Abstract
The eukaryotic exosome is a multi-subunit complex typically composed of a catalytically inactive core and the Rrp44 protein, which contains 3’ to 5’ exo- and endo-ribonuclease activities. RNA substrates have been shown to be recruited through the core to reach Rrp44's exoribonuclease (EXO) site. Using single particle electron microscopy and biochemical analysis, we provide visual evidence that two distinct substrate recruitment pathways exist. In the through-core route, channeling of the single stranded substrates from the core to Rrp44 induces a characteristic conformational change in Rrp44. In the alternative direct-access route, this conformational change does not take place and the RNA substrate is visualized to avoid the core and enter Rrp44's EXO site directly. Our results provide mechanistic explanations for several RNA processing scenarios by the eukaryotic exosome and indicate substrate specific modes of degradation by this complex.
Introduction
The eukaryotic exosome is a multi-subunit protein complex crucial for RNA maturation, surveillance, and turnover1-7. The exosome core is composed of six RNase PH like subunits (Rrp41, Rrp45, Rrp42, Rrp43, Mtr3, and Rrp46), and three capping subunits containing RNA-binding domains (Rrp4, Rrp40, and Csl4)8, in a similar architecture to the archaeal exosome9-11. However, the exonuclease activity of the core has been lost in yeast and humans8,12 but gains its RNase activity by binding a tenth subunit, Rrp44 12,13.
Rrp44 contains multiple functional domains (Fig. 1a). Its PilT N-terminus (PIN) domain exhibits manganese-dependent endonuclease (ENDO) activity14-16. The C-terminal EXO region consists of tandem cold shock domains (CSD1 and CSD2), a magnesium-dependent 3’- to 5’-exoribonuclease RNB domain, and an S1 domain. The overall architecture of Rrp44's EXO region is similar to that of bacterial RNase II, but the recruitment of RNA to the two enzymes differs17-20. Also, while RNase II can only process ssRNA, Rrp44 can unwind and degrade duplex RNA17, likely via elastic based helicase-like activity21.
Figure 1. Rrp44-exosome degrades RNA substrates differently from Rrp44 alone.
(a) Schematics of domain organization in yeast Rrp44, based on the structures of full length (PDB 2WP8)22 and CTD (PDB 2VNU)17 Rrp44 protein. (b) Degradation assay of HDV RNA substrates containing 40 nt AU-rich sequence at the 3’-end. R: RNA only control. RE: Rrp44-exosome reactions. Positions of intermediates and end products are marked by arrows and were determined by hydroxyl radical and RNase T1 ladders (Supplementary Fig. 1a and Supplementary Fig. 1f). (c-d)3D reconstructions of the RE samples incubated with blank buffer (marked as Apo-RE), RNA08, RNA10, RNA12, RNA14, RNA16, RNA24, RNA36, RNA47, and HDV40 (marked on the top of 3D models) in 5 mM EDTA TBS buffer for 15 minutes, respectively. Both the front view and bottom view of each reconstructed model are shown.
Rrp44 associates with the core complex by binding of the PIN domain to subunits Rrp41 and Rrp45 18,22,23. In tune with a similar function of the archaeal exosome, RNA substrates with long single-stranded (ss) 3’-overhangs are first channeled through the eukaryotic exosome core before being degraded by Rrp44 22,24. RNA degradation and protection assays indicate that ~31-33 stretched nucleotides are required to reach Rrp44's EXO site from the top of exosome core's RNA binding subunits22. This through-core route is clearly demonstrated by the crystal structure of the yeast Rrp44-exosome (RE) in complex with an RNA substrate with a 5’-hairpin and long 3’-ss-overhang18. However, degradation of hypomethylated yeast initiator methionine tRNA (tRNA Meti)6 and tRNAs with a double CCA motif at the 3’-end25 by Rrp44 implies the presence of an RNA processing pathway that does not necessarily involve channeling through the core. Recent transcriptome data also suggests the presence of alternative routes bypassing the core for RNA substrates with shorter 3’-ss-overhangs to be processed by the exosome26. The multi-porous structure of the apo-RE complex provides the potential for RNA substrates to take multiple routes including the through-core and direct access pathways to reach Rrp44's EXO site22.
To further dissect the mechanism of RNA recruitment to the exosome, we performed biochemical and single particle electron microscopy (EM) analysis on the Saccharomyces cerevisiae RE in concert with different RNA substrates. Our single particle analysis revealed a substrate induced conformational change of the complex upon RNA binding and substrate-specific alternative routes of RNA recruitment by the exosome complex.
Results
RNase assays for both through-core and direct-access routes
The through-core route and direct-access route predict distinct outcomes in the processing of the 3’-ss-overhang of a highly structured RNA substrate. While the through-core route stalls when the long ss-overhang is trimmed down to ~30 nt, the direct-access route bypasses this restriction and predicts a processed ss-overhang of likely less than 10 nt. We used a set of molecular ruler experiments to distinguish these two pathways, by fusing unstructured AU-rich sequences of various lengths at the 3’-end of the highly structured Hepatitis Delta Virus (HDV) ribozyme (Supplementary Fig. 1a and Supplementary Fig. 1b).
