Abstract
Temporal and spatial control of gene expression is important for studying the molecular and cellular mechanisms of development, physiology, and disease. We used the doxycycline (Dox)-inducible, Tet-On System to develop transgenic zebrafish for inducible, cell specific control of gene expression in the ultraviolet (UV) cone photoreceptors. Two constructs containing the reverse tetracycline-controlled transcriptional transactivator (rtTA) gene driven by the UV opsin-specific promoter (opn1sw1) were used to generate stable transgenic zebrafish lines using the Tol2-based transgenesis method. One construct included a self-reporting GFP (opn1sw1:rtTA, TRE:GFP) and the other incorporated an epitope tag on the rtTA protein (opn1sw1:rtTAflag). UV cone-specific expression of TRE-controlled transgenes was induced by Dox treatment in larvae and adults. Induction of gene expression was observed in 96% of all larval UV cones within 16 hours of Dox treatment. UV cone-specific expression of two genes from a bidirectional TRE construct injected into one-cell Tg(opn1sw1:rtTAflag) embryos were also induced by Dox treatment. In addition, UV cone-specific expression of Crb2aIntraWT was induced by Dox treatment in progeny from crosses of the TRE-response transgenic line, Tg(TRE:HA-Crb2aIntraWT), to the Tg(opn1sw1:rtTA, TRE:GFP) line and the Tg(opn1sw1:rtTAflag) line. These lines can be used in addition to the inducible, rod-specific gene expression system from the Tet-On Toolkit to elucidate the photoreceptor-specific effects of genes of interest in photoreceptor cell biology and retinal disease.
1. Introduction
Inducible, cell-specific expression systems enhance functional studies of the retina by allowing spatial and temporal control of transgenes of interest. Such control has great advantage over global gene disruption for two main reasons. First, global disruption of essential genes often causes embryonic or early postnatal lethality, which can prevent the investigation of gene function at later stages. Second, ubiquitous gene disruption does not allow insights to be made regarding the role of gene expression within particular cell types. For example, inducible transgene expression systems in the mouse have aided in elucidating the role of VEGF in the development and maintenance of retinal vasculature. Loss of even a single VEGF allele (VEGF+/−) causes an embryonic lethal phenotype, thereby eliminating the ability to study the effect of loss of VEGF on the retina (Carmeliet et al., 1996; Ferrara et al., 1996). Tetracycline-inducible gene expression combined with the Cre/lox system was used to knockdown VEGF in the retinal pigment epithelium (RPE) and demonstrate that development of the choroidal vasculature relies on RPE-derived VEGF during organogenesis (Le et al., 2010). A tetracycline-inducible system was also used to demonstrate that VEGF blockade for an extended period of time (up to 7 months) did not cause adverse effects, thereby providing support for the use of VEGF antagonists in the treatment of diseases involving choroidal neovascularization (Ueno et al., 2008). Inducible, cell-specific gene expression systems therefore benefit developmental studies as well as therapeutic studies for common retinal diseases.
The zebrafish has emerged as a powerful system to study eye development and to model the progression of human eye diseases. Zebrafish are easy to maintain in laboratory settings, produce large clutch sizes, and exhibit rapid, external development, which make them particularly suitable for genetic manipulation. Combined with the highly conserved nature of the vertebrate eye (Albalat, 2012; Lamb, 2013), this system is valuable for understanding gene function and disease progression of human blinding diseases. Several large-scale forward genetic screens in zebrafish have led to the identification of mutations that effect the development and function of the vertebrate retina (Brockerhoff et al., 1995; Fadool et al., 1997; Malicki et al., 1996). Unfortunately, many of these mutations are larval lethal and, therefore, severely limit necessary studies of age-related retinal degeneration and late-developing retinal diseases. In order to address this issue we used inducible, cell-specific gene expression in zebrafish rod photoreceptors and built a Tet-On Toolkit to facilitate making further Tet-On transgenics (Campbell et al., 2012). The system employs the reverse tetracycline-controlled transcriptional transactivator (rtTA) under the transcriptional control of a cell-specific promoter to drive expression of a transgene of interest under the control of a tetracycline response element (TRE), which is turned on only in the presence of the inducer, doxycycline (Dox) (Gossen et al., 1995). Similar doxycycline-inducible systems have been used to control transgene expression in zebrafish heart tissue (Huang et al., 2005; Knopf et al., 2010). The Tet-On Toolkit incorporates the flexible Gateway system for rapid cloning and the simple substitution of promoters and genes of interest (Kwan et al., 2007). It is also compatible with Tol2kit vectors for straight-forward generation of inducible, cell-specific transgenic zebrafish lines via Tol2 transgenesis (Kawakami et al., 2004; Kwan et al., 2007).
Here we describe the use of two types of Tet-On driver lines for inducible, cell-specific transgene expression in zebrafish ultraviolet (UV) cone photoreceptors. We used the UV opsin promoter to drive rtTA expression and demonstrated self-reporting GFP expression with Dox treatment. The results demonstrate that additional TRE-controlled transgenes can also be expressed in a UV cone-specific manner by both mosaic and transgenic methods. These UV cone Tet-On driver lines will complement the previously made rod photoreceptor driver lines and support necessary studies seeking to understand the morphological, functional, and disease-related differences between rods and cones.
