Abstract
In prostate and other epithelial cancers, E-cadherin (CDH1) is downregulated inappropriately by DNA methylation to promote an invasive phenotype. Though cancer frequently involves a reawakening of developmental signaling pathways, whether DNA methylation of Cdh1 occurs during organogenesis has not been determined. Here we show that DNA methylation of Cdh1 mediates outgrowth of developing prostate ducts. During the three-day gestational window leading up to and including prostate ductal initiation, Cdh1 promoter methylation increases and its mRNA and protein abundance decreases in epithelium giving rise to prostatic buds. DNA methylation is required for prostate specification, ductal outgrowth, and branching morphogenesis. All three endpoints are impaired by a DNA methylation inhibitor, which also decreases Cdh1 promoter methylation and increases Cdh1 mRNA and protein abundance. A CDH1 function-blocking antibody restores prostatic identity, bud outgrowth, and potentiates epithelial differentiation in the presence of the DNA methylation inhibitor. This is the first study to mechanistically link acquired changes in DNA methylation to the normal process of prostate organogenesis. We propose a novel mechanism whereby Cdh1 promoter methylation restricts Cdh1 abundance in developing prostate epithelium to create a permissive environment for prostatic bud outgrowth. Thus, DNA methylation primes the prostate primordium to respond to developmental cues mediating outgrowth, differentiation and maturation of the ductal network.
Keywords: Prostate, epigenetics, organogenesis, 5-aza-2′-deoxycytidine, urogenital sinus, lower urinary tract
Introduction
E-cadherin (Cdh1) fine tunes cell adhesion, migration, and intracellular signaling dynamics during epithelial development and is a defined mediator of breast, salivary gland, hair follicle, lung and intestinal epithelial morphogenesis (Boussadia et al., 2002; Ewald et al., 2012; Hermiston et al., 1996; Hirai et al., 1989; Hsu et al., 2013; Jamora et al., 2003; Nanba et al., 2001; Reardon et al., 2012; Tinkle et al., 2008; Walker et al., 2008; Young et al., 2003). Though Cdh1 is specifically required for early embryogenesis (Larue et al., 1994), the need for localized control of its activity persists throughout development and adulthood (Boussadia et al., 2002; Hermiston et al., 1996; Reardon et al., 2012). CDH1 also maintains mature epithelial homeostasis and its down-regulation associates with an invasive phenotype in some cancers (van Horssen et al., 2012).
Regulators of CDH1 expression have been identified in developing tissues, including growth factors like TGF-β and FGF, transcriptional repressors such as Snail and Slug and other signaling pathways like NOTCH and WNT (Carraro et al., 2010; Herfs et al., 2008; Jamora et al., 2003; Jamora et al., 2005; Sirour et al., 2011; Wang et al., 2012). These same pathways regulate Cdh1 in epithelial cancers, as do other regulators. One example is DNA methylation, which frequently down-regulates Cdh1 in epithelial tumors to create an invasive phenotype (Graff et al., 1995; Yoshiura et al., 1995). Whether DNA methylation also controls Cdh1 expression and epithelial behavior during organ morphogenesis has not been investigated. Here, for the first time we reveal a novel mechanism whereby DNA methylation of Cdh1 is necessary for prostate morphogenesis.
The prostate develops under the control of androgens in a region of the pelvic urethra known as the urogenital sinus (UGS). Prostate buds initiate as small epithelial projections that elongate into surrounding stroma, undergo branching morphogenesis, and arborize into the mature ductal network. How developing prostate ducts elongate into surrounding stroma during normal development is a particularly intriguing question because it may bear mechanistic similarities to the invasive behavior of prostate cancer. We describe an entirely new mechanistic link between DNA methylation and prostate ductal morphogenesis and the role of Cdh1 in this process. Cdh1 promoter methylation increases and its mRNA and protein abundance decrease in basal epithelial cells giving rise to prostate, and these events are necessary for ductal outgrowth and specification. We propose a priming mechanism whereby DNA methylation restricts Cdh1 abundance in cells that give rise to the prostate, thereby creating a permissive environment for continued development and morphogenesis of prostatic ducts. Our results are the first to reveal a requirement for DNA methylation and CDH1 in prostate development, and describe how these events are mechanistically linked to orchestrate prostate ductal morphogenesis.
Materials and Methods
Animals
C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, ME), housed in polysulfone cages containing corn cob bedding and maintained on a 12 hour light and dark cycle at 25±5°C and 20–50% relative humidity. Feed (Diet 2019 for males and Diet 7002 for pregnant females, Harlan Teklad, Madison, WI. USA) and water were available ad libitum. All procedures were approved by the University of Wisconsin Animal Care and Use Committee and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Females were paired overnight with males to obtain timed-pregnant dams. The next morning was considered 0 days post coitus (dpc). Dams were euthanized by CO2 asphyxiation.
In situ hybridization (ISH)
ISH was conducted on paraformaldehyde (PFA) fixed urogenital sinus (UGS) which were cut into sections with a cryotome or vibrating microtome prior to staining, or stained in whole-mount as described previously (Abler et al., 2011a; Abler et al., 2011b; Keil et al., 2012a; Keil et al., 2012b). Detailed protocols for PCR-based riboprobe synthesis are available at www.gudmap.org. The staining pattern for all riboprobes was assessed in two sections from at least three litter-independent UGSs. Tissue sections were processed in the same tube as a single experimental unit to allow for qualitative comparisons among biological replicates and among treatment groups.
Organ Culture
Male or female UGSs were removed from 14 dpc embryos and cultured for four to seven days as described previously (Vezina et al., 2008). Media was supplemented with 10 nM 5α-dihydrotestosterone (DHT) and one or more of the following treatments; vehicle control (0.1% dimethyl sulfoxide, DMSO), 5 μM 5-aza-2′-deoxycytidine (5AzadC, A3635, Sigma-Aldrich, St. Louis, MO. USA), 100 μg/mL anti-CDH1 function blocking antibody targeting the fifth ectodomain repeat (EC5) of CDH1 (U3254, DECMA-1, Sigma-Aldrich). Media and supplements were changed every 2 days. Sodium azide, which was found to be toxic to explant tissue cultures (results not shown), was removed from the CDH1 blocking antibody prior to culture using a 10,000 molecular weight cutoff dialysis cassette (Pierce, Rockford, IL. USA).
Immunohistochemistry (IHC)
Immunofluorescent staining of ISH-stained and paraffin sections was performed as described previously (Abler et al., 2011a). Primary antibodies were diluted as follows: 1:200 rabbit anti-CDH1 (3195, Cell Signaling Technology, Beverly, MA. USA), 1:200 rabbit anti-KI67 (Ab15580, Abcam, Cambridge, MA. USA), 1:50 mouse anti-KRT14 (ms-115-p0, Thermo Fisher Scientific, Waltham, MA. USA), 1:200 rabbit anti-cleaved caspase 3 (Asp 175) (9661S, Cell Signaling Technology), 1:250 rabbit anti-AR (sc-816, Santa Cruz Biotechnology, Santa Cruz, CA. USA), 1:50 mouse anti-TRP63 (Sc-8431, Santa Cruz Biotechnology), 1:250 rabbit anti-pan cytokeratin (18-0059, Life Technologies, Grand Island, NY. USA). Secondary antibodies were diluted as follows: 1:250 Dylight 549-conjugated goat anti-rabbit IgG (111-507-003, Jackson ImmunoResearch, West Grove, PA. USA), 1:250 AlexaFluor 594-conjugated goat anti-rabbit IgG (A11012, Life Technologies), 1:250 Dylight 488-conjugated goat anti-mouse IgG (115-487-003, Jackson ImmunoResearch), 1:250 Dylight 488-conjugated goat anti-rat IgG (112-486-003, Jackson ImmonoResearch), 1:250 Dylight 488-conjugated goat anti-rabbit IgG (111-487-003, Jackson ImmunoResearch). Labeled tissues were counterstained with 4′,6-diamidino-2-phenylindole, dilactate (DAPI), to label cell nuclei or wheat germ agglutinin (W21505, Life Technologies) to label cell membranes and mounted in anti-fade media (phosphate buffered saline containing 80% glycerol and 0.2% n-propyl gallate). Whole mount immunohistochemistry on ISH stained tissue was performed as described previously (Keil et al., 2012a). Primary antibody was rabbit anti-CDH1 diluted 1:750 and secondary antibody was biotin conjugated goat anti-rabbit IgG diluted at 1:500 (BA-1000, Vector, Burlingame, CA. USA). Following ISH and IHC, some tissues were paraffin embedded and sectioned as described previously (Keil et al., 2012a) and counterstained with nuclear fast red. Brightfield images were captured using an Eclipse 80i compound microscope for sections and a SMZ1000 microscope for whole mount, fluorescent images were captured using an Eclipse E600 compound microscope (Nikon Instruments Inc., Melville, NY. USA) and merged using NIS elements imaging software (Nikon Instruments Inc.). All treatment groups were imaged using the same settings and captured at the same exposure.