With a C75U mutation introduced to prevent the HDV ribozyme cleavage, 5’-end 32P-labeled substrates with a stable tertiary structure27-29 were properly refolded for processing assays (Supplementary Fig. 1c). We carried out RNA processing assays of these substrates by Rrp44 alone and RE complex under single turnover conditions in low Mg2+ (100 μM), where RE was more active12. The low Mg2+ condition, however, inadvertently resulted in the partial destabilization of the 3’-portion of the HDV structure beyond the tightly folded pseudo-knot (after nucleotide G40), increasing the length of the 3’-ss-overhang by 33-nt. All HDV RNAs were efficiently processed by both Rrp44 and RE, producing ~5-nt completely digested products as well as one or two predominant processing intermediates (Fig. 1b and Supplementary Fig. 1d). The EXO active site mutant (D551N, EX−) but not the ENDO active site mutant (D171N, EN−) resulted in the loss of degradation, similar to the behavior of the double mutant (D551N and D171N, DM) (Supplementary Fig. 1e), suggesting predominant exonuclease activity from Rrp44 and RE in our assays. Mapping of the HDV ribozyme with RNase T1 and OH ladders (Supplementary Fig. 1a and Supplementary Fig. 1f) indicated that the single processing intermediate in Rrp44-alone reactions represents a stalling event near residue G40 of HDV RNA, presumably due to the blockage of the direct-access pathway by the tightly folded 5’-pseudoknot. Interestingly, the same processing intermediate was also present in RE reactions, suggesting that a similar, direct-access route is present in the complex. The short processing intermediate in RE reactions is unlikely due to excess free Rrp44s because our purification procedure ensures a 1:1 molar ratio between Rrp44 and the exosome core (Supplementary Fig. 1g). The complex's integrity was further supported by EM (Supplementary Fig. 1h). In the RE reactions, an additional processing intermediate ~26 nt longer than the Rrp44 reaction intermediate was evident for all HDV RNAs, consistent with a processing event by the through-core route (Fig. 1b and Supplementary Fig. 1d). The above results suggest that while the through-core route is predominant (~70%), a direct-access route is simultaneously present for the HDV RNA degradation.
Two distinct RE conformations with different RNA substrates
We further performed 3D reconstruction of RE incubated with ssRNA substrates with different lengths (Supplementary Table 1) under non-digesting conditions (with 5 mM EDTA present to sequester the catalytic metal ions), where all RNA substrates were capable to bind RE without detectable degradation (Supplementary Fig. 2). Interestingly, the series of 3D reconstructions revealed two distinct conformations of RE, depending on the ssRNA length (Fig. 1c and Fig. 1d). When incubated with RNA substrates shorter than 12 nt, the resulting EM reconstruction of the complexes revealed a conformation (RE-short) very similar to that of apo-RE (Fig. 1c). By contrast, when the RNA substrates were longer than 14 nt, RE adopted a distinct conformation (RE-long) (Fig. 1d). The major difference occurred within the Rrp44 protein, which is more compact in RE-long than in RE-short conformation, but the structure of the core complex is almost unchanged between the two conformers (Table 1, Supplementary Table 2, and Supplementary Table 3). This RNA-induced conformational change is not necessarily restricted to purely ssRNAs as the same conformation of RE-long was also observed in the RE-HDV40 complex (Fig. 1d).
Table 1.
Cross-correlation coefficients among different 3D models
| Apo-RE | RNA08-RE | RNA10-RE | RNA12-RE | SA-RNA47-RE | |
|---|---|---|---|---|---|
| RNA47-RE Core | 0.9323 | 0.9265 | 0.9287 | 0.9135 | 0.8892* |
| RNA47-RE Rrp44 | 0.6822 | 0.6443 | 0.6573 | 0.6573 | 0.6849 |
The upper row is the comparison among different models at the core; the bottom row is the comparison among different models at the Rrp44 portion.
The comparison is calculated at the core region only, with SA density removed.
We docked atomic models into the two conformations to rationalize the observed conformational changes. For the RE-short 3D reconstruction, the crystal structure of the human core exosome (PDB 2NN6)8 fits nicely in the map as a rigid body, while the crystal structures of Rrp44's PIN domain and C-terminal EXO region from the Rrp44-Rrp41-Rrp45 subcomplex structure (PDB 2WP8)22 fit well in the map as separate rigid bodies (Fig. 2a and Supplementary Video 1). In the docked model, the overall spatial relationship among Rrp44, Rrp41, and Rrp45 is very similar as in the crystal structure of the sub-complex except that the PIN domain moves ~10 Å away from Rrp44's EXO region. For the RE-long 3D reconstruction, we took the recently published crystal structure of the yeast RE in complex with a partial hairpin RNA substrate (PDB 4IFD)18 and docked it in the map as a rigid body. The crystal structure fit with good agreement into the 3D envelope of the RE-long structure, except that the PIN domain of Rrp44 needed to be shifted by about 10 Å closer to the EXO region (Fig. 2b and Supplementary Video 2). The high level of agreement between the two independently determined structures indicates the strong biological relevance of these two conformations. Compared to the crystal structure, the EM reconstructions all had a substantially weaker density for Csl4 on top of the core, in consistence with previous observations that Csl4 is prone to dissociation from the complex during purification23,26.
Figure 2. Comparison and analysis of the RE-short and RE-long 3D models.
(a-b) Docking of crystal structures into the RE-short (a) and RE-long (b) 3D models, respectively. The front, back and bottom views of both docking models are shown. The color coding of the different subunits in the core complex follows the name list on the left of panel a. The cleft between CSD and RNB domains of Rrp44 that could recruit RNA substrate is marked with an arrow in a. (c-d) Front views of the RE-short and RE-long docking models, respectively, with CSD1 domain (green), CSD2 domain (purple), RNB domain (blue) and S1 domain (orange) labeled. The orange arrows in c show the rotation orientation for Rrp44's C-terminal EXO region around the pivotal point (circled) from the RE-short to RE-long conformation. (e-f) Vertical section of the RE-short and RE-long docking models, respectively. The orange arrow in e indicates the CSD2 movement from the RE-short to RE-long state.