2. Results
2.1 Ultraviolet Cone-Specific, Doxycycline-Induced Green Fluorescent Protein Expression
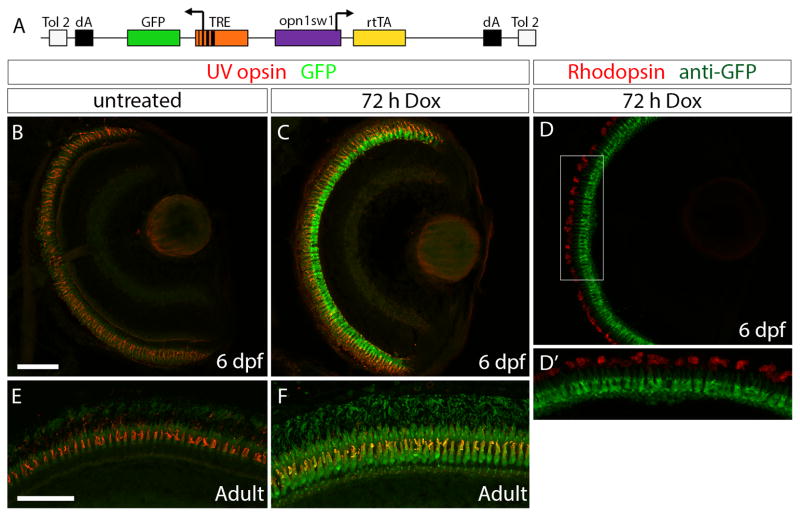
In order to induce gene expression in cone photoreceptors we designed a construct with the zebrafish UV opsin promoter (opn1sw1) (Takechi et al., 2003) driving the reverse tetracycline-controlled transcriptional transactivator (rtTA) and the tetracycline response element (TRE) upstream of green fluorescent protein (GFP) (Fig. 1A). Using the pTol transgenesis system (Kawakami et al., 2004), we generated the stable, transgenic line Tg(opn1sw1:rtTA, TRE:GFP). Retinal sections of untreated Tg(opn1sw1:rtTA, TRE:GFP) larvae at 6 days post fertilization (dpf) showed no GFP fluorescence (Fig. 1B). UV cone photoreceptors were identified by anti-UV opsin immunofluorescence. No GFP expression was seen elsewhere in the larvae although some background auto-fluorescence of outer segments was visible (e.g. Fig. 1B, E). Retinal sections of 6 dpf Tg(opn1sw1:rtTA, TRE:GFP) larvae treated with Dox for 72 hours (h) showed strong GFP expression in cones (Fig. 1C) and no overlap with the anti-Rhodopsin immunofluorescence that labeled the rod outer segments (Fig. 1D, D′). Treatment with Dox for 72 h also induced GFP expression in Tg(opn1sw1:rtTA, TRE:GFP) adults (Fig. 1F), while untreated Tg(opn1sw1:rtTA, TRE:GFP) adult UV cones showed no GFP expression (Fig. 1E).
Figure 1. Generation of an UV cone-specific, doxycycline-inducible, self-reporting gene expression system.
(A) Diagram of the construct used to generate the stable, transgenic line Tg(opn1sw1:rtTA, TRE:GFP), which expresses doxycycline (Dox)-inducible green fluorescent protein (GFP) specifically in UV cone photoreceptors. The UV opsin promoter (opn1sw1) drives expression of the reverse tetracycline-controlled transcriptional activator (rtTA) gene while the tetracycline responsive element (TRE) is upstream of GFP. (B, C) Confocal z-projections of retinal sections from 6 dpf Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larvae labeled with an anti-UV opsin antibody (red). (B) GFP fluorescence (green) is not visible in the UV cones of larval transgenic zebrafish in the absence of Dox treatment. (C) GFP fluorescence is clearly visible in UV cones in transgenic larvae after 72 hours of Dox treatment (3–6 dpf). (D) Confocal z-projection of retinal section from 6 dpf Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larva labeled with an anti-GFP antibody (green) and an anti-Rhodopsin antibody (red). (D′) Corresponds to boxed area in D. Anti-GFP immunofluorescence (green) is visible in UV opsin cone photoreceptors while anti-Rhodopsin immunofluorescence (red) is visible in the outer segments of rod photoreceptors. (E, F) Confocal z-projections of the photoreceptor layer of retinas from adult Tg(opn1sw1:rtTA, TRE:GFP) zebrafish labeled with anti-UV opsin antibody (red). (E) GFP fluorescence (green) is not visible in the adult transgenic UV cones in the absence of Dox treatment. (F) GFP fluorescence is clearly visible in transgenic adult UV cones after 72 hours of Dox treatment. dA, polyadenylation signal; Tol2, pTol integration site. Scale bars (B, E), 50 μm.
2.2 Time-Course of Doxycycline-Induced Gene Expression
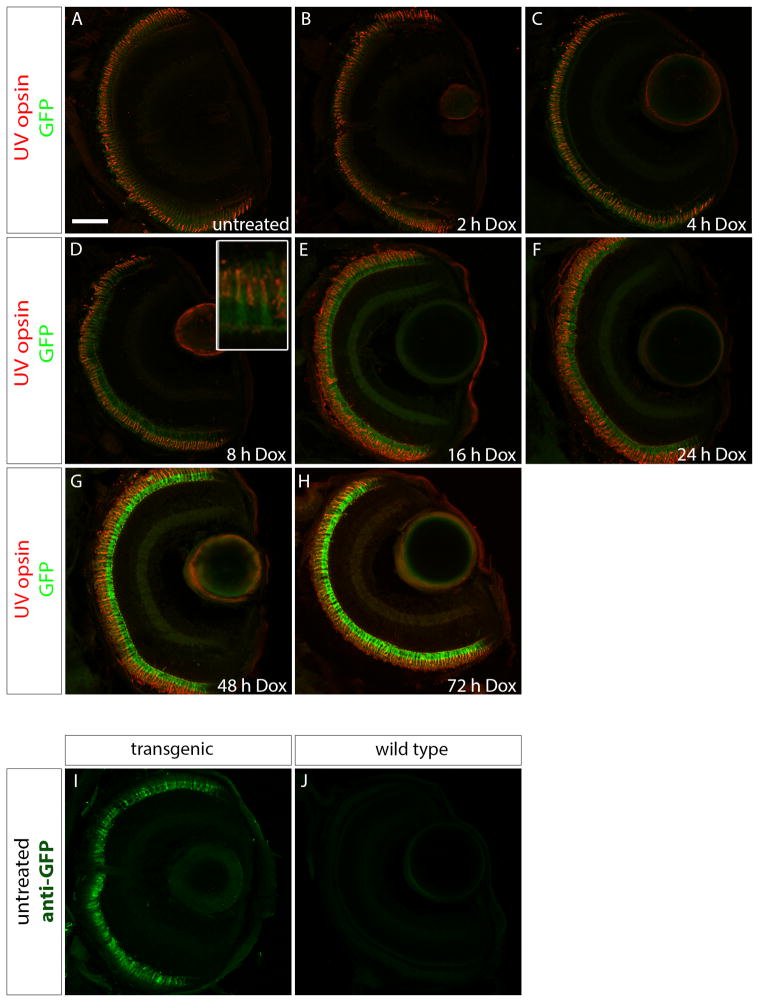
In order to examine the time-course of the onset of GFP expression with Dox treatment, Tg(opn1sw1:rtTA, TRE:GFP) larvae were treated with Dox for 2, 4, 8, 16, 24, 48, or 72 h prior to fixation at 6 dpf. Treated larval fish were genotyped, resulting in approximately 50% of the offspring containing the transgene, suggesting single copy insertion. Transgenic larvae were embedded and labeled with anti-UV opsin antibody. GFP fluorescence was seen only in rare cones in the untreated, 2 h, or 4 h Dox treatment groups (Fig 2 A–C). After 8 h of Dox treatment (Fig. 2D), GFP expression was apparent in a majority of the cones, although fluorescence appeared faint (Fig. 2D inset). Large numbers of UV cones expressed GFP following Dox treatment for 16, 24, 48, and 72 h (Fig. 2E–H). Although no GFP fluorescence was observed in the untreated Tg(opn1sw1:rtTA, TRE:GFP) larval UV cones, a positive immunofluorescence signal was detected when labeled with anti-GFP antibody, indicating that a low level of Dox-independent GFP is expressed in this transgenic line (Fig. 2I). No anti-GFP immunofluorescence signal was detected in wild-type larval fish (Fig. 2J).