Methylated DNA Immunoprecipitation (MeDIP)
Genomic DNA was isolated using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA. USA) according to manufacturer’s instructions. Two micrograms of genomic DNA was diluted into a 50 μl volume with phosphate buffered saline + 0.1% Tween 20 (PBS) and was fragmented by 30 cycles of 30 seconds on, 30 seconds off in an ice cold sonicating water bath (Chicago Electric 120v/60Hz/35W, model 3305). Agarose gel electrophoresis was used to confirm efficient DNA fragmentation to an average size of 200–1000 base pairs. Fragmented DNA was boiled for 10 minutes and cooled on ice for 10 minutes. Each sample was divided into two fractions with 10% of the divided fraction saved as input control. 2 μg anti-mouse IgG antibody (ab18413, Abcam) was added to one fraction, 2 μg anti-5-methyl cytidine antibody (33D3 ab10805, Abcam) was added to the other fraction and both fractions were incubated overnight at 4°C. The conjugated antibodies were captured during a 2 hour incubation at 4°C with 30 μL/mL of Protein A Agarose/Salmon Sperm DNA Beads (#16-157, Millipore, Billerica, MA. USA). Beads were washed with PBS, resuspended in 100 μL of digestion buffer (50 mM Tris pH8, 10 mM EDTA, 0.5% SDS) containing 0.4 mg/mL Proteinase K and incubated overnight on a rotating platform at 50°C to release the DNA. Samples were washed twice with digestion buffer and supernatant collected. MeDIP DNA was purified using the QIAquick PCR Purification Kit (Qiagen, #28104). Real time quantitative PCR (QPCR) was performed as described previously (Keil et al., 2012b) using gene specific primers for Cdh1: 5′-CACGGAGGGAGAACAATGTAAG-3′ and 5′-TATGCATCCCACATCCATCAG-3′ (Entrez gene ID 12550, NC_000074.6). Results were analyzed using the ΔCt method and expressed as enrichment over IgG (Livak and Schmittgen, 2001). Results are representative of at least three litter independent pools of DNA with at least five tissues per pool.
Flow Cytometry
Single cell suspensions from pooled intact male 17 dpc UGSs (3–5 UGSs/pool) were generated as described previously (Lukacs et al., 2010). UGSs were dissected to remove seminal vesicle rudiments and the bladder was trimmed at the bladder neck. Cells were labeled with the following fluorescently conjugated primary antibodies: rat anti-CD49f-PE, rat anti-CD31-FITC, rat anti-CD45-FITC and rat anti-TER119-FITC (clones: GoH3, 390, 30-F11, and TER-119 respectively, eBioscience, San Diego, CA. USA) as described previously (Lukacs et al., 2010), washed in 0.5% BSA/PBS, fixed in 2% PFA for 10 minutes and permeabilized with PBS + 0.5% Tween20 at room temperature for 15 minutes. Cells were then incubated with unconjugated rabbit anti-CDH1 primary antibody diluted at 1:200 (3195, Cell Signaling Technology) at room temperature for 30 minutes, followed by incubation with APC-conjugated goat anti-rabbit secondary antibody diluted at 1:100 (Sc-3846, Santa Cruz) at room temperature for 30 minutes. Cells were washed in PBS and stored in 2% PFA protected from light until analyzed on a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA. USA). Single antibody staining controls were used for each sample. Flow cytometry data were analyzed using FlowJo software (TreeStar Inc. Ashland, OR. USA).
Renal Grafting
Female UGSs removed at 14 dpc were cultured for seven days as described above in media containing 10 nM DHT and either vehicle control (0.1% DMSO) or 5 μM 5AzadC. Tissues were then placed under the renal capsule of intact male syngeneic (C57BL/6J) host mice and grown for 1 month. Graft volume was calculated using the modified ellipsoid formula of (Euhus et al., 1986; Tomayko and Reynolds, 1989).
Statistical analyses
Prostatic buds were counted as described previously (Keil et al., 2012a). Prostatic bud length, from the base to the distal tip, was measured for at least 6 buds per UGS in at least three UGSs per treatment group. The frequency of immunolabeled Ki-67 positive, KRT14 positive and double-positive cells was determined in at least two sections per UGS in at least three UGSs per treatment group. Image J software was used to determine the relative integrated density, a measure of the sum of converted grayscale values of the pixels in a field, for red and green channels captured from 3 basal epithelial fields per UGS in at least three litter independent samples per group. Statistical analysis was performed using R version 2.13.1. Homogeneity of variance was determined using Levene’s test. Student’s t-test, ANOVA or Kruskal-Wallis test followed by Tukey’s Honestly Significant Difference (HSD) were used to identify significant differences (p ≤ 0.05) between or among treatment groups.
Results
Cdh1 mRNA and protein expression are diminished in a prostate epithelial cell subpopulation during male prostatic bud initiation
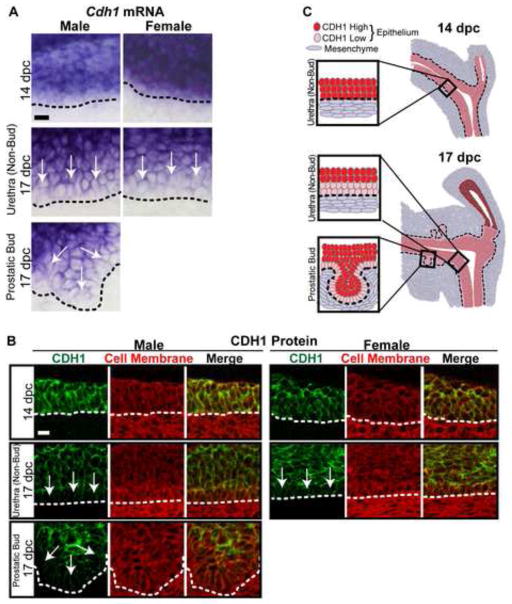
In situ hybridization (ISH) and immunohistochemistry (IHC) were used to visualize Cdh1 mRNA and protein expression patterns within the male and female UGS prior to (14 dpc) and during (17 dpc) prostatic bud outgrowth in males. Prostate develops only in male C57BL/6J mice under normal hormonal conditions (Price and Williams-Ashman, 1961), though both sexes are competent to form prostatic buds in response to androgens during this time period (Bard et al., 1979; Price and Williams-Ashman, 1961; Lasnitzki and Mizuno, 1980). IHC sections were counterstained with the cell membrane marker wheat germ agglutinin to visualize the epithelial-mesenchymal boundary and to evaluate epithelial membrane density. Cdh1 is present in all epithelial cells at the stages examined, but differs in its relative abundance among epithelial layers (Fig. 1). Cdh1 mRNA and protein staining is abundant and ubiquitously expressed across male and female UGS epithelium at 14 dpc, but a graded pattern emerges in both sexes by 17 dpc when prostatic buds are forming in C57BL/6J male mice (Fig. 1A,B). The graded staining pattern features strong Cdh1 staining in epithelial cells closest to the urethral lumen and weak staining in cells closest to the basement membrane (Fig. 1C). The weakest Cdh1 mRNA and protein staining is in prostatic bud tips (Fig. 1). These results reveal a change in Cdh1 expression during the window when the UGS becomes competent to form prostate. Additional staining was used to determine that CDH1 diminishes specifically in KRT14+ basal epithelium, the epithelial cell type present in developing prostatic buds (Fig. 2A,B), and that is required for prostatic bud formation (Kurita et al., 2004; Signoretti et al., 2000). Because this change is observed across male and female UGS, it is not likely to be reliant on the presence of testicular androgens.