Close examination of the two models revealed that the major structural change occurring in Rrp44 concerns a rigid body rotation (~120 degrees) of its EXO region from the RE-short to RE-long state (RMSD of 4.3 Å between the two atomic models using the ProFit program, Martin, A.C.R., http://www.bioinf.org.uk/software/profit/) (Fig. 2c). In the RE-short conformation, there are two solvent exposed openings in Rrp44: the opening between the PIN and the CSD domains, and a major cleft between the CSD1 and the RNB domain that makes the EXO site exposed to solvent (Fig. 2a, arrow). This cleft has been shown to bind ssRNA substrates that enter the EXO site for degradation17,18. The two openings are separated by the CSD domains in the center, with the CSD2 domain blocking in way from the core channel's bottom exit to Rrp44's EXO site (Fig. 2c and Fig. 2e). In contrast, in the RE-long conformation, the Rrp44's region undergoes a dramatic rotation as it rolls against the bottom of the core and pivots around the CSD1 domain, so that the CSD2 domain moves away from the core channel while the RNB domain moves closer to the core channel (Fig. 2d, Fig. 2e and Supplementary Video 3). As a result, the cleft between the CSD1 and the RNB domain moves just to the core channel's bottom exit, thus forming a continuous route for ssRNA substrates to thread from the core to Rrp44's EXO site. In agreement with this notion, the RNA molecule from the crystal structure fits nicely within the central channel (Fig. 2f).
Our series of 3D reconstructions revealed that only ssRNAs longer than 14 nt induce the dramatic conformational change in Rrp44 within RE, while ssRNAs shorter than 12 nt maintain the same apo-RE conformation. Such a dramatic structural transition in response to the ssRNA's length agrees with the crystal structure of the RNA-RE complex, in which interpretable electron density for the RNA substrate's very 3’-end extends from the bottom exit of the core to Rrp44's EXO site18. In the crystal structure, nucleotides numbered −13 ~ −15 of the RNA interact with the Rrp45 and Rrp41 subunits, while −1 ~ −12 interact entirely with Rrp44. This indicates that ssRNA substrates with enough length to connect the core to Rrp44 are ready to induce Rrp44's dramatic rearrangement.
EM of RE in complex with RNAs with long 3’-ss-overhangs
In order to visualize the RNA recruitment pathways to the exosome, we pre-formed a complex between streptavidin (SA) and 5’-biotinylated ss-RNA oligos of various lengths (SA-RNA, Supplementary Table 1 and Supplementary Fig. 2b), incubated the SA-RNAs with RE under non-digesting condition, and sought to locate the SA density in the resulting negative stain EM reconstructions as an indicator of the ss-RNA substrates’ 5’-end. Single particle 2D classification of such samples allowed us to detect additional globular densities attached to the RE (Fig. 3a), the size of which agrees well with a SA tetramer. There were substantially more class averages showing the additional SA densities for the ssRNA substrates longer than 36 nt, but very few with SA densities for the ssRNA substrates shorter than 24 nt (Fig. 3b). This result was in agreement with pull-down assays using RE as bait to detect horseradish peroxidase (HRP)-conjugated SA-RNA binding (Supplementary Fig. 3a). The observation reflects the fact that SA prevents RNAs with short 3’-ss-overhangs (<24 nt) from forming stable complex with RE but does not affect those with long enough 3’-ss-overhangs (>36 nt) to thread through the core's entry site to Rrp44's EXO site to form a stable complex. We performed 2D classification and 3D reconstruction of the SA-RNA47-RE and SA-RNA50-RE and revealed clear shape of RE with a globular density attached to the entry side of core complex in close proximity to the Rrp4 protein (Fig. 3c and Supplementary Fig. 3). The dimension of the globular density agrees very well with the size of a SA tetramer so that we can unambiguously dock a SA atomic model within it (Fig. 3d and Fig. 3e). Interestingly, the 3D model shows the same conformational state of Rrp44 seen in the RE-long conformation (Fig. 3f and Fig. 3g), as verified by comparing the 2D class averages with reprojections from the 3D models of RE-short and RE-long (Supplementary Fig. 3b and Supplementary Fig. 3c). This further proves that RNAs going through the core channel and reaching Rrp44 are responsible for RE's conformational change. The fact that RNAs with very different sequences as the 50-mer and 47-mer behave similarly in their interactions with RE underscores the importance of the single stranded nature of RNA substrates over their specific sequences.
Figure 3. Analysis of the RE incubated with streptavidin labeled RNAs (SA-RNA).
(a) Representative 2D class averages of the RE incubated with different SA-RNA substrates (SA-RNA08, SA-RNA16, SA-RNA24, SA-RNA36, SA-RNA47 and SA-RNA50). Additional SA densities compared to Apo-RE class-averages are marked with red circles. The scale bar represents 20 nm. (b) Distribution of particles belonging to the class-average views with or without SA among particles for each sample set. (c-f) Front (c-d), back(e) and bottom (f) views of the SA-RNA47-RE 3D model, where d and e both have a tetrameric streptavidin crystal structure (PDB 1NDJ)37 docked into the electron densities. (g-h) 3D models of the RE-long (g) and RE-short (h) conformations in bottom view, respectively.