Figure 2. Time course of doxycycline-induced GFP expression in Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larvae.
Confocal z-projections of retinal sections from 6 dpf Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larvae that were (A) untreated or treated with 10 μg/mL Dox for (B) 2, (C) 4, (D) 8, (E) 16, (F) 24, (G) 48, or (H) 72 h. (A–H) Anti-UV opsin immunofluorescence (red) labels UV cone outer segments. GFP fluorescence (green) is apparent after 8 h of treatment (D, D inset) and continues to increase by number of cones with the increase in Dox exposure time (E–H). (I) Anti-GFP immunofluorescence (green) in untreated transgenic larvae indicates GFP expression that is not present in untreated, wild type larvae (J). Scale bar (A), 50 μm.
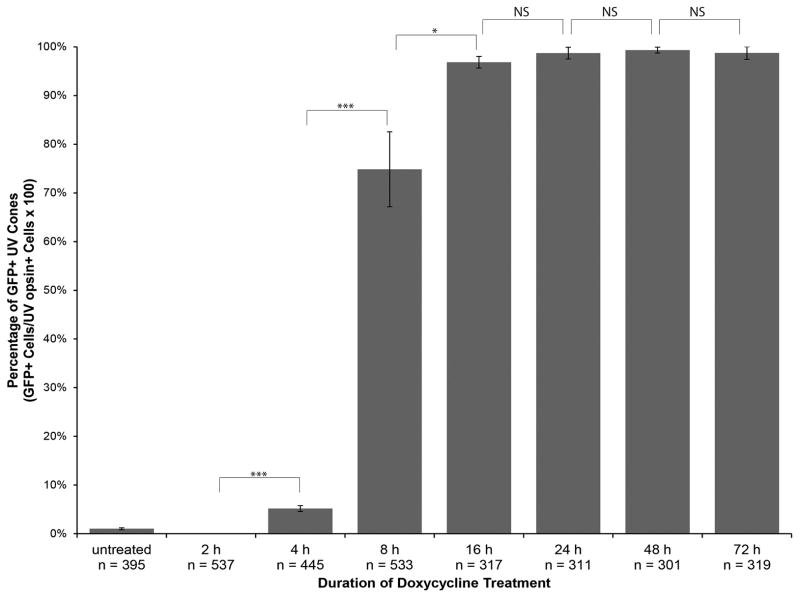
We quantified Dox-dependent GFP expression in UV cones by counting the number of cells expressing GFP as well as the number of cells that were positive for anti-UV opsin immunofluorescence. The ratio of the number of GFP-positive cones to anti-UV opsin positive cells was calculated from analysis of three confocal z-projections for each time point. The ratios were plotted as percentages of GFP positive UV cones against the duration of Dox treatment (Fig. 3). The graph indicates an increase in the number of GFP-positive cones with the increase in Dox treatment duration until a plateau is reached after approximately 16 h Dox, at which point over 96% of UV cones show expression. Statistically significant increases in GFP-positive cells occur with each doubling of Dox treatment time between 2 and 16 h of treatment. Changes in the percentage of GFP-positive UV cones was not significant between 16 and 24 h, 24 and 48 h, and 48 and 72 h of Dox treatment. Based on these results, we can conclude that the shortest amount of time to induce Dox-dependent gene expression in nearly all Tg(opn1sw1:rtTA, TRE:GFP) UV cones is approximately 16 h and the minimum amount of time necessary to induce gene expression with Dox treatment is 4 h.
Figure 3. Quantitative analysis of the time course of doxycycline-induced GFP expression in Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larvae.
Tg(opn1sw1:rtTA, TRE:GFP); alb−/− larvae were treated with 10 μg/mL Dox for the indicated durations prior to fixation at 6 dpf. Fixed larval tissues were processed for anti-UV opsin immunofluorescence to identify UV cones. UV opsin-positive cells (n) and GFP-positive cells were counted over 3 confocal z-projections from separate individuals for each treatment group. The percentage of UV cones that were also GFP-positive is plotted for each treatment group. Error bars represent standard deviation. Statistical comparisons by Student’s t-test are indicated by brackets. ***, p≤0.001; *, p≤0.05; NS, not significant.