Fig. 1.
E-cadherin (Cdh1) mRNA and protein expression are diminished in a specific UGS epithelial cell subpopulation during the period of prostatic bud formation. UGS sections were stained to visualize Cdh1 abundance within urethra and prostatic bud epithelium. (A) Male and female sagittal sections (50 μm) were stained by ISH to visualize Cdh1 mRNA expression (purple). (B) Male and female sagittal sections (5 μm) were immunofluorescently labeled to detect CDH1 (green) and cell membranes (wheat germ agglutinin, red). (C) Schematic representation of E-cadherin mRNA and protein expression within UGS epithelium prior to (14 dpc) and during (17 dpc) prostatic bud formation. A dotted line indicates the interface between epithelium and mesenchyme, as determined by cell morphology. Arrows indicates areas of diminished E-cadherin mRNA and protein abundance. Scalebar represents 10 μm.
Fig. 2.
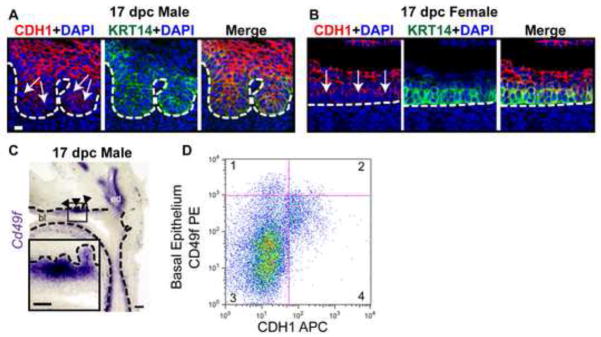
Diminished E-cadherin (Cdh1) mRNA and protein abundance occurs in the subpopulation of basal epithelial cells during prostatic bud outgrowth. (A-B) 17 dpc male and female UGS sagittal sections (5 μm) were immunofluorescently labeled to detect E-cadherin (CDH1, red) and basal epithelial cell marker keratin 14 (KRT14, green). Cell nuclei were stained with DAPI (blue). Scalebar 10 μm. (C) 17 dpc male UGS sagittal sections (50 μm) were stained by ISH to confirm localization of Cd49f mRNA (purple) in basal epithelium. Scalebar 100 μm, inset 50 μm. (D) Flow cytometry plot of 17 dpc UGS single cell suspensions labeled with CD49f-PE/CDH1-APC in the Lin (CD31, TER-119, CD45) negative population. A dotted line indicates the interface between epithelium and mesenchyme, as determined by cell morphology. Arrows indicate areas of diminished CDH1 abundance. Closed arrowheads indicate prostatic buds. Abbreviations: bl, bladder; ed, ejaculatory duct..
We used independent markers and methodology to further investigate the relationship between basal epithelial cell identity and CDH1 abundance. CD49f (also known as integrin alpha 6, Itga6) marks mature prostate basal epithelium (Lukacs et al., 2008; Lukacs et al., 2010; Lawson et al., 2007) and we confirmed it also marks fetal prostate basal epithelium within the urethra and developing prostate buds (Fig. 2C, inset). We conducted flow cytometry on lineage (Ter119, CD45, CD31) negative intact 17 dpc male UGS cells to exclude erythrocytes (TER-119), hematopoetic (CD45) and endothelial (CD31) cells. The cells were then sorted on the basis of CD49f and CDH1 abundance. The CD49fHigh, CDH1Low (quadrant 1) illustrate that basal epithelial cells have low CDH1 expression (Fig. 2D), consistent with our results in Figure 1. The CD49fHigh, CDH1High (quadrant 2) cells likely represent proximal rudiments of ejaculatory duct which remain even after removal of seminal vesicle and maintain high CDH1 and CD49f expression at this stage (Fig. 2C). The CD49fLow, CDH1Low cells (quadrant 3) represent stromal cells and the CD49fLow, CDH1High cells (quadrant 4) represent the non-basal cells within the epithelium. These results in conjunction with those in Figure 1 support the hypothesis that during prostate ductal initiation, Cdh1 mRNA and protein expression are decreased in basal epithelium giving rise to prostatic buds.
Cdh1 promoter methylation increases in UGS epithelium during the period of prostatic bud outgrowth
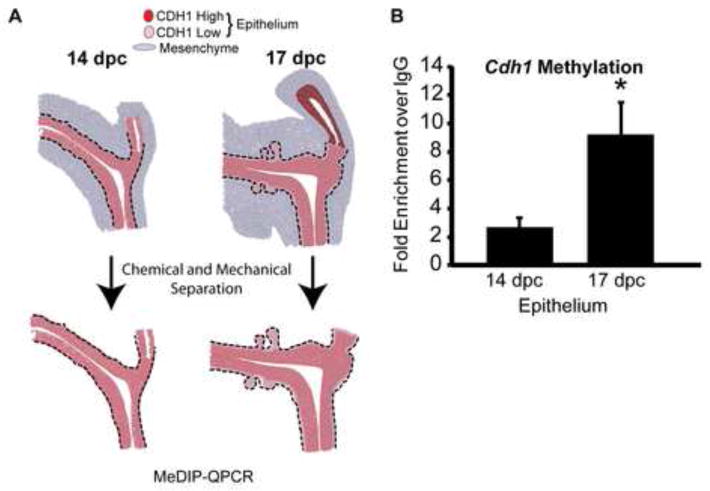
Our next objective was to determine why Cdh1 abundance diminishes in UGS basal epithelium during prostate development. Two factors led us to consider DNA methylation as a possible mechanism: DNA methylation represses Cdh1 expression in prostate and other cancers (Yoshiura et al., 1995, Graff et al., 1995) and the enzymes responsible for catalyzing DNA methylation, the DNA methyltransferases, increase in abundance at the same time and in the same basal epithelial cell population where Cdh1 abundance diminishes during prostate development (Keil et al., 2013). We compared Cdh1 methylation prior to (14 dpc) and during (17 dpc) prostatic bud formation. UGSs at both stages were treated with trypsin, microdissected to separate epithelium from stroma (Fig. 3A) and the purified epithelium was analyzed by MeDIP-QPCR. Cdh1 DNA methylation significantly increased in UGS epithelium between 14 and 17 dpc (Fig. 3B). Therefore, at the same time Cdh1 abundance decreases in UGS epithelium, Cdh1 methylation increases.
Fig. 3.
Cdh1 promoter methylation increases in UGS epithelium during the period of prostatic bud outgrowth. (A) Schematic representation of isolating UGS epithelium. (B) MeDIP-QPCR was conducted to quantify E-cadherin (Cdh1) methylation in isolated UGS epithelium prior to (14 dpc) and during (17 dpc) prostatic bud initiation. Results are mean ± s.e.m, n ≥ 3 litter independent UGS tissue pools per group. Asterisk indicates a significant difference between 17 dpc and 14 dpc UGS epithelium (p < 0.05).