EM of RE in complex with RNAs with short 3’-ss-overhangs
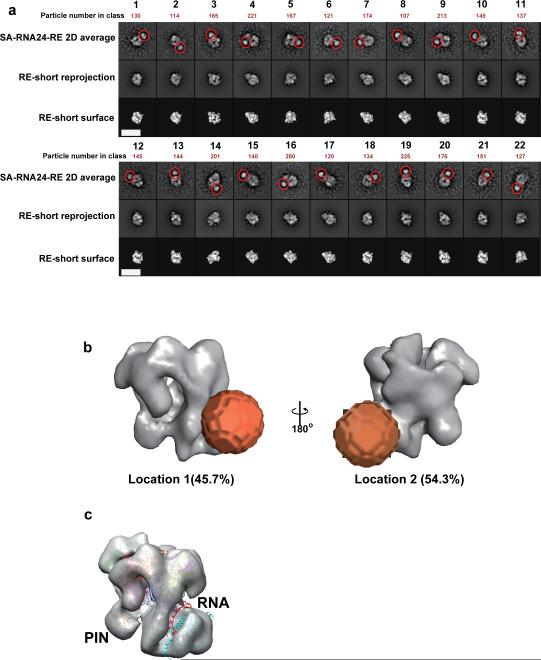
As we have revealed, in contrast to the longer RNA substrates, SA labeled RNA substrates shorter than 24 nt do not seem to form a very stable complex with RE in the non-digestive condition in the presence of 5 mM EDTA (Supplementary Fig. 3a), because SA prevents the relatively short ss-overhang to reach Rrp44's EXO site through the core. However, in the presence of 2 mM Mg2+, when the RE is active for RNA processing, we found that SA-RNA substrates with a 3’-ss-overhang shorter than 24 nt can also form reasonably stable complexes with RE that can be characterized by EM (Fig. 4a). We were able to detect additional densities corresponding to the dimension of a SA in a portion of class averages from the SA-RNA24-RE sample, albeit fewer than those in the SA-RNA47-RE. The RE portion of 2D class averages of SA-RNA24-RE compared reasonably well with reprojections of the 3D model of apo-RE, allowing us to assign each class average to a specific Euler angle in 3D space. The comparison between the 2D class averages and corresponding reprojections showed almost no SA additional density appearing at the top of the core complex, in contrast to what was seen for the SA-RNA47-RE sample, but more occasional SA densities attached to other parts of RE. Unfortunately, our efforts to get a 3D reconstruction from these 2D class averages were not successful, presumably due to two reasons: 1) The 2D class averages with clear SA densities were not enough to cover the full set of views necessary for 3D reconstruction; 2) There may exist more than one SA labeling sites on RE. Based on the Euler angle assigned to each 2D class average, we back-projected the 2D class averages into 3D space with their SA density as a rod (Supplementary Fig. 4). This allowed us to capture two major cross points of SA back-projection rods around the RE 3D model, thus allowing us to locate the most probable SA locations attached to RE in 3D space (Fig. 4b).
Figure 4. RNA substrates with short 3’ single stranded overhangs can be recruited to the exosome via direct-access routes.
(a) Representative 2D class-averages (Row 1) with additional densities (marked with red circles) sorted from RE incubated with SA-RNA24 under 2mM MgCl2 for 5 minutes in comparison with the corresponding reprojections (Row 2) and surface-renders of the RE-short 3D model (Row 3). The scale bars represent 20 nm. (b) The two most probable locations of SA (orange spheres) attached to the RE model estimated using back-projection procedure as described in the Online Methods and Supplementary Fig. 5. The relative proportion of particles contributing to location 1 is 45.7%, and location 2 is 54.3%. (c) Docking of the 13nt-ssRNA substrate of the atomic model of Rrp44 RNB domain in complex with ssRNA (PDB: 4IFD)18 into the RE-short 3D model, mimicking the direct-access route.
Compared with the SA-RNA47-RE sample, the SA density appeared mostly close to Rrp44 in the SA-RNA24-RE sample. The locations identified are around the RNB domain, facing its front and back sides, respectively. Both locations are close to the cleft between Rrp44's CSD and RNB domains which has been shown by crystallography to be able to recruit short ssRNAs17,18,22 (Fig. 4c). Therefore, the SA densities directly attached to these two sites provide strong evidence of RNA substrates’ direct access to Rrp44 bypassing the core. In correlation with the RNase assays of the HDV ribozyme substrates by Rrp44 and RE, this observation suggests that Rrp44 maintains the capability to directly recruit RNAs when in complex with the core. We also detected a minor population of 2D class averages with the SA density near Rrp44's PIN domain, implying that PIN domain's endonuclease site may also recruit RNA substrates directly.
Time-resolved EM of RNA degradation by RE
To directly monitor exosome's RNA processing reaction, we mixed RE with SA-labeled RNA47 under the processing condition (2 mM Mg2+ in buffer) and took samples at different reaction time points for single particle EM analysis as done above. This revealed 10~30% of 2D class averages with clear additional densities corresponding to RE-bound SAs. Based on the locations of SA in the 2D class averages, we grouped the actively processing SA-RNA-RE complex into three major families (Fig. 5): 1) Particles with a single SA density at the top of RE, corresponding to the complex recruiting RNA substrates via the through-core pathway (Fig. 5a); 2) Particles with one single SA density around the bottom part of RE, corresponding to the complex recruiting RNA substrates by direct-access pathway (Fig. 5b); and 3) Particles with two SA densities attached to RE at the top and bottom, respectively, corresponding to the complex recruiting RNA substrates via both pathways simultaneously (Fig. 5c).
Figure 5. Alternative RNA substrate recruitment routes by RE during the degradation process.
(a-c) Representative 2D class-averages from samples of RE incubated with SA coupled RNA47 under 2 mM MgCl2 buffer and their corresponding reprojections and surface renders of 3D models. For (a) and (c), the 2D class averages were compared with the reprojections and surface renders of the RE-long model. For (b), the 2D class averages were compared with the reprojections and surface renders of the RE-short model. In all surface renders, the core of RE was marked with golden color and Rrp44 with blue color. The SA densities in the class averages are marked with red circles. The scale bars represent 20 nm. (d) The distribution of three types of particles over the reaction time. Standard deviation for each point is labeled. The columns for the “both” type at time point 40 min, 90 min and 3 hr are missing due to the negligible detection of it.
We examined the distribution of particles within each family during the reaction (Fig. 5d). Because RNA47 has a long ss-overhang, it is expected to be mainly recruited to the EXO site of Rrp44 via the through-core route. Indeed, in all the reaction time points, family 1 was the most populated among the three families. In agreement with the processive degradation, the population of family 1 particles decreased as the reaction time went on, indicative of the release of shortened SA-RNA substrates from the core channel. In contrast, family 2, albeit less populated than family 1, existed and kept at roughly the same percentage at all reaction time points. Together with a similar assay of SA-RNA24 substrates (Supplementary Fig. 6 and Supplementary Note), this suggests that the direct-access route of RNA substrate recruitment is less competitive than the through-core route on RNAs with long 3’-ss-overhangs but works as a distinct route for RNA substrates with short 3’-ss-overhangs towards deeper processing. The very minor population of family 3 particles at the beginning of the reaction may correspond to some RE complexes competing the through-core and direct-access routes for RNA recruitment.
