteoarthritis and Cartilage. 2002;10:253–63. doi: 10.1053/joca.2001.0508. [DOI] [PubMed] [Google Scholar]
- 50.Min J, Meulenbelt I, Riyazi N. Association of matrilin-3 polymorphisms with spinal disc degeneration and osteoarthritis of the first carpometacarpal joint of the hand. Annals of the Rheumatic Diseases. 2006;65:1060–6. doi: 10.1136/ard.2005.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pullig O, Tagariello A, Schweizer A, et al. MATN3 (matrilin-3) sequence variation (pT303M) is a risk factor for osteoarthritis of the CMC1 joint of the hand, but not for knee osteoarthritis. Annals of the Rheumatic Diseases. 2007;66:279–80. doi: 10.1136/ard.2006.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch M, Laub F, Zhou P, et al. Collagen XXIV, a Vertebrate Fibrillar Collagen with Structural Features of Invertebrate Collagens: Selective Expression in Developing Cornea and Bone. J Biol Chem. 2003;278:43236–44. doi: 10.1074/jbc.M302112200. [DOI] [PubMed] [Google Scholar]
- 53.Skagen P, Horn T, Kruse H, et al. Osteochondritis Dissecans (Ocd), an Endoplasmic Reticulum Storage Disease?: A Morphological and Molecular Study of Ocd Fragments Scand. J Med Sci Sports. 2011;21:e17–33. doi: 10.1111/j.1600-0838.2010.01128.x. [DOI] [PubMed] [Google Scholar]
- 54.Matsuo N, Tanaka S, Yoshioka H, et al. Collagen XXIV (Col24a1) gene expression is a specific marker of osteoblast differentiation and bone formation. Connect Tissue Res. 2008;49:68–75. doi: 10.1080/03008200801913502. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Olson D, Liang G, et al. Collagen XXIV (Col24α1) promotes osteoblastic differentiation and mineralization through TGF-β/Smads signaling pathway. Int J Biol Sci. 2012;8:1310–22. doi: 10.7150/ijbs.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suleman F, Gualeni B, Gregson H, et al. A novel form of chondrocyte stress is triggered by a COMP mutation causing pseudoachondroplasia. Hum Mutat. 2012;33:218–31. doi: 10.1002/humu.21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nundlall S, Rajpar M, Bell P, et al. An unfolded protein response is the initial cellular response to the expression of mutant matrilin-3 in a mouse model of multiple epiphyseal dysplasia. Cell Stress Chaperones. 2010;15:835–49. doi: 10.1007/s12192-010-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y, Han S, Zhao H, et al. Comparative analysis of serum proteomes of degenerative scoliosis. J Orthop Res. 2011;29:1896–903. doi: 10.1002/jor.21466. [DOI] [PubMed] [Google Scholar]
- 59.Huan C, Greene K, Shui B, et al. mCLCA4 ER processing and secretion requires luminalsorting motifs. Am J Physiol Cell Physiol. 2008;295:C279–87. doi: 10.1152/ajpcell.00060.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muraoka M, Kawakita M, Ishida N. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 2001;495:87–93. doi: 10.1016/s0014-5793(01)02358-4. [DOI] [PubMed] [Google Scholar]
- 61.Hiraoka S, Furuichi T, Nishimura G, et al. Nucleotide-sugar transporter SLC35D1 is critical to chondroitin sulfate synthesis in cartilage and skeletal development in mouse and human. Nat Med. 2007;13:1363–7. doi: 10.1038/nm1655. [DOI] [PubMed] [Google Scholar]
- 62.Mannstadt M, Juppner H, Gardella T. Receptors for PTH and PTHrP: their biological importance and functional properties. Am J Physiol. 1999;277:F665–75. doi: 10.1152/ajprenal.1999.277.5.F665. [DOI] [PubMed] [Google Scholar]
- 63.Abou-Samra A, Uneno S, Jueppner H, et al. Non-homologous sequences of parathyroid hormone and the parathyroid hormone related peptide bind to a common receptor on ROS 17/2. 8 cells. Endocrinology. 1989;125:2215–7. doi: 10.1210/endo-125-4-2215. [DOI] [PubMed] [Google Scholar]
- 64.Amizuka N, Henderson J, White J, et al. Recent studies on the biological action of parathyroid hormone (PTH)-related peptide (PTHrP) and PTH/PTHrP receptor in cartilage and bone. Histol Histopathol. 2000;15:957–70. doi: 10.14670/HH-15.957. [DOI] [PubMed] [Google Scholar]
- 65.Lee K, Deeds J, Segre G. Expression of parathyroid hormone-related peptide and its receptor messenger ribonucleic acids during fetal development of rats. Endocrinology. 1995;136:453–63. doi: 10.1210/endo.136.2.7835276. [DOI] [PubMed] [Google Scholar]