DNA methylation is required for prostatic bud specification and outgrowth of prostate epithelium
In prostate cancer, CDH1 abundance and activity are inversely associated with cell invasiveness and motility (Graff et al., 1995; Yoshiura et al., 1995; van Horssen et al., 2012). We surmised that these same cell behaviors guide prostatic bud outgrowth into surrounding stroma. To test this hypothesis, we examined prostate development in the presence of a DNA methylation inhibitor, which we expected to increase CDH1 abundance and impair prostatic bud outgrowth.
The DNA methylation inhibitor 5-aza-2′-deoxycytidine (5AzadC) interferes with testicular development (Cisneros and Branch, 2003; Choi et al., 2013). Since associated changes in testicular androgen synthesis would confound our study, we used an in vitro approach that recapitulates many features of in vivo prostate development (Doles et al., 2005) but isolates the UGS from influences of testes and other tissues. Female 14 dpc UGSs were used since they are naïve to high levels of circulating androgen prior to the culture period (vom Saal, 1989), are capable of forming prostate in the presence of androgens (Lasnitzki and Mizuno, 1980), and form the same number of prostatic buds as male UGSs grown under equivalent conditions in androgen-containing medium (Keil et al., 2012a). UGS explants were grown for seven days in a defined serum-free culture medium containing androgen, 5α-dihydrotestosterone (DHT, 10 nM) and graded concentrations of 5AzadC.
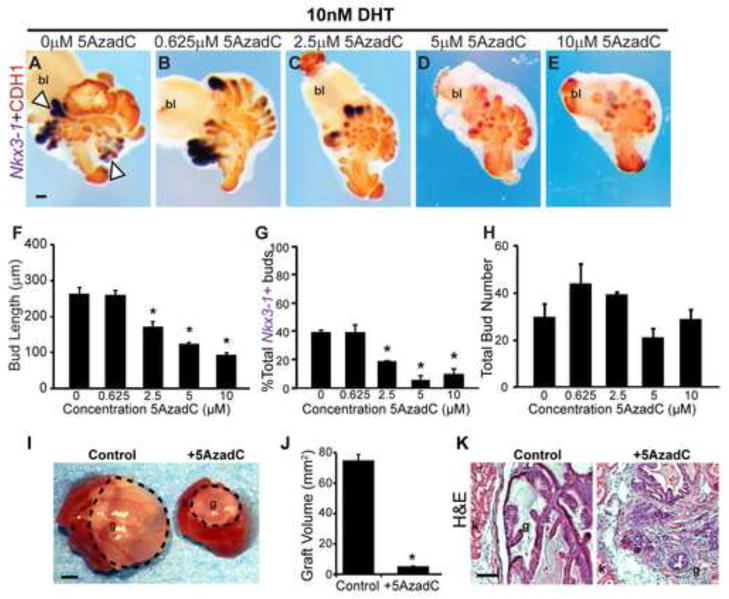
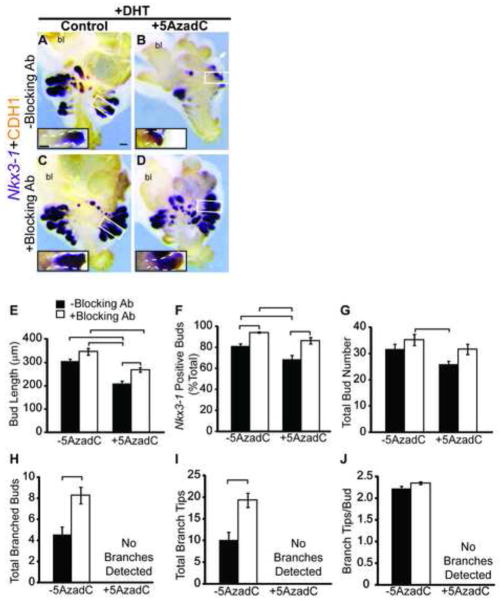
Tissue explants were stained to visualize CDH1 and the earliest marker of prostate identity expressed by prostatic buds, NK-3 transcription factor, locus 1 (Nkx3-1) (Bhatia-Gaur et al., 1999; Bieberich et al., 1996; Keil et al., 2012a; Sciavolino et al., 1997) (Fig. 4A–E). This dual staining approach enabled us to objectively assess whether buds expressed Nkx3-1 and were therefore appropriately specified as prostate, to quantify the number and length of buds formed in culture, and to determine whether buds underwent branching morphogenesis, a normal aspect of continuing ductal development. 5AzadC decreases prostatic bud length (Fig. 4F) and reduces the number of buds with detectable Nkx3-1 staining (Fig. 4G) without changing overall number of buds formed per UGS (Fig. 4H). 5AzadC also blocks branching morphogenesis (Fig. 4A–E). We used an independent marker of developing prostate identity, ectodysplasin A receptor (Edar) (Keil et al 2012a) to confirm the results observed with 5AzadC and Nkx3-1. 5AzadC also reduces the number of buds with detectable Edar staining (Fig. S1). Together these results indicate that the DNA methylation inhibitor, interferes with prostatic bud specification (expression of Nkx3-1 and Edar), elongation and branching morphogenesis.
Fig. 4.
DNA methylation is required for proper prostate bud specification, outgrowth, and branching in UGS explant cultures. (A-E) 14 dpc female UGSs were cultured for 7 days in media containing 5α-dihydrotestosterone (DHT, 10 nM) and increasing concentrations of the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5AzadC, 0–10 μM). At the end of the culture period, tissues were stained by ISH to visualize the prostate marker NK-3 transcription factor, locus 1 (Nkx3-1, purple), counterstained by IHC with CDH1 (red) to visualize all epithelium and imaged in a sagittal orientation. Scalebar 100 μm. Quantification of (F) prostate bud length from tip to base, (G) percent total buds expressing the prostate marker Nkx3-1 and (H) total prostatic bud number. (I) Following 7 day organ culture control and tissues treated with 5 μM 5AzadC were grafted under the renal capsule of an intact male syngeneic host mouse for 1 month. Scalebar 10 mm. (J) Quantification of graft volume in control and 5AzadC treated tissues after 1 month under the renal capsule. (K) Renal capsule grafts were paraffin embedded and sectioned into 5 μm sagittal sections and stained with hematoxylin and eosin. Scalebar 100 μm. Results are presented as mean ± s.e.m, n ≥ 3 litter independent UGSs per group. Asterisks indicate a significant difference from control (p < 0.05). Arrowheads indicate branching buds. A dotted line surrounds graft tissue. Abbreviations; bl, bladder; g, graft; k, kidney.
High concentrations of 5AzadC are capable of decreasing proliferation, inducing apoptosis, and altering differentiation status of cells grown in vitro (Konieczny and Emerson, 1984; Patra et al., 2011). We determined that 5AzadC impaired bud outgrowth without causing obvious changes in formation of basal epithelium (Fig. S2A,B), percentage of proliferating cells within basal epithelium, stroma or the distal most tips of buds (Fig. S2C,D), cell death (Fig. S3A–C), androgen receptor (AR) protein expression, or distribution of androgen responsive steroid 5 alpha reductase 2 (Srd5a2) (Abler et al., 2011a; Matsui et al., 2002) (Fig. S3D–G).
To determine if inhibition of DNA methylation during prostatic outgrowth impairs continued glandular development, we grew UGS explants for seven days in a defined serum-free culture medium containing androgen (10 nM DHT) and either 5μM 5AzadC or its vehicle (control), and then grew them for one month under the renal capsule of intact adult male syngeneic (C57BL/6J) host mice. The host mice were untreated during this period. Control grafts develop extensive, organized prostate ductal structures with evident secretory products (Fig. 4I). 5AzadC treated tissues are significantly smaller (Fig. 4J) and feature smaller, disorganized ducts with few secretions (Fig. 4K). These results suggest that DNA methylation during the initial phases of prostate outgrowth is required for continued prostate ductal maturation.