The results above suggest that RE may adopt a mixture of states during the processing of RNA substrates. Agreeing with this, 3D reconstruction from a full set of particle images taken of RE in the presence of both RNA and Mg2+ showed a much weaker density of the Rrp44 region (Supplementary Fig. 5), indicative of a highly mobile nature of the Rrp44 during RNA processing. Supervised classification of the above dataset revealed the coexistence of roughly 30% RE-short and 70% of RE-long conformers (Supplementary Fig. 5), verifying the coexistence and possible dynamic transition between the two recruitment pathways.
Cryo-EM reconstruction of RE in complex with tRNA substrates
tRNAs are major substrates under the quality control by the exosome and related complexes such as the TRAMP complex, which adds short poly A tail (6 to 15 nt) to the 3’-end of RNAs before processing30-32. To examine RE's transactions with tRNA substrates, we produced various tRNAs with different 3’-end polyA tail from 0 nt to 20 nt using in vitro transcription (lacking post-transcriptional modifications), and performed degradation assays with Rrp44 and RE. tRNAs with tails of at least 10 nt were efficiently processed by Rrp44, while the tail-less tRNA only produced faint degradation products (Fig. 6a). Note that these results differ slightly from another published report6 and this is likely because our assays used a four-fold higher concentration of Mg2+ to prevent unfolding of the tRNA. The removal of the 3’-ss-overhang from the tRNA by RE occurs only when the overhang is 20 nt or longer (Fig. 6a). Because an intermediate overhang length of 20 nt is too short for channeling through the core but longer than required for degradation by Rrp44 alone, it suggests that direct-access products can be generated by RE, and that the core slightly modifies the pathway of the RNA to Rrp44.
Figure 6. tRNA with short 3’ end overhang was degraded through the direct route.
(a) Degradation assay of unmodified tRNAMet with 3’ end tail lengths of 0 (CCA), 10, and 20 nucleotides. Lanes labeled as “R” contain no protein in the reaction, while “Rrp44” and “RE” labeled lanes represent degradation reactions of Rrp44 or RE, respectively. Lanes labeled as “T” are tRNAMet-CCA that is used solely as a size marker. (b) 3D reconstructions of frozen-hydrated Rrp44-Exosome complex incubated with tRNAMet substrate. The classified models of Apo-RE and tRNA-bound RE are shown in the left and right columns, respectively. A clear additional density (labeled with golden color) corresponding to the size and shape of a tRNA can be seen in the tRNA-RE reconstruction. (c) Docking of crystal structures into the Apo-RE (upper row) and RE-tRNA (lower row) 3D models. The crystal structure of the human core exosome (PDB 2NN6)8, the Rrp44 protein of Rrp44-Rrp41-Rrp45 sub-complex structure (PDB 2WP8)22 and Phe tRNA (PDB 4NTA)34 were rigidly docked into Apo-RE or RE-tRNA with Chimera. An ~1.5 nm shift-down of the CSD1-CSD2-RNB-S1 domain for the Rrp44 protein relative to the apo-RE is marked by the orange arrow.
In order to directly visualize tRNA's interaction with RE, we performed cryo-EM 3D analysis of a frozen-hydrated specimen of RE in complex with unmodified yeast tRNAMet. Using single particle 3D maximum-likelihood classification method33, we classified the dataset and reconstructed two major conformers, one as a model of apo-RE and the other as a model of RE with clear additional density attached to the pocket formed by Rrp44, Rrp45 and Rrp43 at the predicted direct-access route (Fig. 6b). The additional density of the latter model can be fitted with the atomic model of a tRNA (PDB ID 4TNA)34 with its 3’-end facing the tunnel to the EXO site of Rrp44 (Fig. 6c). Compared to the apo-RE, the tRNA-bound RE has its C-terminal EXO domain shifting down with about 20 Angstroms likely due to the tRNA insertion (Fig. 6c). These results further support the direct-access route for biologically relevant structured RNA substrates within RE.
Discussion
Compared to its bacterial and archaeal homologues, the eukaryotic exosome core complex has lost the phosphorolytic activity but evolves a function to channel the 3’-end of long ssRNA substrates into the hydrolytic exonuclease Rrp44 for processive degradation. Such a through-core route has been supported by biochemical data, EM analysis, and, most recently, X-ray crystallography18,22,24. To date, a few crystal structures of the RE complex, its sub-complexes, or individual proteins are available but don't fully agree: Rrp44's EXO region adopts very different orientations in the 5’-hairpin-RNA-bound RE complex26 and the Rrp44-Rrp41-Rrp45 sub-complex22. Our work shows that, depending on the RNA substrates, RE adopts at least two conformations, in which Rrp44's orientations agree to the two crystal structures, respectively. Furthermore, the 3D reconstructions of RE bound with a series of ssRNA oligos illustrate that Rrp44's conformational change observed in the crystal structure of the holo-RE complex with RNA substrate is solely induced by the 3’-ss portion of the RNA substrate rather than the presence of the 5’-hairpin or crystallographic packing.
Another agreement between our single particle EM reconstruction analysis and the crystal structure of RE with RNA substrates is the identification of the minimum length of 3’-ss portion as 14 nt to induce the conformational change of Rrp44. RNAs longer than 13 nt would form a continuous linker bridging Rrp45 and Rrp40 of the core and Rrp44. Thus, the continuity of the long ssRNA chain and its multiple interaction sites with RE is essential in holding Rrp44's EXO region in a new orientation so that the cleft between the CSD1 and RNB domains aligns to the exit of core channel (Fig. 7). The fact that such a conformation exists only in the presence of long 3’-ss-overhangs indicates that this conformation is unstable upon the loss of RNA bridging between the core and Rrp44. Rrp44 has the tendency to resume its apo-state, when the ssRNA becomes too short to hold Rrp44's EXO region with the core. We therefore hypothesize that RE breathes between the apo-RE and RE-long conformations so that it can generate a Brownian Ratchet-like pulling force on the 3’-ss-overhang inside the core towards Rrp44's EXO site (Fig. 7). This force, together with the intrinsic elasticity of Rrp44 21, probably contributes to the processivity and weak RNA duplex unwinding activity of the entire complex. Furthermore, this suggests a mechanism of how RE releases RNA substrates with stable secondary or ternary structure at the 5’-end that cannot be unwound by the complex. When the 3’-end of such RNA substrates is processed to a length that can barely hold Rrp44 in the active processing conformation, the tension caused by resuming the apo-conformation could release the RNA's 3’-end from the protein and eventually release those stable RNA substrates from RE for downstream processing and vacate the complex for recruiting new RNA substrates (Fig. 7).