2.3 Generation of a Stable Transgenic Line that Expresses an Epitope-Tagged rtTA in UV Cone Photoreceptors
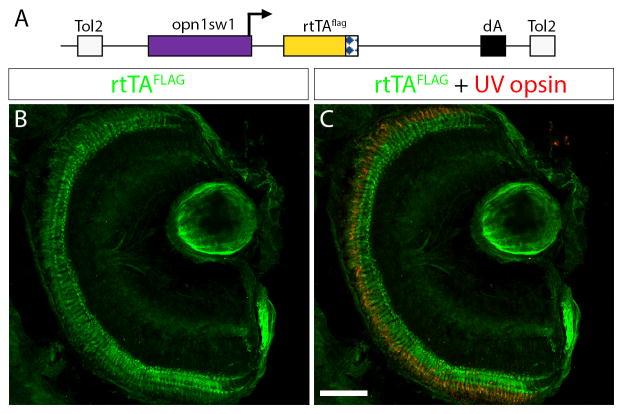
Expression of GFP in Tg(opn1sw1:rtTA, TRE:GFP) is useful to reflect rtTA activity with Dox treatment. In some experiments it may be more useful to have a fluorescent protein, like GFP, directly reflect the expression of the transgene of interest in the response plasmid or transgenic line. In this case a transgenic driver line that expresses only the rtTA protein is needed. We generated a construct where the opn1sw1 promoter drives expression of a C-terminal FLAG epitope-tagged rtTA protein (Fig. 4A). This construct was used to create a stable transgenic line, Tg(opn1sw1:rtTAflag), using the pTol transgenesis method (Kawakami et al., 2004). We examined retinal sections of 6 dpf Tg(opn1sw1:rtTAflag) larvae labeled with an anti-FLAG antibody and observed rtTAFLAG expression in the photoreceptor layer (Fig. 4B). In addition, anti-UV opsin immunofluorescence confirmed that rtTAFLAG expression was limited to UV cones (Fig. 4C). Over 93% of the UV cones that were positive for the anti-UV opsin signal were also positive for the anti-FLAG immunofluorescence signal.
Figure 4. Generation of UV-cone-specific, epitope-tagged, rtTA-expressing transgenic line for doxycycline-inducible gene expression.
(A) Diagram of the construct used to generate the stable transgenic zebrafish line, Tg(opn1sw1:rtTAflag); alb−/−, which expresses a FLAG-tagged rtTA protein specifically in UV cone photoreceptors. The UV opsin promoter (opn1sw1) drives expression of the flag-epitope tagged reverse tetracycline-controlled transcriptional activator (rtTAflag) gene. (B, C) Confocal z-projection of a retinal section from a 6 dpf Tg(opn1sw1:rtTAflag); alb−/− larva labeled with anti-FLAG and anti-UV opsin antibodies. (B) Anti-FLAG immunofluorescence (green) is visible in the photoreceptor layer of the retina. (C) Anti-UV opsin immunofluorescence (red) colocalizes with the anti-FLAG immunofluorescence (green) in UV opsin cone photoreceptors. dA, polyadenylation signal; Tol2, pTol integration site. Scale bar (C), 50 μm.
2.4 Bidirectional Transactivation of the Tetracycline Response Element
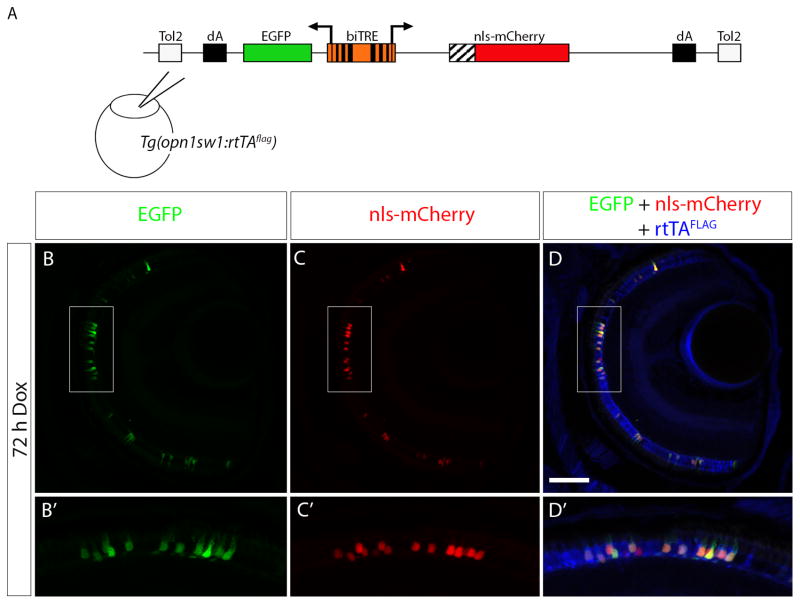
The epitope-tagged rtTA eliminates the need for a fluorescent protein reporter of rtTA activity and consequently allows for generation of response vectors or response transgenic lines that express both a fluorescent protein and the transgene of interest. We used the previously described bidirectional response construct, biTRE:EGFP, nls-mCherry, that drives expression of EGFP and nls-mCherry in the presence of rtTA and Dox (Campbell et al., 2012). The biTRE construct was injected into Tg(opn1sw1:rtTAflag) embryos at the one cell stage (Fig. 5A). In the absence of Dox, there was no EGFP or nls-mCherry fluorescence observed, but UV cones were positive for anti-FLAG immunofluorescence (data not shown). When the injected transgenic larvae were treated with Dox for 72 h prior to fixation at 6 dpf, EGFP fluorescence and nls-mCherry fluorescence were visible (Fig. 5B, C) in the photoreceptor layer along with anti-FLAG immunofluorescence (Fig. 5D). A higher magnification shows EGFP and nls-mCherry co-localized with the anti-FLAG immunofluorescence (Fig. 5 B′, C′, D′) in the UV cones. While in some instances one of the fluorescent proteins would appear brighter than the other, UV cones showed expression of both EGFP and nls-mCherry, indicating that the UV cone-specific FLAG-tagged rtTA protein, in the presence of Dox, was able to activate the expression of both genes that were driven by the biTRE.
Figure 5. Bidirectional transactivation of an injected biTRE-containing plasmid into Tg(opn1sw1:rtTAflag).