DNA methylation is required for appropriate CDH1 expression in basal epithelium
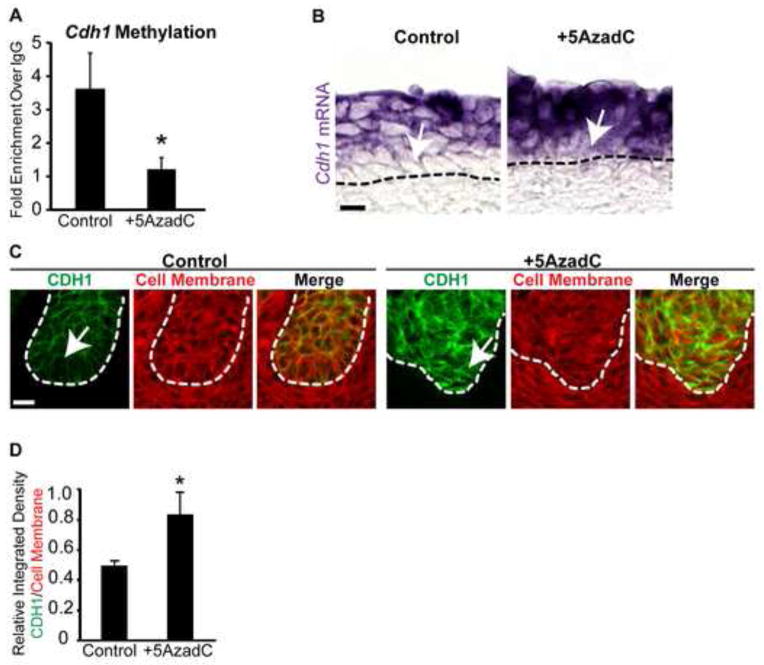
We next tested if 5AzadC reduced Cdh1 DNA methylation and increased Cdh1 abundance in UGS epithelium. To assess Cdh1 DNA methylation, MeDIP-QPCR was conducted on isolated UGS epithelium from 14 dpc female mouse UGS tissues cultured in the presence of androgen ± 5AzadC (5μM). 5AzadC significantly reduces Cdh1 promoter methylation (Fig. 5A) and increases Cdh1 mRNA staining intensity in basal epithelium (Fig. 5B,C). Also within basal epithelium, 5AzadC increases relative integrated density (sum of converted grayscale pixel values) of CDH1 protein (green) when normalized to cell membrane density (red, unaltered by treatment) in 5AzadC versus control tissues (Fig. 5D). These results reveal that 5AzadC reduces CDH1 promoter methylation and increases relative CDH1 abundance in basal epithelium.
Fig. 5.
DNA methylation is required for appropriate E-cadherin mRNA and protein expression in basal epithelium of UGS explant cultures. (A) MeDIP-QPCR was used to quantify E-cadherin (Cdh1) methylation in epithelium from 14 dpc UGSs cultured for 4 days in media containing 5α-dihydrotestosterone (DHT, 10 nM) alone or DHT plus the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5AzadC, 5 μM). Female 14 dpc UGSs cultured for 7days in media containing DHT (10 nM) alone or DHT plus 5AzadC were cut into 5 μm sagittal sections and (B) stained by ISH to visualize Cdh1 mRNA expression (purple) and (C) IHC to visualize CDH1 expression (green) and cell membranes (wheat germ agglutinin, red). (D) Quantification of relative integrated density for CDH1 (green) versus cell membrane (wheat germ agglutinin, red) within the basal-most layer in control and 5AzadC treated UGSs. Results are presented as mean ± s.e.m, n ≥ 3 litter independent UGS sections and tissues per group. Asterisks indicate a significant difference from control (p < 0.05). A dotted line indicates the interface between epithelium and mesenchyme, as determined by cell morphology. Arrows indicate cell layers with altered E-cadherin mRNA and protein abundance. Scalebar 10 μm.
A CDH1 function-blocking antibody bypasses the requirement for DNA methylation in prostatic bud elongation
Because 5AzadC is a non-specific DNA methylation inhibitor and likely to reduce DNA methylation at many sites across the genome, we next wanted to pinpoint whether Cdh1 hypomethylation and an associated increase in Cdh1 abundance are responsible for its impairment of prostatic bud morphogenesis. Female 14 dpc UGS explants were grown in culture media containing androgen and either 5AzadC, a CDH1 function-blocking antibody or both 5AzadC and antibody (Fig. 6A–D). This antibody is directed against the extracellular fifth ectodomain repeat (EC5) of CDH1 which disrupts homotypic interactions (Ozawa et al., 1990; Vestweber and Kemler, 1985) and has been used successfully to impair CDH1 function in salivary gland and lung explant cultures (Tsao et al., 2008; Walker et al., 2008). CDH1 function-blocking antibody penetrates UGS stroma and localizes to its intended target in UGS epithelium (Fig. S4). 5AzadC reduces prostatic bud length by 31% when added to culture medium without the CDH1 function blocking antibody (Fig. 6E). Though the antibody does not independently change prostatic bud number, length or number of branched tips per bud in UGSs grown without 5AzadC (Fig. 6), it restores prostatic bud outgrowth in 5AzadC-treated UGSs (compare Fig. 6B to 6D) and increases Nkx3-1-positive bud percentage in tissues treated with or without 5AzadC, without changing total bud number (Fig. 6E-G). These results show that 5AzadC impairs prostatic bud specification and outgrowth through a CDH1 dependent mechanism. This mechanism is specific to the initial phases of prostatic development because the CDH1 function-blocking antibody does not restore branching morphogenesis in 5AzadC-treated UGSs (Fig. 6H-J).
Fig. 6.
A CDH1 function-blocking antibody restores prostatic bud specification and outgrowth in UGS explant cultures treated with a DNA methylation inhibitor. (A–D) Female 14 dpc UGSs were cultured for 6 days in media containing 5α-dihydrotestosterone (DHT, 10 nM) and either vehicle, the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5AzadC, 5 μM), an E-cadherin (CDH1) function-blocking antibody (Blocking Ab, 100 μg/mL) or both 5AzadC and Blocking Ab. Following culture, tissues were stained by ISH for the prostatic bud marker NK-3 transcription factor, locus 1 (Nkx3-1, purple), immunohistochemically stained for CDH1 (red) to label all epithelium and imaged in a sagittal orientation. Scalebars 100 μm. Quantification of (E) prostatic bud length, (F) percent total Nkx3-1 positive buds (G) total bud number, (H) total number of buds with branches, (I) total number of branch tips and (J) branch tips per bud was conducted on cultured UGSs. Results are presented as mean ± s.e.m, n ≥ 6 litter independent UGS tissues per group. Insets are magnified images across the length of a representative prostatic bud, a white dotted line outlines the epithelium. Lines above bar graphs indicate a significant difference between values (p < 0.05). Abbreviations; bl, bladder.
