Abstract
Isolation of cells from heterogeneous mixtures is critically important in both basic cell biology studies and clinical diagnostics. Cell isolation can be realized based on physical properties such as size, density and electrical properties. Alternatively, affinity binding of target cells by surface-immobilized ligands, such as antibodies, can be used to achieve specific cell isolation. Microfluidics technology has recently been used in conjunction with antibody-based affinity isolation methods to capture, purify and isolate cells with higher yield rates, better efficiencies and lower costs. However, a method that allows easy release and collection of live cells from affinity surfaces for subsequent analysis and detection has yet to be developed. This paper presents a microfluidic device that not only achieves specific affinity capture and enrichment, but also enables non-destructive, temperature-mediated release and retrieval of cells. Specific cell capture is achieved using surface-immobilized aptamers in a microchamber. Release of the captured cells is realized by a moderate temperature change, effected via integrated heaters and a temperature sensor, to reversibly disrupt the cell-aptamer interaction. Experimental results with CCRF-CEM cells have demonstrated that the device is capable of specific capture and temperature-mediated release of cells, that the released cells remain viable and that the aptamer-functionalized surface is regenerable.
1. Introduction
Isolation of cells from biological samples involves the separation and retrieval of cell subpopulations from a heterogeneous mixture in blood or other body fluids, and is widely used in both fundamental cell biology research and clinical diagnostics. For example, the ability to detect and characterize cancer cells from blood or other body fluids is essential for detecting cancer in the early stages and understanding cancer development and progression mechanisms, such as metastasis, which would significantly improve survival rates.1 In addition, studies of phenotypically pure subpopulations of human lymphocytes can provide valuable information concerning immune responses to injury and disease.2 To enable these applications, target cells must be selectively captured, and in some instances, such as tissue engineering3 and cell-based therapeutics,4 retrieved nondestructively without any mechanical or biochemical damage.
Isolation of cells can be based on the size or volume, density, electrical properties or surface characteristics, using methods such as filtration, centrifugation, dielectrophoresis or affinity binding.5–7 Among these methods, affinity binding, which recognizes cells by binding of ligands to biomarkers on cell membranes, is highly attractive due to its high specificity to target cells. The most commonly employed ligands for affinity cell isolation are antibodies, which are generated in vivo against target antigens found on cell membranes. Antibody-based cell isolation techniques have been implemented using methods such as magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS).7,8 MACS uses a magnetic field to manipulate antibody-coated microbeads specifically bound to the cells and is amenable to high-throughput operation.9 Relying on a single physical parameter (i.e., the presence or absence of magnetic force) to distinguish between different cell types, MACS is in general a single-parameter cell isolation method. In comparison, FACS usually uses multiple species of fluorescently labeled antibodies and allows sorting of multiple cell types simultaneously.8 Thus, FACS is a multi-parameter method, although it is relatively low-yield and typically requires more complex and expensive experimental instrumentation.