- 66.Lee K, Lanske B, Karaplis A, et al. Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology. 1996;137:5109–18. doi: 10.1210/endo.137.11.8895385. [DOI] [PubMed] [Google Scholar]
- 67.Elliott K, Chapman K, Day-Williams A, et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Ann Rheum Dis. 2013;72:935–941. doi: 10.1136/annrheumdis-2012-202081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lanske B, Karaplis A, Lee K, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–66. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- 69.Harrington E, Lunsford L, Svoboda K. Chondrocyte terminal differentiation, apoptosis, and type X collagen expression are downregulated by parathyroid hormone. Anat Rec A Discov Mol Cell Evol Biol. 2004;281:1286–95. doi: 10.1002/ar.a.20129. [DOI] [PubMed] [Google Scholar]
- 70.Vortkamp A, Lee K, Lanske B, et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–22. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 71.Hilal G, Massicotte F, Martel-Pelletier J, et al. Endogenous prostaglandin E2 and insulin-like growth factor 1 can modulate the levels of parathyroid hormone receptor in human osteoarthritic osteoblasts. J Bone Miner Res. 2001;16:713–21. doi: 10.1359/jbmr.2001.16.4.713. [DOI] [PubMed] [Google Scholar]
- 72.Westacott C, Webb G, Warnock M, et al. Alteration of cartilage metabolism by cells from osteoarthritic bone. Arthritis Rheum. 1997;40:1282–91. doi: 10.1002/1529-0131(199707)40:7<1282::AID-ART13>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 73.Kohno H, Shigeno C, Kasai R, et al. Synovial fluids from patients with osteoarthritis and rheumatoid arthritis contain high levels of parathyroid hormone-related peptide. J Bone Miner Res. 1997;12:847–54. doi: 10.1359/jbmr.1997.12.5.847. [DOI] [PubMed] [Google Scholar]
- 74.Becher C, Szuwart T, Ronstedt P, et al. Decrease in the expression of the type 1 PTH/PTHrP receptor (PTH1R) on chondrocytes in animals with osteoarthritis. Journal of Orthopaedic Surgery and Research. 2010;5:28. doi: 10.1186/1749-799X-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kulkarni N, Halladay D, Miles R, et al. Effects of Parathyroid Hormone on Wnt Signaling Pathway in Bone. J Cell Biochem. 2005;95:1178–90. doi: 10.1002/jcb.20506. [DOI] [PubMed] [Google Scholar]
- 76.Semevolos S, Brower-Toland B, Bent S, et al. Parathyroid Hormone-Related Peptide and Indian Hedgehog Expression Patterns in Naturally Acquired Equine Osteochondrosis. J Orthop Res. 2002;20:1290–7. doi: 10.1016/S0736-0266(02)00055-4. [DOI] [PubMed] [Google Scholar]
- 77.Dateki M, Horii T, Kasuya Y, et al. Neurochondrin negatively regulates CaMKII phosphorylation, and nervous system-specific gene disruption results inepileptic seizure. J Biol Chem. 2005 May 27;280:20503–8. doi: 10.1074/jbc.M414033200. [DOI] [PubMed] [Google Scholar]
- 78.Ishiduka Y, Mochizuki R, Yanai K, et al. Induction of hydroxyapatite resorptive activity in bonemarrow cell populations resistant to bafilomycin A1 by a factor with restricted expression to bone and brain, neurochondrin. Biochim Biophys Acta. 1999;1450:92–8. doi: 10.1016/s0167-4889(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 79.Li Y, Ahrens M, Wu A, et al. Calcium/calmodulin-dependent protein kinase II activity regulates the proliferative potential of growth plate chondrocytes. Development. 2011;138:359–70. doi: 10.1242/dev.052324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhai L, Zhao K, Wang Z, et al. Mesenchymal stem cells display different gene expression profiles compared to hyaline and elastic chondrocytes. Int J Clin Exp Med. 2011;4:81–90. [PMC free article] [PubMed] [Google Scholar]