(A) Diagram of the bidirectional tetracycline response element (biTRE)-containing construct injected into Tg(opn1sw1:rtTAflag) one-cell embryos. EGFP and mCherry with a nuclear localization sequence (nls-mCherry) flank the biTRE. (B–D) Confocal z-projection of a retinal section from an injected 6 dpf Tg(opn1sw1:rtTAflag) larva treated with Dox for 72 hours and labeled with anti-FLAG antibody (blue). GFP fluorescence (B, B′; green) and nls-mCherry fluorescence (C, C′; red) are visible in the photoreceptor layer and co-localize with anti-FLAG immunofluorescence (D, D′; blue) in UV cones. Boxed regions in B, C, and D correspond to B′, C′, and D′, respectively. dA, polyadenylation sequence; Tol2, pTol integration site. Scale bar (D), 50μm.
2.5 Transactivation of a Stable Transgenic Tetracycline Response Element
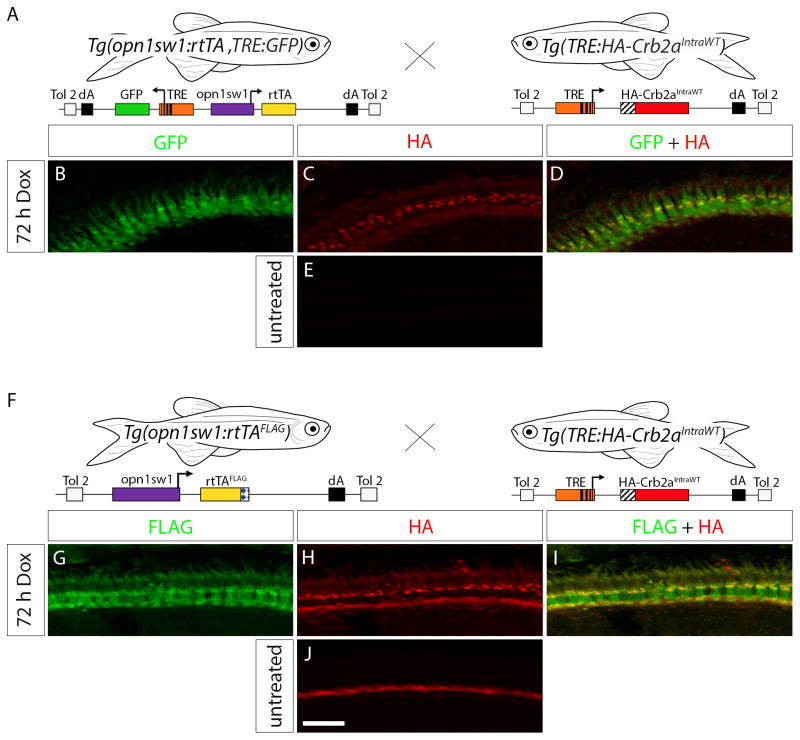
Injection of a construct containing a TRE-driven gene of interest into one-cell stage Tg(opn1sw1:rtTA, TRE:GFP) or Tg(opn1sw1:rtTAflag) allows for mosaic analysis where the transgene is incorporated into a subset of cones that can be compared to non-transgenic neighboring cells. Alternatively, generation of a stable transgenic line allows for transgenic analysis of inducible gene expression in the entire UV cone population. In order to demonstrate UV-specific expression of a stable transgenic, inducible gene of interest, we generated the stable transgenic line, Tg(TRE:HA-Crb2aIntraWT), and crossed to the Tg(opn1sw1:rtTA, TRE:GFP) line (Fig. 6A) and to the Tg(opn1sw1:rtTAflag) line (Fig. 6F). The TRE:HA-Crb2aIntraWT construct used for transgenic line generation encodes a protein comprising the Crb2b signal peptide and an HA-epitope tag fused to the transmembrane and intracellular domains of the zebrafish Crumbs Homolog 2a (Crb2a) protein under control of a TRE (Hsu and Jensen, 2010). Retinal sections of 6 dpf, 72 h Dox-treated Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) were labeled with anti-HA antibody. GFP and the HA-Crb2aIntraWT were both expressed in cone photoreceptors as shown by GFP fluorescence (Fig. 6B) and HA immunofluorescence (Fig. 6C) in the photoreceptor layer. The HA-Crb2aIntraWT protein appears to be predominantly localized to the base of the myoid region of the UV cone inner segments (Fig. 6D). Anti-HA immunofluorescence was not observed in untreated Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) retinas (Fig. 6E), but anti-GFP immunofluorescence signal was still present as a result of low level Dox-independent GFP expression from the Tg(opn1sw1:rtTA, TRE:GFP) line (data not shown). Retinal sections of 6 dpf, 72 h Dox-treated Tg(opn1sw1:rtTAflag; TRE:HA-Crb2aIntraWT) were labeled with anti-FLAG and anti-HA antibodies. FLAG immunofluorescence (Fig. 6G) and HA immunofluorescence (Fig. 6H) signals were detected in cone photoreceptors where the HA-Crb2aIntraWT was again localized to the inner segment myoid region (Fig. 6I). Anti-HA immunofluorescence was not observed in untreated Tg(opn1sw1:rtTAflag; TRE:HA-Crb2aIntraWT) retinas (Fig. 6J), indicating tight control of the inducible transgene. The rtTA protein can, therefore, transactivate a stable transgenic TRE and drive Dox-dependent expression of a gene of interest for transgenic analysis.
Figure 6. Transactivation in the Tg(TRE:HA-Crb2aIntraWT) line.