Figure 7. A hypothetical model for the degradation routes of RNA substrates with 3’ single-stranded overhangs in the Rrp44-Exosome.

RNAs with long 3’end ss-overhangs can be recruited via both the through-exosome route (1) and direct-access route (5). The through-exosome recruited RNA substrates can be fully degraded if its 5’ portion does not form a stable structure (2) or will be stalled in the central channel of the core if its 5’ portion has a stable structure (3). A stalled product with short 3’ ss-overhang may be released from the central channel due to the tension built in the complex (4). It may be further processed via the direct-access route just like RNAs with short 3’ end ss-overhangs (5). The direct-access route leads to either further cleaving the 3’ end tail and releasing the processed RNA or fully degrading the RNA substrate (6).
The physiological role of the through-core route for RNA degradation has been firmly established from both in vitro and in vivo assays. The most recent yeast genetics analysis revealed that the central channel of the exosome core is essential for function and viability of budding yeast35,36. But this does not preclude the presence of an alternative RNA recruitment route, which could be very important in processing a subset of RNA substrates. The existence of a possible direct-access pathway for RNA recruitment and processing by RE was first hypothesized based on our EM reconstruction of the RE complex23, as well as implied from genetic and biochemical work from others6,25, but no direct observation of such a route was available. Our data here provides the first direct visual evidence of such an RNA recruitment pathway that bypasses the core channel. Compared to the through-core route, the direct-access route has a weaker affinity for RNA, yet contributes significantly to the RNA processing even at the beginning of a reaction. The direct-access route likely plays a more dominant role in the processing or degradation of RNA substrates with 3’-ss-overhangs shorter than 30 nt, for both single stranded and structured RNAs. This agrees with the transcriptome analysis that reported structured RNAs with short 3’-ss tails contacting directly with Rrp6 and Rrp44 while bypassing the core complex26. The same work also revealed significant substrate spectrum overlap between Rrp6 and Rrp44 with each enzyme being responsible for the processing of different regions of the same substrate. It is plausible that evolutionary pressure preserves the two conformations of RE observed by us for different purposes of RNA processing. While the through-core pathway may be mainly for RNA degradation, the direct-access pathway might be for RNA processing, which removes the 3’-ss-overhangs from structured RNAs. It is also possible that a structured RNA with a long 3’-ss-overhang may first be processed via the through-core pathway, then further cleaved by the direct-access pathway after retracting from the core. In vivo, activities of the two recruitment pathways may be regulated by multiple factors such as the RNA substrate themselves, as for tRNA in this work, and cofactors such as the Rrp6, TRAMP, and SKI complexes. Indeed, Rrp6 has been shown to deviate certain RNA substrates from the core directly to Rrp44 36. Future research needs to be conducted to elucidate the mode of RNA recruitment to the exosome in the context of multiple cofactors to more holistically define the role of the exosome in RNA metabolism in the eukaryotic cell.
Online Methods
Tandem affinity purification (TAP) and reconstitution of S. cerevisiae exosomes
Purification of the core and Rrp44-exosome was done as described with minor modification23. Briefly, ~500 gram of TAP-Rrp46ΔRrp6 yeast cells were lysed with bead-beaters, and after ultracentrifugation the supernatant was loaded onto 3 ml of Rabbit IgG resin (Sigma-Aldrich). The resin was extensively washed with high salt buffer (500 M NaCl), and the Rrp44-exosome was eluted by on-column TEV cleavage to remove the protein A tag. For purifying the core exosome, the Rrp44 was removed from the complex by washing with 40 ml of 1.2 M MgCl2. The mutant Rrp44-exosome complexes were formed by incubating a 50% excess amount of purified recombinant mutant Rrp44 (EX−: D551N, EN−: D117N, or DM: D551N D117N) with the core exosome at 4 °C overnight. All Rrp44-exosome complexes used in the biochemical assays, wild-type or mutant, were further purified on an ion-exchange Mono Q 5/50 GL column (GE Healthcare), where any excess Rrp44 and endogenous nucleic acids were well-separated from the Rrp44-exosome complex (Supplementary Fig. 1g). Samples were flash frozen in liquid nitrogen and stored at −80 °C until later use.
RNA degradation assays
HDV and tRNA substrates were produced by RNA polymerase T7 based in vitro transcription. RNA was separated on a 10% polyacrylamide/8 M urea gel and RNA visualized by UV shadowing. The RNA band was excised from the gel, crushed, and eluted in nuclease free deionized water at 4 °C overnight.
The purified RNAs were dephosphorylated with FastAP Alkaline Phosphotase (Fermentas), phenol extracted, ethanol precipitated, and dissolved in nuclease free water. RNA was 5’ end labeled with Polynucleotide Kinase (New England Biolabs) and 2 μl of 3000 Ci/mmol 32γP-ATP (Perkin Elmer), purified on 10% polyacrylamide/8 M gels, eluted and ethanol precipitated. RNA pellets were washed with 80% ethanol, air dried, dissolved in a final volume of 30 μl nuclease free water, and stored at −20°C until use.
Immediately before use, RNA substrates were refolded in 2X reaction buffer (20 mM HEPES pH 7.5, 100 mM NaCl, and twice the concentration of MgCl2 used in the assay) by heating to 80 °C for 5 minutes followed by snap cooling at 4 °C. All reactions were performed in 1X reaction buffer, where the molar ratio of protein to RNA was 2:1, and incubated at 30 °C for indicated time points. The reactions were terminated by adding an equal volume of formamide plus 10 mM EDTA and heating the solution at 65°C for 10 minutes. The reaction products were analyzed on 16% acrylamide/8M Urea sequencing gels in 0.5X TBE and quantified by a Typhoon Phosphorimager (GE Healthcare).