Cdh1 controls the rate of prostate ductal maturation
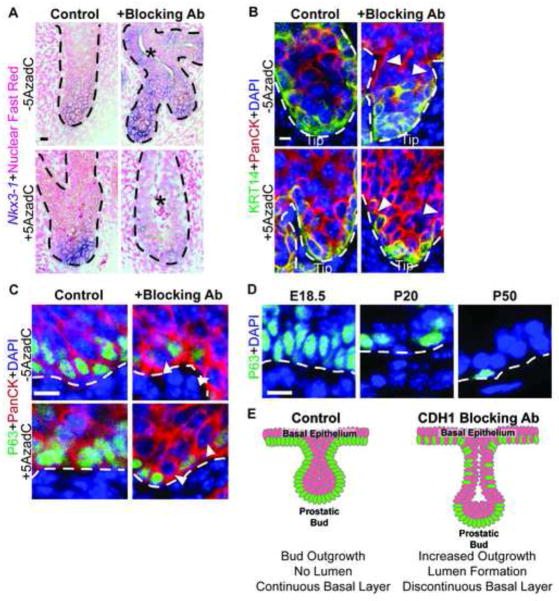
In control UGS tissues, the CDH1 function-blocking antibody increases the number of buds undergoing branching morphogenesis (Fig. 6H-J), suggesting additional roles for CDH1 activity in later stages of prostate development. The CDH1 function-blocking antibody accelerates several hallmarks of prostate ductal maturation. It causes precocious bud lumen formation (Fig. 7A, asterisks). Prostatic ducts do not typically form lumens until several days after birth and are not typically observed in 14 dpc explants cultured for seven days (Doles et al., 2005; Keil et al., 2012a; Sugimura et al., 1986). The CDH1 function-blocking antibody also changes the pattern of basal epithelium from a uniform distribution around the bud and extending into the urethra, to a localized concentration in prostatic bud tips and discontinuous single cell layer extending into the urethra (Fig. 7B,C; arrowheads). The formation of a discontinuous basal epithelium is especially striking since this pattern typically occurs postnatally in normal prostate (Fig. 7D) at a stage much older than that achieved by the six day organ culture. A schematic representation of how the CDH1 function-blocking antibody alters prostate bud epithelial organization is illustrated in Fig. 7E. Together these results suggest that CDH1 not only impacts prostate bud specification and outgrowth but also the maturation and differentiation of prostatic bud epithelium. Therefore Cdh1 DNA methylation not only primes UGS epithelium for prostatic bud specification and outgrowth but, permits continued development and morphogenesis of prostatic ducts.
Fig. 7.
E-cadherin (CDH1) controls the rate of prostate ductal maturation. (A–D) Female 14 dpc UGSs were cultured for 6 days in media containing 5α-dihydrotestosterone (DHT, 10 nM) and either vehicle, the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5AzadC, 5 μM), a CDH1 function-blocking antibody (Blocking Ab, 100 μg/mL) or both 5AzadC and Blocking Ab. (A) Whole mount ISH was used to visualize NK-3 transcription factor, locus 1 (Nkx3-1, purple) tissues were then cut into sagittal sections (5 μm) and counterstained with nuclear fast red. (B-D) IHC was used to detect basal epithelial markers keratin 14 (KRT14, green) or transformation-related protein, 63 (TRP63, green), and general epithelial marker Pan-cytokeratin (PanCK, red) in urethra and developing prostatic buds in sagittal sections (5 μm). Cell nuclei were stained with DAPI (blue) (D) 18.5 dpc, postnatal day P20 and P50 mouse prostate sagittal sections (5 μm) were immunofluorescently labeled to visualize the basal epithelial marker TRP63 (green). (E) Schematic representation of observed changes in basal epithelial organization in UGSs cultured in the presence of CDH1 function blocking antibody. Asterisks indicate the presence of lumens. A dotted line indicates the interface between epithelium and mesenchyme, as determined by cell morphology and epithelial staining. Closed arrowheads indicate areas of discontinuous basal epithelium. Scalebar 10 μm.
Discussion
Our results are the first to establish a mechanistic link between DNA methylation and prostate morphogenesis in addition to providing evidence for the role of CDH1 during prostate ductal development and maturation. We reveal Cdh1 DNA methylation during prostate initiation is a priming event that creates a permissive environment for outgrowth and continued morphogenesis of prostatic ducts during later stages of development (Fig. 8). Our results suggest that a temporal reduction of Cdh1 activity in basal epithelium is necessary for prostatic bud maturation. Prostatic bud outgrowth is impaired under conditions of elevated CDH1 abundance (the presence of a DNA methylation inhibitor) and restored under conditions that neutralized CDH1 activity (the presence of a blocking antibody). Prostatic buds can form without Cdh1 methylation. However, we show that the specific role of Cdh1 methylation is to promote bud elongation, maintenance and acquisition of prostate identity through the reduction of CDH1 activity. We also show that CDH1 activity influences prostatic bud lumen formation and arrangement of ductal epithelium. These events are needed to establish the architecture of mature prostate glandular epithelium. Thus, fine tuning of Cdh1 expression by DNA methylation during development, at least in part, controls the timing and sequence of several prostate ductal maturation events.
Fig. 8.
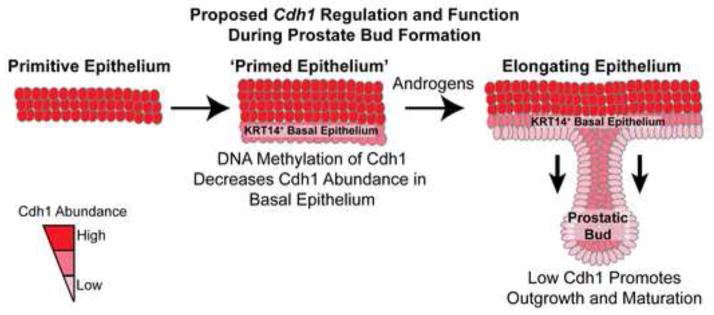
Proposed mechanism of E-cadherin regulation and function during mouse prostatic bud formation. Methylation of E-cadherin (Cdh1) decreases mRNA and protein abundance in basal epithelial cells that give rise to prostatic buds. This event primes prostatic buds for the subsequent actions of androgens and other factors that are needed for outgrowth, differentiation and maturation of prostatic buds.
We also provide evidence that appropriate DNA methylation patterns are required for prostatic ductal branching morphogenesis. Approximately 20% of buds form at least one branch in UGSs cultured for seven days in basal medium containing androgen, but branching formation is completely inhibited by a DNA methylation inhibitor and cannot be restored by neutralizing CDH1 activity (Fig. 4,6). These results indicate a CDH1-independent role of DNA methylation in branching morphogenesis. Focused studies on the identity of methylated genes during prostatic branching morphogenesis, and the developmental timeframe in which methylation is required, will improve understanding of how DNA methylation functions to coordinate organogenesis of the prostate and other developing organs.
A question raised by our results is whether DNA methylation intersects or functions in parallel with other known Cdh1 transcriptional regulators. Cdh1 has previously been shown to be down-regulated by localized actions of beta-catenin (CTNNB1) and the Snail family of genes (Jamora et al., 2003, Jamora et al., 2005). Though Ctnnb1 is required for formation of UGS basal epithelium and prostatic buds, conditional deletion or activation of Ctnnb1 signaling during prostatic bud formation does not appear to alter Cdh1 abundance in basal epithelium (Francis et al., 2013; Mehta et al., 2013). These results indicate that down-regulation of Cdh1 expression in fetal prostate epithelium is likely to happen independently of CTNNB1 function. Similarly, the Snail family of transcriptional repressors do not localize exclusively to developing prostate basal epithelium (gudmap.org), however some are present in this cell population and elsewhere during prostatic bud formation. One function of Snail family members is to down-regulate Cdh1 through chromatin modifications (Espada et al., 2011; Herranz et al., 2008; Hou et al., 2008; Lin et al., 2010; Peinado et al., 2004). An intriguing avenue of future study will be to identify whether factors such as the Snail family members recruit chromatin modifiers to the Cdh1 promoter during prostate development, to fine tune epithelial morphogenesis by acting in concert with or upstream of DNA methylation.
Cdh1 promoter methylation occurs inappropriately in several cancers, including prostate cancer, where it is associated with tumor invasiveness (Behrens and Birchmeier, 1994; Costa et al., 2010; Takeichi, 1993; van Horssen et al., 2012). Why CDH1 becomes methylated in cancer is not known and difficult to determine in part because of the need to capture tumor cells precisely as DNA methylation occurs. We have revealed a specific window during mouse prostate development (14 dpc to 17 dpc) during which Cdh1 methylation increases and Cdh1 mRNA and protein become less abundant in cells giving rise to the prostate. Having defined the window, we are now poised to uncover mechanisms controlling Cdh1 methylation, which may inform future studies of how CDH1 methylation occurs inappropriately in prostate and other cancers.