(A) Diagram indicating the cross of Tg(opn1sw1:rtTA, TRE:GFP) to Tg(TRE:HA-Crb2aIntraWT) for double-transgenic embryo production. (B–D) Confocal z-projection of a retinal section from a 6 dpf Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) larva treated with Dox for 72 h and labeled with anti-HA antibody. (B) GFP fluorescence (green) and (C) anti-HA immunofluorescence (red) indicate that HA-Crb2aIntraWT is expressed in UV cones (D, merge, yellow). (E) Confocal z-projection of an untreated 6 dpf Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) retinal section labeled with anti-HA antibody. (F) Diagram indicating the cross of Tg(opn1sw1:rtTAflag) to Tg(TRE:HA-Crb2aIntraWT) for double-transgenic embryo production. (G–I) Confocal z-projection of a retinal section from a 6 dpf Tg(opn1sw1:rtTAflag; TRE:HA-Crb2aIntraWT) larva treated with Dox for 72 h and labeled with anti-FLAG and anti-HA antibodies. (G) Anti-FLAG immunofluorescence (green) and (H) anti-HA immunofluorescence (red) indicate HA-Crb2aIntraWT is expressed in the UV cones (I, merge, yellow). (J) Confocal z-projection of an untreated retinal section from a 6 dpf Tg(opn1sw1:rtTAflag; TRE:HA-Crb2aIntraWT) larva labeled with anti-HA antibody. Scale bar (J), 10 μm.
3. Discussion
We created two types of transgenic lines for inducible, UV cone-specific gene expression in transgenic zebrafish using our Tet-On Toolkit (Campbell et al., 2012). We previously made transgenic lines for inducible, rod-specific gene expression, however there is also a need for investigations of cone-specific morphology, function, and disease. UV-sensitive cones are the first cones to mature in zebrafish (Fadool and Dowling, 2008) and their cell bodies remain below the outer limiting membrane adjacent to rod photoreceptor cell bodies (Raymond et al., 1993). Given these factors we chose to make transgenic lines to enable conditional gene expression in UV-sensitive cones. The Tet-On Toolkit (Campbell et al., 2012) was used with Gateway cloning (Kwan et al., 2007) to make two constructs with the reverse tetracycline-controlled transcriptional transactivator, rtTA, under the control of the ultraviolet opsin promoter, opn1sw1, (Takechi et al., 2003) for pTol-based transgenesis (Kawakami et al., 2004) in zebrafish. One construct included the green fluorescent protein (GFP) gene driven by a tetracycline response element (TRE) for self-reporting GFP activity. The other construct included an epitope tag on the rtTA, which eliminated the need for the self-reporting GFP. Colocalization data with anti-UV opsin immunofluorescence indicate that genes preceded by a TRE can be conditionally expressed with doxycycline (Dox) treatment in a UV cone-specific manner within the transgenic rtTA-expressing zebrafish.
There are several factors to consider in analyzing the inducible, UV cone-specific gene expression system including potential pathology of the rtTA protein and Dox treatment as well as the effectiveness of Dox treatment by bath application. Our confocal microscopy studies with GFP expression from the self-reporting rtTA line and anti-FLAG immunofluorescence in the epitope-tagged rtTA line provide full morphological view of the UV cones and demonstrate that rtTA protein expression and Dox treatment for up to 72 hours do not affect the morphology or survival of the UV cones. This is consistent with previous reports that rtTA protein and Dox treatment do not cause negative effects on cell and tissue morphology in zebrafish (Campbell et al., 2012; Huang et al., 2005; Knopf et al., 2010). Another factor of consideration was whether Dox treatment by bath application was sufficient for conditional gene expression in adult zebrafish. As shown previously with the rod-specific Tet-On system (Campbell et al., 2012), GFP expression from the self-reporting, UV cone-specific, rtTA transgenic line was induced in adults after 72 hours of submersion in Dox-treated water, thereby making intraperitoneal injection unnecessary.
The pharmacokinetics of Dox delivery to the zebrafish eye has not been studied in extensive detail, however in previous studies Dox was able to activate rtTA expression in approximately 90% of rod photoreceptors within 24 hours of bath application (Campbell et al., 2012). The data here indicate slightly faster rtTA activation in UV cones since rtTA-driven gene expression was induced by Dox in 96% of UV cones within 16 hours of treatment. This result was determined by self-reporting GFP expression from the Tg(opn1sw1:rtTA, TRE:GFP) line. Additional independently-integrated Dox-inducible transgenes may exhibit different time courses of expression, so individual characterization of transgenes will be necessary. Another factor of consideration was the tightness of the system in the absence of Dox treatment. Our results illustrate that GFP fluorescence from the self-reporting rtTA line is not apparent in the absence of Dox, however, labeling with a polyclonal anti-GFP antibody suggests that the system exhibits a low level of self-reporting activity in the absence of treatment. Further investigation of untreated, transgenic fish shows that additional, independently-integrated transgenes do not express without Dox induction, suggesting that the uninduced GFP expression is specific to the TRE:GFP transgene within the self-reporting rtTA transgenic line, Tg(opn1sw1:rtTA, TRE:GFP). As demonstrated by the diagram in Figure 1A, the TRE:GFP was a component of the same construct as the opn1sw1-driven rtTA. We suspect that the proximity of the TRE:GFP to the opn1sw1 promoter may contribute to the uninduced GFP expression seen in the absence of Dox treatment. Given that uninduced GFP expression was only an issue with the self-reporting rtTA driver line and not with other independently incorporated TRE-driven transgenes, we do not anticipate expression issues with transgenes of interest in the untreated state as shown with anti-HA labeling in untreated Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) larvae in Figure 6E. Independent characterization will be required to confirm this.
We find that expression of the TRE:HA-Crb2aIntraWT transgene is Dox dependent in Tg(opn1sw1:rtTA, TRE:GFP; TRE:HA-Crb2aIntraWT) and Tg(opn1sw1:rtTAflag; TRE:HA-Crb2aIntraWT) larval UV cones as HA-Crb2aIntraWT expression was not observed in the absence of Dox. Previous studies with transgenic HA-Crb2aIntraWT expression in rods demonstrated that the HA-Crb2aIntraWT protein localized to the outer segments of rod photoreceptors (Campbell et al., 2012; Hsu and Jensen, 2010). However, here we find that UV cone-specific HA-Crb2aIntraWT protein localizes to the myoid region of the UV cone inner segment, illustrating an interesting protein localization difference between these cell types.