Preparation of exosome coupled IgG resin beads
An amount of 100 ml supernatant of TAP-Rrp46-ΔRrp6 yeast lysate was incubated with rabbit IgG affinity resin beads (Sigma-Aldrich, St. Louis, MO) at 4 °C for 2 hours with constant stirring. The resin was then centrifuged at 500 rpm for 2 minutes and washed three times each with 10 ml washing buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10% glycerol with 2mM DTT, 2mM EGTA and 1 tablet of complete EDTA-free protease inhibitor mixture from Roche Applied Science). The resin was then washed three times each with 10 ml of high-salt buffer (washing buffer plus 250 mM NaCl) and the beads were re-equilibrated three times with 5 ml washing buffer.
Gel shifting and pull down assays
Streptavidin (SA) was purchased from Sigma and HRP-coupled streptavidin (HRP-SA) was from Abicom. For gel shifting assays, proteins and RNAs were mixed together in a 4:1 molar ratio in nuclease free water and incubated on ice for 5 minutes, then run on a 6% acrylamide native gel in 0.5X TBE buffer. The gels were subsequently stained with ethidium bromide and imaged in a regular UV illuminated gel scanner.
For pull down assays, HRP-SA and RNAs were mixed together in a 4:1 molar ratio and incubated on ice for 5 minutes. 1μl of HRP-SA-RNA complex (2.5uM) was incubated with 30μl of exosome coupled IgG resin in 500 μl of reaction buffer (50 mM Tris·HCl pH 8.0, 150 mM NaCl, 5mM EDTA, 1mM DTT) at room temperature (~22 °C) for 15 minutes with occasional stirring. The mixture was centrifuged at 500 rpm for 2 minutes, and the resin was washed three times each with 1.5 ml reaction buffer. 2 μl supernatant or beads were loaded on nitrocellose film. The film was subsequently incubated in ECA agent for HRP signal development as in normal Western Blotting.
EM sample preparation
The RNA-RE complex for EM analysis was prepared by mixing RNA and RE in a 1.2:1 molar in a non-digestive buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5mM EDTA, and 1mM DTT at room temperature for 15 minutes. For preparing SA-RNA-RE sample in non-digestive condition, SA and biotinylated RNAs were first mixed together in nuclease free water and incubated on ice for 15 minutes in a 2:1 molar ratio and diluted to a final concentration of 6 μM of RNA. Subsequently, the SA-RNA mixture was incubated with RE complex in non-digestive buffer as described above in a molar ratio of 1.2:1. All samples were diluted at a final concentration of ~80 nM of the exosome in the non-digestive buffer and negatively stained in 2% (w/v) uranyl acetate solution following the standard deep stain procedure on holey-carbon coated EM copper grids covered with a thin layer of continuous carbon 38. For SA-RNA-RE samples in magnesium condition, everything followed the same procedure except for that the non-digestive reaction buffer was replaced by a magnesium buffer (50 mM Tirs-HCl pH 8.0, 150 mM NaCl, and 2 mM MgCl2) and the incubation time of SA-RNA and RE was 5 minutes.
Time-resolved EM sample preparation
To monitor the structure of SA-RNA being processed by RE in a degradation reaction, we first mixed 4 μl of SA-RNAs produced as described above with 2 μl of 3X reaction buffer (150 mM Tirs-HCl pH 8.0, 450 mM NaCl, 6 mM MgCl2) at 30 °C. Subsequently, we added 4 μl of RE (2.5 μM dissolved in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, and 2 mM MgCl2) to the reaction system to initiate the reaction. At given reaction time points, the reaction was terminated with the addition of 10 μl termination buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 20 mM EDTA) and immediate cooled on ice. 4 μl of the final solution was then negatively stained.
Electron microscopy and image processing
We examined all the negative stained specimens on an FEI Tecnai-F20 electron microscope operated at 200 kV acceleration voltage at 50,000 nominal magnification with a range of defocus from 0.8 to 1.2 μm. The electron micrographs were taken on a Gatan Ultrascan4000 4k X 4k CCD camera.
The digital micrographs taken from CCD camera was then processed using EMAN2 package 39 to semi-automatically pick the individual particle images of the complexes and using IMAGIC-4D package40 to determine and correct the contrast transfer functions using phase-flip method. For each sample, 20,000-50,000 negatively stained particle images were analyzed two-dimensionally using IMAGIC-4D. Briefly, we performed iterative multi-variant statistical analysis (MSA) and multi-reference alignment (MRA) cycles on the particle stacks using IMAGIC-4D to obtain 2D class averages as described previously38. We performed 3D reconstruction of all the samples using projection-matching refinement of particle stacks against the same initial model of apo-RE (EMD1439 23) low-pass filtered to 70 Å in SPIDER41 as described previously38. The resolution of final reconstruction was estimated using Fourier Shell Correlation (FSC) algorithm at a criteria of 0.5. The number of particles contributing to and the resolution of each 3D reconstruction (measured by FSC 0.5 criteria) are summarized in Supplementary Table 4. The distinct differences among the final reconstructions indicated that our results are free of model bias. This was also verified by using a totally different initial model of an elongated bar to converge to the same structures of RE sample after about 30 rounds of projection-matching refining iterations. The model-bias free nature of the RE sample is very likely due to its strong intrinsic structural features in the high contrast negative stained images.
The docking of atomic models were performed by UCSF-Chimera42 using fit-model-in-map functions. All the 3D models in the figures are rendered in UCSF-Chimera.