Though Cdh1 abundance diminishes in early tumor progression, it often increases in remote sites of metastases to re-establish cell adhesion (Graff et al., 2000). This raises the question of whether methyl marks are removed from CDH1 once prostate development is completed. Determining if there is plasticity in Cdh1 methylation during all stages of prostate development will be critical in determining regulatory mechanisms for Cdh1 DNA methylation.
The identified role of DNA methylation in priming prostate development may also shed light on mechanisms by which endocrine disrupting chemicals perturb prostate development in utero. Bisphenol A and others are capable of altering prostate development (Anway et al., 2006; Dolinoy et al., 2007; Gupta, 2000; Ho et al., 2006; Timms et al., 2005; Walker, 2011) and have been shown to alter DNA methylation in prostate and elsewhere (Ho et al., 2006; Tang et al., 2012; Walker, 2011). The critical window during which CDH1 methylation occurs overlaps the known window of action for some of these chemicals (Dolinoy et al., 2007; Gupta, 2000; Ho et al., 2006; Tang et al., 2012), raising questions about whether their teratogenic actions are mediated in part through DNA methylation.
Supplementary Material
Highlights.
E-cadherin abundance is temporally down-regulated in epithelial cells giving rise to the prostate
DNA methylation restricts Cdh1 abundance in the prostate primordium
Restricted Cdh1 abundance and activity is crucial for bud outgrowth
Acknowledgments
Grant Sponsors: National Institutes of Health Grants DK083425, DK070219, DK096074 and ES001332, and National Science Foundation Grant DGE-0718123. We thank Robert Lipinski and Deborah Anderson for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abler LL, Keil KP, Mehta V, Joshi PS, Schmitz CT, Vezina CM. A high-resolution molecular atlas of the fetal mouse lower urogenital tract. Dev Dyn. 2011a;240:2364–2377. doi: 10.1002/dvdy.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abler LL, Mehta V, Keil KP, Joshi PS, Flucus C, Hardin HA, Schmitz CT, Vezina CM. A high throughput in situ hybridization method to characterize mrna expression patterns in the fetal mouse lower urogenital tract. J Vis Exp. 2011b;54:2912. doi: 10.3791/2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard DR, Lasnitzki I, Mizuno T. Testosterone metabolism in the urogenital epithelium and mesenchyme of the foetal rat [proceedings] J Endocrinol. 1979;83:35P. [PubMed] [Google Scholar]
- Behrens J, Birchmeier W. Cell-cell adhesion in invasion and metastasis of carcinomas. Cancer Treat Res. 1994;71:251–266. doi: 10.1007/978-1-4615-2592-9_13. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, et al. Roles for nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich CJ, Fujita K, He WW, Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J Biol Chem. 1996;271:31779–31782. doi: 10.1074/jbc.271.50.31779. [DOI] [PubMed] [Google Scholar]
- Boussadia O, Kutsch S, Hierholzer A, Delmas V, Kemler R. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech Dev. 2002;115:53–62. doi: 10.1016/s0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- Carraro G, del Moral P, Warburton D. Mouse embryonic lung culture, a system to evaluate the molecular mechanisms of branching. J Vis Exp. 2010;30:40. doi: 10.3791/2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Lee S, Hwang S, Jo SA, Kim M, Kim YJ, Pang MG, Jo I. Histone H3 lysine 27 and 9 hypermethylation within the Bad promoter region mediates 5-Aza-2′-deoxycytidine-induced Leydig cell apoptosis: implications of 5-Aza-2′-deoxycytidine toxicity to male reproduction. Apoptosis. 2013;18:99–109. doi: 10.1007/s10495-012-0768-4. [DOI] [PubMed] [Google Scholar]
- Cisneros FJ, Branch S. 5-AZA-2′-deoxycytidine (5-AZA-CdR): a demethylating agent affecting development and reproductive capacity. J Appl Toxicol. 2003;23:115–20. doi: 10.1002/jat.898. [DOI] [PubMed] [Google Scholar]
- Costa VL, Henrique R, Ribeiro FR, Carvalho JR, Oliveira J, Lobo F, Teixeira MR, Jerónimo C. Epigenetic regulation of wnt signaling pathway in urological cancer. Epigenetics. 2010;5:343–351. doi: 10.4161/epi.5.4.11749. [DOI] [PubMed] [Google Scholar]
- Doles JD, Vezina CM, Lipinski RJ, Peterson RE, Bushman W. Growth, morphogenesis, and differentiation during mouse prostate development in situ, in renal grafts, and in vitro. Prostate. 2005;65:390–399. doi: 10.1002/pros.20321. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Espada J, Peinado H, Lopez-Serra L, Setién F, Lopez-Serra P, Portela A, Renart J, Carrasco E, Calvo M, Juarranz A, et al. Regulation of snail1 and e-cadherin function by dnmt1 in a dna methylation-independent context. Nucleic Acids Res. 2011;39:9194–9205. doi: 10.1093/nar/gkr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Huebner RJ, Palsdottir H, Lee JK, Perez MJ, Jorgens DM, Tauscher AN, Cheung KJ, Werb Z, Auer M. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci. 2012;125:2638–2654. doi: 10.1242/jcs.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JC, Thomsen MK, Taketo MM, Swain A. β-catenin is required for prostate development and cooperates with pten loss to drive invasive carcinoma. PLoS Genet. 2013;9:e1003180. doi: 10.1371/journal.pgen.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the e-cadherin 5′ cpg island are unstable and reflect the dynamic, heterogeneous loss of e-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by dna hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- Gupta C. Reproductive malformation of the male offspring following maternal exposure to estrogenic chemicals. Proc Soc Exp Biol Med. 2000;224:61–68. doi: 10.1046/j.1525-1373.2000.22402.x. [DOI] [PubMed] [Google Scholar]
- Herfs M, Hubert P, Kholod N, Caberg JH, Gilles C, Berx G, Savagner P, Boniver J, Delvenne P. Transforming growth factor-beta1-mediated slug and snail transcription factor up-regulation reduces the density of langerhans cells in epithelial metaplasia by affecting e-cadherin expression. Am J Pathol. 2008;172:1391–1402. doi: 10.2353/ajpath.2008.071004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Wong MH, Gordon JI. Forced expression of e-cadherin in the mouse intestinal epithelium slows cell migration and provides evidence for nonautonomous regulation of cell fate in a self-renewing system. Genes Dev. 1996;10:985–996. doi: 10.1101/gad.10.8.985. [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for e-cadherin repression by the snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai Y, Nose A, Kobayashi S, Takeichi M. Expression and role of e- and p-cadherin adhesion molecules in embryonic histogenesis. i lung epithelial morphogenesis. Development. 1989;105:263–270. doi: 10.1242/dev.105.2.263. [DOI] [PubMed] [Google Scholar]
- Ho S, Tang W, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol a increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, Ayyanathan K, Yan K, Langer EM, Longmore GD, Rauscher FJ., 3 The lim protein ajuba recruits protein arginine methyltransferase 5 to mediate snail-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JC, Koo H, Harunaga JS, Matsumoto K, Doyle AD, Yamada KM. Region-specific epithelial cell dynamics during branching morphogenesis. Dev Dyn. 2013;242:C1. doi: 10.1002/dvdy.24000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, Lee P, Kocieniewski P, Azhar M, Hosokawa R, Chai Y, Fuchs E. A signaling pathway involving tgf-beta2 and snail in hair follicle morphogenesis. PLoS Biol. 2005;3:e11. doi: 10.1371/journal.pbio.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil KP, Altmann HM, Mehta V, Abler LL, Elton EA, Vezina CM. Catalog of mrna expression patterns for dna methylating and demethylating genes in developing mouse lower urinary tract. Gene Expr Patterns. 2013;13:413–424. doi: 10.1016/j.gep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil KP, Mehta V, Abler LL, Joshi PS, Schmitz CT, Vezina CM. Visualization and quantification of mouse prostate development by in situ hybridization. Differentiation. 2012a;84:232–239. doi: 10.1016/j.diff.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil KP, Mehta V, Branam AM, Abler LL, Buresh-Stiemke RA, Joshi PS, Schmitz CT, Marker PC, Vezina CM. Wnt inhibitory factor 1 (wif1) is regulated by androgens and enhances androgen-dependent prostate development. Endocrinology. 2012b;153:6091–6103. doi: 10.1210/en.2012-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny SF, Emerson CPJ. 5-azacytidine induction of stable mesodermal stem cell lineages from 10t1/2 cells: evidence for regulatory genes controlling determination. Cell. 1984;38:791–800. doi: 10.1016/0092-8674(84)90274-5. [DOI] [PubMed] [Google Scholar]