The data shown here describe an ultraviolet-sensitive cone-specific system for conditional gene expression in zebrafish. These lines add to the rod photoreceptor Tet-On driver lines and are the first system for inducible, cone-specific gene expression. Genes of interest for investigation of cone-specific morphology, function, or disease can be incorporated into the system by recombining genes downstream of the TRE and introducing into one of the rtTA transgenic lines. The rtTA line with self-reporting GFP activity provides fluorescent protein expression to give full view of cone morphology. Alternatively, the epitope-tagged line eliminates the use of GFP to allow green fluorescence to be reserved for immunofluorescence purposes or for creating a GFP fusion with a protein of interest. In addition, a bidirectional TRE can be used to simultaneously express two genes of interest, perhaps if a transgene of interest cannot be epitope tagged.
As demonstrated in this study, inducible transgenes can be expressed mosaically by injection into the one-cell stage transgenic rtTA zebrafish embryo or by an independently-generated transgenic line crossed to one of the rtTA driver lines. In mosaic analysis, cones expressing the transgene of interest can be compared to non-expressing neighboring cells, whereas in transgenic analysis the transgene-expressing cones can be compared to cones expressing a benign transgene, such as a fluorescent protein, or to cones from an untreated sibling. The expansion of the Tet-On Toolkit means that UV cone morphology and function can be investigated independently of other photoreceptors and also that complementary studies between zebrafish rods and UV cones can be pursued.
4. Experimental Procedures
4.1 Animals
Wild-type AB, albinob4/+ (alb−/+) (Dooley et al., 2013; Tsetskhladze et al., 2012), Tg(opn1sw1:rtTA, TRE:GFP), Tg(opn1sw1:rtTAflag), and Tg(TRE:HA-Crb2aIntraWT) zebrafish lines were maintained and bred by standard procedures (Westerfield, 1995). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the University of Massachusetts IACUC (Protocol Number: 2012-0020).
4.2 Plasmid Constructs
A XhoI-SacII fragment from pEGFP-SWS1 containing the UV opsin promoter (opn1sw1) (Takechi et al., 2003) was cloned into the Gateway entry vector, p5′E, to create p5′E-SWS1. The p5′E-SWS1 was recombined using LR Clonase® II Plus (Invitrogen, Carlsbad, California) in three-way recombinations with pL1L2-rtTA (AJ3) and pTolDestR4-R2pA-TRE:GFP (AJ6) to create opn1sw1:rtTA, TRE:GFP or with pL1L2-rtTAflag (AJ13) and pTolDestR4-R2pA (NL#465) to create opn1sw1:rtTAflag. The Tet-On components were from the Tet-On Toolkit (Campbell et al., 2012) and the Lawson Laboratory reagents (Villefranc et al., 2007). The SV40 early T antigen polyadenylation signal was used for the dA sequence. Tg(TRE:HA-Crb2aIntraWT) was described previously (Campbell et al., 2012). Information and details about Tet-On Toolkit components are available online at http://www.bio.umass.edu/biology/jensen/node/5 (Campbell et al., 2012).
4.3 Transgenesis and Genotyping
Transgenic zebrafish lines were generated using the pTol system (Kawakami et al., 2004). A mixture containing Tg(opn1sw1:rtTA, TRE:GFP) or Tg(opn1sw1:rtTAflag) plasmid DNA (40 ng/μL), pTol transposase mRNA (40 ng/μL), and phenol red was injected into one-cell stage wild-type AB embryos. Injected embryos were reared to adulthood and out-crossed with alb−/+. The offspring were used to identify germline transgenic founder (F0) fish via PCR. Forward primer, CCTTTTCATCCCCGCCTTTTCACACC, and reverse primer, GAAAAGGAAGGCAGGTTCGGCTC, were used for genotyping both the Tg(opn1sw1:rtTA, TRE:GFP) and Tg(opn1sw1:rtTAflag) transgenic lines. Offspring from identified F0 fish were reared to adulthood to generate F1 transgenic fish that were genotyped using fin clip PCR with the above primers.
4.4 Doxycycline Treatment
Doxycycline hyclate (Sigma-Aldrich, St. Louis, Missouri) dissolved in ethanol at a stock concentration of 10 mg/mL was added to fish water for a final concentration of 10 μg/mL. Larval and adult fish were submerged by bath application in the Dox-treated water for a time ranging from two to 72 hours.
4.5 Immunofluorescence and Microscopy
Larval fish were fixed in 4% paraformaldehyde for one hour and adult fish were fixed in 4% paraformaldehyde for four hours. Fixed specimens were rinsed with phosphate buffered saline (PBS). Fixed fish heads were embedded in a 1.5% agar/5% sucrose media and corresponding bodies were used for genotyping. Embedded tissues were equilibrated in a 30% sucrose solution and then sectioned at 25 μm using a Leica CM1850 cryostat (Solms, Germany).
Tissue slides were rehydrated with PBS for fifteen minutes and incubated for several hours with a blocking solution of 20% goat serum in PBS with 0.1% Triton X-100. Primary antibodies were diluted in 0.1% Tween in PBS (PBS-Tw) and incubated on the slides at 4°C overnight. Slides were washed with PBS-Tw for one hour and then incubated with the appropriate secondary antibody diluted in PBS-Tw for 5 hours at room temperature. Slides were washed with PBS-Tw for one hour and mounted with ProLong® Gold Antifade Reagent (Invitrogen). Primary antibodies and dilutions include rabbit anti-GFP (Molecular Probes®/Invitrogen) at 1:1,000, rabbit anti-UV opsin (gift from David Hyde, University of Notre Dame, Notre Dame, Indiana) at 1:200, monoclonal mouse IgG1 anti-FLAG® M2 (Sigma) at 1:500, monoclonal mouse IgG2a anti-Rhodopsin R6-5 (Röhlich et al., 1989) at 1:200, monoclonal mouse IgG1 anti-HA (Covance, Princeton, New Jersey) at 1:1,000, and monoclonal mouse IgG3 anti-HA (Upstate®/Millipore, Billerica, Massachusetts) at 1:500. Secondary antibodies and dilutions include Alexa Fluor® 488-conjugated goat anti-mouse (Invitrogen) at 1:200, Alexa Fluor® 488-conjugated goat anti-rabbit (Invitrogen) at 1:1,000, Alexa Fluor® 546-conjugated goat anti-rabbit (Invitrogen) at 1:200, Cy5-conjugated goat anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania) at 1:50, FITC-conjugated goat anti-mouse IgG1 (Jackson ImmunoResearch Laboratories) at 1:200, and Rhodamine Red-conjugated goat anti-mouse IgG3 (Jackson ImmunoResearch Laboratories) at 1:100. Imaging was performed using a Zeiss LSM 700 confocal microscope (Okerkochen, Germany).