- 81.Thompson H, Griffiths J, Jeffery G, et al. The retinal pigment epithelium of the eye regulates the development of scleral cartilage. Dev Biol. 2010;347:40–52. doi: 10.1016/j.ydbio.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baker-Lepain J, Lynch J, Parimi N, et al. Variant alleles of the WNT antagonist FRZB are determinants of hip shape and modify the relationship between hip shape and osteoarthritis. Arthritis Rheum. 2012;64:1457–65. doi: 10.1002/art.34526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Velasco J, Zarrabeitia M, Prieto J, et al. Wnt pathway genes in osteoporosis and osteoarthritis: differential expression and genetic association study. Osteoporos Int. 2010;21:109–18. doi: 10.1007/s00198-009-0931-0. [DOI] [PubMed] [Google Scholar]
- 84.Luyten F, Tylzanowski P, Lories R. Wnt signaling and osteoarthritis. Bone. 2009;44:522–7. doi: 10.1016/j.bone.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 85.Corr M. Wnt-beta-catenin signaling in the pathogenesis of osteoarthritis. Nat Clin Pract Rheumatol. 2008;4:550–6. doi: 10.1038/ncprheum0904. [DOI] [PubMed] [Google Scholar]
- 86.Pacholsky D, Vakeel P, Himmel M, et al. Xin repeats define a novel actin-binding motif. J Cell Sci. 2004;117:5257–68. doi: 10.1242/jcs.01406. [DOI] [PubMed] [Google Scholar]
- 87.Caverzasio J, Bonjour J. Characteristics and regulation of Pi transport in osteogenic cells for bone metabolism. Kidney Int. 1996;49:975–80. doi: 10.1038/ki.1996.138. [DOI] [PubMed] [Google Scholar]
- 88.Sugita A, Kawai S, Hayashibara T, et al. Cellular ATP synthesis mediated by type III sodium-dependent phosphate transporter Pit-1 is critical to chondrogenesis. J Biol Chem. 2011;286:3094–103. doi: 10.1074/jbc.M110.148403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colnot C, Sidhu S, Balmain N, et al. Uncoupling of chondrocyte death and vascular invasion in mouse galectin 3 null mutant bones. Dev Biol. 2001;229:203–14. doi: 10.1006/dbio.2000.9933. [DOI] [PubMed] [Google Scholar]
- 90.Uezono Y, Bradley J, Min C, et al. Receptors that couple to 2 classes of G proteins increase cAMP and activate CFTR expressed in Xenopus oocytes. Receptors Channels. 1993;1:233–41. [PubMed] [Google Scholar]
- 91.Sarneveld, Barneveld A, van Weeren P. Conclusions regarding the influence of exercise on the development of the equine musculoskeletal system with special reference to osteochondrosis. Equine Vet J Suppl. 1999;31:112–9. doi: 10.1111/j.2042-3306.1999.tb05323.x. [DOI] [PubMed] [Google Scholar]
- 92.Delgado-Calle J, Sanudo C, Bolado A, Fernandez A, et al. DNA methylation contributes to the regulation of sclerostin expression in human osteocytes. J BoneMiner Res. 2012;27:926–37. doi: 10.1002/jbmr.1491. [DOI] [PubMed] [Google Scholar]
- 93.Delgado-Calle J, Sanudo C, Fernandez A, et al. Role of DNA methylation in the regulation of the RANKL-OPG system in human bone. Epigenetics. 2012;7:83–91. doi: 10.4161/epi.7.1.18753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Delgado-Calle J, Fernández A, Sainz J, et al. Genome-wide profiling of bone reveals differentially methylated regions in osteoporosis and osteoarthritis. Arthritis Rheum. 2013;65:197–205. doi: 10.1002/art.37753. [DOI] [PubMed] [Google Scholar]
- 95.Reynard L, Loughlin J. Genetics and epigenetics of osteoarthritis. Maturitas. 2012;71:200–4. doi: 10.1016/j.maturitas.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 96.Barter M, Bui C, Young D. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthritis Cartilage. 2012;20:339–49. doi: 10.1016/j.joca.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 97.Chiang H, Schoenfeld L, Ruby J, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mendell J. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–84. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 99.Kapinas K, Delany A. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–85. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012;8:543–52. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCoy AM, Toth F, Dolvik NI, Ekman S, Ellermann J, Olstad K, Ytrehus B, Carlson CS. Articular Osteochondrosis: A Comparison of Naturally-Occurring Human and Animal Disease. Osteoarthritis and Cartilage. 2013 doi: 10.1016/j.joca.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moher D, Liberati A, Tetzlaff J, et al. The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013 Jan;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]