- Kurita T, Medina RT, Mills AA, Cunha GR. Role of p63 and basal cells in the prostate. Development. 2004;131:4955–4964. doi: 10.1242/dev.01384. [DOI] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasnitzki I, Mizuno T. Prostatic induction: interaction of epithelium and mesenchyme from normal wild-type mice and androgen-insensitive mice with testicular feminization. J Endocrinol. 1980;85:423–428. doi: 10.1677/joe.0.0850423. [DOI] [PubMed] [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase lsd1 in snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs RU, Lawson DA, Xin L, Zong Y, Garraway I, Goldstein AS, Memarzadeh S, Witte ON. Epithelial stem cells of the prostate and their role in cancer progression. Cold Spring Harb Symp Quant Biol. 2008;73:491–502. doi: 10.1101/sqb.2008.73.012. [DOI] [PubMed] [Google Scholar]
- Matsui D, Sakari M, Sato T, Murayama A, Takada I, Kim M, Takeyama K, Kato S. Transcriptional regulation of the mouse steroid 5alpha-reductase type ii gene by progesterone in brain. Nucleic Acids Res. 2002;30:1387–1393. doi: 10.1093/nar/30.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V, Abler LL, Keil KP, Schmitz CT, Joshi PS, Vezina CM. Atlas of wnt and r-spondin gene expression in the developing male mouse lower urogenital tract. Dev Dyn. 2011;240:2548–2560. doi: 10.1002/dvdy.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin T, Taketo MM, Sun X, Vezina CM. Beta-catenin (ctnnb1) induces bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Dev Biol. 2013;376:125–135. doi: 10.1016/j.ydbio.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba D, Nakanishi Y, Hieda Y. Changes in adhesive properties of epithelial cells during early morphogenesis of the mammary gland. Dev Growth Differ. 2001;43:535–544. doi: 10.1046/j.1440-169x.2001.00596.x. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Hoschützky H, Herrenknecht K, Kemler R. A possible new adhesive site in the cell-adhesion molecule uvomorulin. Mech Dev. 1990;33:49–56. doi: 10.1016/0925-4773(90)90134-8. [DOI] [PubMed] [Google Scholar]
- Patra A, Deb M, Dahiya R, Patra SK. 5-aza-2′-deoxycytidine stress response and apoptosis in prostate cancer. Clin Epigenetics. 2011;2:339–348. doi: 10.1007/s13148-010-0019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates e-cadherin repression by the recruitment of the sin3a/histone deacetylase 1 (hdac1)/hdac2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D, Williams-Ashman HG. The accessory reproductive glands of mammals. In: Young WC, editor. Sex and Internal Secretions. Baltimore: Williams & Wilkins Co; 1961. pp. 366–448. [Google Scholar]
- Reardon SN, King ML, MacLean JA, 2, Mann JL, DeMayo FJ, Lydon JP, Hayashi K. Cdh1 is essential for endometrial differentiation, gland development, and adult function in the mouse uterus. Biol Reprod. 2012;86:141, 1–10. doi: 10.1095/biolreprod.112.098871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. P63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirour C, Hidalgo M, Bello V, Buisson N, Darribère T, Moreau N. Dystroglycan is involved in skin morphogenesis downstream of the notch signaling pathway. Mol Biol Cell. 2011;22:2957–2969. doi: 10.1091/mbc.E11-01-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura Y, Cunha GR, Donjacour AA. Morphogenesis of ductal networks in the mouse prostate. Biol Reprod. 1986;34:961–971. doi: 10.1095/biolreprod34.5.961. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- Tang W, Morey LM, Cheung YY, Birch L, Prins GS, Ho S. Neonatal exposure to estradiol/bisphenol a alters promoter methylation and expression of nsbp1 and hpcal1 genes and transcriptional programs of dnmt3a/b and mbd2/4 in the rat prostate gland throughout life. Endocrinology. 2012;153:42–55. doi: 10.1210/en.2011-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms BG, Howdeshell KL, Barton L, Bradley S, Richter CA, vom Saal FS. Estrogenic chemicals in plastic and oral contraceptives disrupt development of the fetal mouse prostate and urethra. Proc Natl Acad Sci. 2005;102:7014–7019. doi: 10.1073/pnas.0502544102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkle CL, Pasolli HA, Stokes N, Fuchs E. New insights into cadherin function in epidermal sheet formation and maintenance of tissue integrity. Proc Natl Acad Sci USA. 2008;105:15405–15410. doi: 10.1073/pnas.0807374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- Tsao P, Chen F, Izvolsky KI, Walker J, Kukuruzinska MA, Lu J, Cardoso WV. Gamma-secretase activation of notch signaling regulates the balance of proximal and distal fates in progenitor cells of the developing lung. J Biol Chem. 2008;283:29532–29544. doi: 10.1074/jbc.M801565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D, Kemler R. Identification of a putative cell adhesion domain of uvomorulin. EMBO J. 1985;4:3393–3398. doi: 10.1002/j.1460-2075.1985.tb04095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina CM, Allgeier SH, Fritz WA, Moore RW, Strerath M, Bushman W, Peterson RE. Retinoic acid induces prostatic bud formation. Dev Dyn. 2008;237:1321–1333. doi: 10.1002/dvdy.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JL, Menko AS, Khalil S, Rebustini I, Hoffman MP, Kreidberg JA, Kukuruzinska MA. Diverse roles of e-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Dev Dyn. 2008;237:3128–3141. doi: 10.1002/dvdy.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Fan L, Wei J, Weng Y, Zhou L, Shi Y, Zhou W, Ma D, Wang C. Akt mediates metastasis-associated gene 1 (mta1) regulating the expression of e-cadherin and promoting the invasiveness of prostate cancer cells. PLoS One. 2012;7:e46888. doi: 10.1371/journal.pone.0046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the e-cadherin invasion-suppressor gene by cpg methylation in human carcinomas. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Boussadia O, Halfter H, Grose R, Berger P, Leone DP, Robenek H, Charnay P, Kemler R, Suter U. E-cadherin controls adherens junctions in the epidermis and the renewal of hair follicles. EMBO J. 2003;22:5723–5733. doi: 10.1093/emboj/cdg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Horssen R, Hollestelle A, Rens JAP, Eggermont AMM, Schutte M, Ten Hagen TLM. E-cadherin promotor methylation and mutation are inversely related to motility capacity of breast cancer cells. Breast Cancer Res Treat. 2012;136:365–377. doi: 10.1007/s10549-012-2261-8. [DOI] [PubMed] [Google Scholar]
- vom Saal FS. Sexual differentiation in litter-bearing mammals: influence of sex of adjacent fetuses in utero. J Anim Sci. 1989;67:1824–1840. doi: 10.2527/jas1989.6771824x. [DOI] [PubMed] [Google Scholar]
- Walker CL. Epigenomic reprogramming of the developing reproductive tract and disease susceptibility in adulthood. Part A Clin Mol Teratol. 2011;91:666–671. doi: 10.1002/bdra.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.