4.6 Cell Counting
To determine the ratio of Dox-induced, GFP-expressing UV cones in relation to the total UV cone population, z-projections of whole eye sections were obtained and the UV cones were counted using Volocity 3D imaging software (PerkinElmer, Waltham, Massachusetts). Three z-projections, each from a separate individual, were counted for each treatment group. The total number of UV cones was determined by counting the number of cells with anti-UV opsin signal. UV-opsin positive cells that were GFP-negative were subtracted from the total UV cone number to give the number of Dox-induced, GFP-expressing UV cones. The percentage of GFP-positive UV cones was plotted against the duration of Dox treatment. Standard deviation was calculated for each treatment group and statistical significance between treatment groups was determined using Student’s t-test (Microsoft Excel, Redmond, Washington).
*Highlights.
Two transgenic zebrafish lines have been created for inducible transgene expression in cones.
Induction by doxycycline is robust, rapid and UV-cone specific.
These transgenic lines will facilitate the study of cone development and function.
Acknowledgments
The authors thank Judy Bennett for fish care, David Hyde (University of Notre Dame) for the UV opsin antibody, Shoji Kawamura (The University of Tokyo) for the SWS1 (opn1sw1) promoter, and Nathan Lawson (University of Massachusetts Medical School), Chi-Bin Chien, and Kristen Kwan (University of Utah) for generously sharing Gateway vectors.
Sources of Funding: EY015420
Footnotes
None of the authors have any proprietary/financial interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Megan C. West, Email: mcwest@bio.umass.edu.
Leah J. Campbell, Email: leahc@bio.umass.edu.
John J. Willoughby, Email: johnw@bio.umass.edu.
References
- Albalat R. Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol Biol Evol. 2012;29:1461–1469. doi: 10.1093/molbev/msr313. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Hurley JB, Janssen-Bienhold U, Neuhauss SC, Driever W, Dowling JE. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc Natl Acad Sci U S A. 1995;92:10545–10549. doi: 10.1073/pnas.92.23.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LJ, Willoughby JJ, Jensen AM. Two types of Tet-On transgenic lines for doxycycline-inducible gene expression in zebrafish rod photoreceptors and a gateway-based tet-on toolkit. PLoS ONE. 2012;7:e51270. doi: 10.1371/journal.pone.0051270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Dooley CM, Schwarz H, Mueller KP, Mongera A, Konantz M, Neuhauss SC, Nusslein-Volhard C, Geisler R. Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Melanoma Res. 2013;26:205–217. doi: 10.1111/pcmr.12053. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Brockerhoff SE, Hyatt GA, Dowling JE. Mutations affecting eye morphology in the developing zebrafish (Danio rerio) Developmental Genetics. 1997;20:288–295. doi: 10.1002/(SICI)1520-6408(1997)20:3<288::AID-DVG11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Dowling JE. Zebrafish: A model system for the study of eye genetics. Progress in Retinal and Eye Research. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Jensen A. Multiple domains in the Crumbs Homolog 2a (Crb2a) protein are required for regulating rod photoreceptor size. BMC Cell Biol. 2010;11:60. doi: 10.1186/1471-2121-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Jou TS, Ho YL, Lee WH, Jeng YT, Hsieh FJ, Tsai HJ. Conditional expression of a myocardium-specific transgene in zebrafish transgenic lines. Dev Dyn. 2005;233:1294–1303. doi: 10.1002/dvdy.20485. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Knopf F, Schnabel K, Haase C, Pfeifer K, Anastassiadis K, Weidinger G. Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proc Natl Acad Sci U S A. 2010;107:19933–19938. doi: 10.1073/pnas.1007799107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: A multisite Gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lamb TD. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 2013 doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Le YZ, Bai Y, Zhu M, Zheng L. Temporal requirement of RPE-derived VEGF in the development of choroidal vasculature. J Neurochem. 2010;112:1584–1592. doi: 10.1111/j.1471-4159.2010.06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Mutations affecting development of the zebrafish retina. Development. 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Rounsifer ME, Sullivan SA, Knight JK. Expression of rod and cone visual pigments in goldfish and zebrafish: A rhodopsin-like gene is expressed in cones. Neuron. 1993;10:1161–1174. doi: 10.1016/0896-6273(93)90064-x. [DOI] [PubMed] [Google Scholar]
- Röhlich P, Adamus G, Hugh McDowell J, Hargrave PA. Binding pattern of anti-Rhodopsin monoclonal antibodies to photoreceptor cells: An immunocytochemical study. Exp Eye Res. 1989;49:999–1013. doi: 10.1016/s0014-4835(89)80022-3. [DOI] [PubMed] [Google Scholar]
- Takechi M, Hamaoka T, Kawamura S. Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett. 2003;553:90–94. doi: 10.1016/s0014-5793(03)00977-3. [DOI] [PubMed] [Google Scholar]
- Tsetskhladze ZR, Canfield VA, Ang KC, Wentzel SM, Reid KP, Berg AS, Johnson SL, Kawakami K, Cheng KC. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS ONE. 2012;7:e47398. doi: 10.1371/journal.pone.0047398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Pease ME, Wersinger DM, Masuda T, Vinores SA, Licht T, Zack DJ, Quigley H, Keshet E, Campochiaro PA. Prolonged blockade of VEGF family members does not cause identifiable damage to retinal neurons or vessels. J Cell Physiol. 2008;217:13–22. doi: 10.1002/jcp.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (Danio rerio) 3. Univ. of Oregon Press; Eugene: 1995. The zebrafish book. [Google Scholar]