Cryo-EM of RE in complex with tRNA
We incubated 250 nM RE with 1 μM unmodified yeast tRNAMet in reaction buffer (50mM Tris-Hcl, 150mM NaCl, 2mM MgCl2) on ice for 10 min. 4 μl of the reaction solution was then applied to glow-discharged C-flat grids (1.2/1.3) covered with a layer of continuous carbon with a thickness of ~4 nm. The grids were then blotted and plunged into liquid ethane in a FEI Vitrobot Mark IV. The frozen-hydrated grids were subsequently examined under a FEI Titan Krios electron microscope operated at 120 kV acceleration voltage. Micrographs of RE were collected using the AutoEMation software 43 installed on the microscope at low-dose condition with a dose of ~20 electron/Å2 and a defocus value ranging from −1.2 to −4 μm. The micrographs were collected on a FEI Eagle CCD camera with a pixel size of 1.5 Å. We used EMAN2 package to perform raw micrograph screening, CTF determination, and semi-automatic particle picking39 and obtained a dataset of about 61,000 particle images. We then performed 3D refinement and classification of the particle images using the Relion package44. After the first 25 iterations of classification, we identified two major classes of particles (accounting for more than 56,000 images) with good quality of reconstruction out of six classes. We further performed 3D refinement of the two classes and got a final reconstruction of the RE-tRNA complex from about 28,800 particles and of the apo-RE complex from about 27,200 particles, respectively. The number of particles contributing to and the resolution of each 3D reconstruction (measured by FSC 0.5 criteria) are summarized in Supplementary Table 4.
Back-projection analysis of SA-RNA24-RE complex
For back-projection analysis of the SA-RNA24-RE complex, we calculated the Euler angle of the 2D class averages by firstly masking off the SA densities in the 2D class averages and then matching them to reprojections of the 3D model of apo-RE. The corresponding view of the 3D model of apo-RE matching to a 2D class average was then displayed in UCSF-Chimera. A 3D model of rod perpendicular to the view was added in UCSF-Chimera to represent the back-projection of the corresponding SA density in the 2D class average. By applying this procedure to all the views of the 3D model matching to the 2D class averages with SA density, we finally visualize the cross-point of all the rods, assuming that the most crossed points have a higher probability to represent the location of SA in the 3D model.
Supplementary Material
Acknowledgement
We thank J.S. Butler of University of Rochester for providing the yeast strain of TAP-Rrp46 deltaRrp6, J.-L. Lei and Y. Xu for the EM support, J. Wang for EM image processing scripting help, the Wang group members at Yale University and Tsinghua University and the Sui group members at Tsinghua University for their helpful discussions, Prof. E. Nogales, S. Wolin, and S. Sui for their helpful comments on the manuscript. We acknowledge the China National Center for Protein Sciences Beijing and “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology for providing the facility support. This study was funded by the National Basic Research Program of China grant 2010CB912401 (to H.-W.W.), US NIH grant GM-086766 (to A.K.). H.-W.W. is supported as a principle investigator of the Tsinghua-Peking Joint Center for Life Sciences and the awardee of Youth One-Thousand Talent Program by the State Council of China. J.-J.L. is supported as the awardee of Gatan Scholarship. M.A.B. was partially supported by the US NIH Molecular Biophysics Training Grant (GM-008267).
Footnotes
Author Contributions
J.-J.L., X.L., C.-Y.N., H.-W.W. performed EM and single-particle analysis. J.-J.L., M.A.B., X.L., C.-Y.N. purified exosome complex. M.A.B. and A.K. performed RNA degradation assays of wild-type and mutant complexes. J.-J.L., M.A.B., X.L., A.K. and H.-W.W. planned the experiments. J.-J.L., M.A.B., A.K. and H.-W.W. wrote the manuscript.
Accession Codes
The three-dimensional reconstruction electron density maps of RNA08-RE, RNA10-RE, RNA12-RE, RNA14-RE, RNA16-RE, RNA24-RE, RNA36-RE, RNA47-RE, SA-RNA47-RE and SA-RNA50-RE by negative stain EM have been deposited into the Electron Microscopy Data Bank under accession codes EMD-2491, EMD-2492, EMD-2493, EMD-2494, EMD-2495, EMD-2496, EMD-2497, EMD-2498, EMD-2499 and EMD-2500, respectively. The three-dimensional reconstruction electron density maps of tRNA-RE and apo-RE by cryo-EM have been deposited into the Electron Microscopy Data Bank under accession codes of EMD-2451 and EMD-2452, respectively.
References
- 1.Allmang C, et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–91. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 4.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–52. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadaba S, et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–40. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–31. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–75. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–71. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Lorentzen E, Conti E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell. 2005;20:473–81. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Lorentzen E, et al. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–81. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 12.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 13.Allmang C, et al. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–6. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer D, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–40. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Makino DL, Baumgartner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–5. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- 19.Zuo Y, et al. Structural basis for processivity and single-strand specificity of RNase II. Mol Cell. 2006;24:149–56. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Frazao C, et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 21.Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science. 2012;336:1726–9. doi: 10.1126/science.1216848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–59. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Wang HW, et al. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci U S A. 2007;104:16844–9. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malet H, et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–42. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science. 2011;334:817–21. doi: 10.1126/science.1213671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide analysis of exosome targets. Mol Cell. 2012;48:422–33. doi: 10.1016/j.molcel.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferre-D'Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–74. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 28.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–5. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 29.Rosenstein SP, Been MD. Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry. 1990;29:8011–6. doi: 10.1021/bi00487a002. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–16. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia H, et al. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheres SH. Classification of structural heterogeneity by maximum-likelihood methods. Methods Enzymol. 2010;482:295–320. doi: 10.1016/S0076-6879(10)82012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hingerty B, Brown RS, Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978;124:523–34. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- 35.Drazkowska K, et al. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–58. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell. 2012;48:133–44. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Trong I, et al. Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin. Acta Crystallogr D Biol Crystallogr. 2003;59:1567–73. doi: 10.1107/s0907444903014562. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Wang HW. Single particle electron microscopy reconstruction of the exosome complex using the random conical tilt method. J Vis Exp. 2011 doi: 10.3791/2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 41.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Lei J, Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol. 2005;150:69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.