Microfluidic technology can potentially enable low-cost, automated and portable cell isolation systems with improved sensitivity, resolution and efficiency as well as minimized sample and reagent consumption.10 Microfluidic cell isolation devices have implemented the aforementioned separation principles, for instance exploiting differences in the size,11 acoustic response,12 dielectrophoretic characteristics13 and affinity of cells to ligands.1 In particular, microfluidic affinity assays using ligands with high specificity to cell membrane proteins have shown great promise for cell isolation.1,14 For example, by using anti-epithelial cell adhesion molecule (anti-EpCAM) antibodies immobilized on microposts, the isolation of rare circulating tumor cells (CTCs) from samples of whole blood has been demonstrated.1 Unfortunately, antibodies generally have limited stability, are expensive and time-consuming to develop, and antibody-coated surfaces are susceptible to biofouling due to non-specific interactions between antibodies and other proteins.15 Moreover, rapid and non-destructive release of antibody-bound cells is generally difficult,16 as the antibody-antigen binding is practically irreversible under normal physiological conditions.17
Aptamers are oligonucleotides or peptides that bind specifically to target molecules. Isolated from a randomized oligonucleotide or peptide library using a synthetic selection process called systematic evolution of ligands by exponential enrichment (SELEX),18 aptamers can recognize a large variety of target biomolecules, such as small molecules,19 peptides20 and proteins,21 via unique three-dimensional conformations formed through interactions with the targets. Recently, aptamers have also been developed for cellular targets, such as acute lymphoblastic leukemia (ALL) precursor T cells,22 liver cancer cells23 and even stem cells.24 Since aptamers are produced using synthetic processes and are stable and amenable to chemical modifications,25 they offer an attractive alternative to antibodies as affinity ligands for isolation of rare cells. Although most aptamers use cell lines as their targets, they are often capable of targeting the more general population of diseased cells from real patient samples. For example, the aptamer sgc8c generated for a type of human ALL T cell line, CCRF-CEM cells, is capable of specifically targeting ALL T cells in patient blood.22,26,27 Thanks to ongoing research efforts to develop improved SELEX methods and instruments, it is expected that aptamers will become readily available to recognize an increasingly broad collection of biological targets.28
Aptamers have been explored in microfluidic systems as affinity ligands for cell isolation.29–32 For example, surface-immobilized aptamers targeting prostate-specific membrane antigen (PSMA) and aptamers targeting protein tyrosine kinase 7 (PTK7) have been used to separate LNCaP cells and CCRF-CEM cells, respectively, from heterogeneous cell mixtures.29,30,32,33 However, there has been very limited work on releasing the captured cells from aptamer-functionalized surfaces. Attempts to rapidly and non-destructively release cells using methods, such as tryptic digestion of target proteins,32 exonuclease degradation of aptamers34 and hydrodynamic shear by infused air bubbles,30 have been hindered by several issues. Trypsin is able to digest only a small portion of biomarkers involved in affinity cell capture35 and may negatively affect cell viability and phenotypic property,36,37 while the use of exonucleases is limited by inefficient diffusive transport of the enzymes, slow enzymatic reaction rates and the destruction of the cell recognition surfaces. Similarly, air bubbles, which release cells from aptamer surfaces by shear force, may cause physical damage to cells, while also generating dead volumes in microfluidic devices that lead to low cell release efficiency. There is hence a strong need for methods that allow specific capture and efficient, non-destructive release of subpopulations of cells in microfluidic devices.
Aptamer-target interactions are significantly influenced by environmental conditions; in particular, the affinity binding between aptamers and biomolecules can be strongly temperature dependent, as has been shown in our previous work.38–40 Cell-specific aptamers also target biomolecules, in this case proteins expressed on the cell membrane.33 Therefore, it is reasonable to expect that interactions between aptamers and cells are also highly temperature sensitive. We have previously demonstrated the use of aptamer-coated solid surfaces for thermally controlled purification and enrichment of molecules,15,38–41 and conducted preliminary experiments to extend this approach to the manipulation of cells.42,43 In the current work, we build on these efforts to investigate the specific capture and subsequent temperature-mediated release of cells in an aptamer-functionalized microfluidic device. Target cells can be captured with high specificity by a surface-immobilized aptamer, enriched by repeated introduction of multiple samples, and purified by washing with buffer. This is followed by a moderate temperature change of the surface, produced on-chip, to reversibly disrupt the cell-aptamer interaction, allowing release and elution of viable target cells for downstream analysis and regeneration of the aptamer-functionalized surface for device reuse. The device is applied to CCRF-CEM cells to demonstrate its potential utility for specific capture and non-destructive release of cells in basic biological research and clinical diagnostics.
2. Principle and design
This section describes the principle and design of our microfluidic device. We first describe the specific cell capture and thermally controlled cell release approach, and then present the device design.
2.1. Cell Capture and release principle
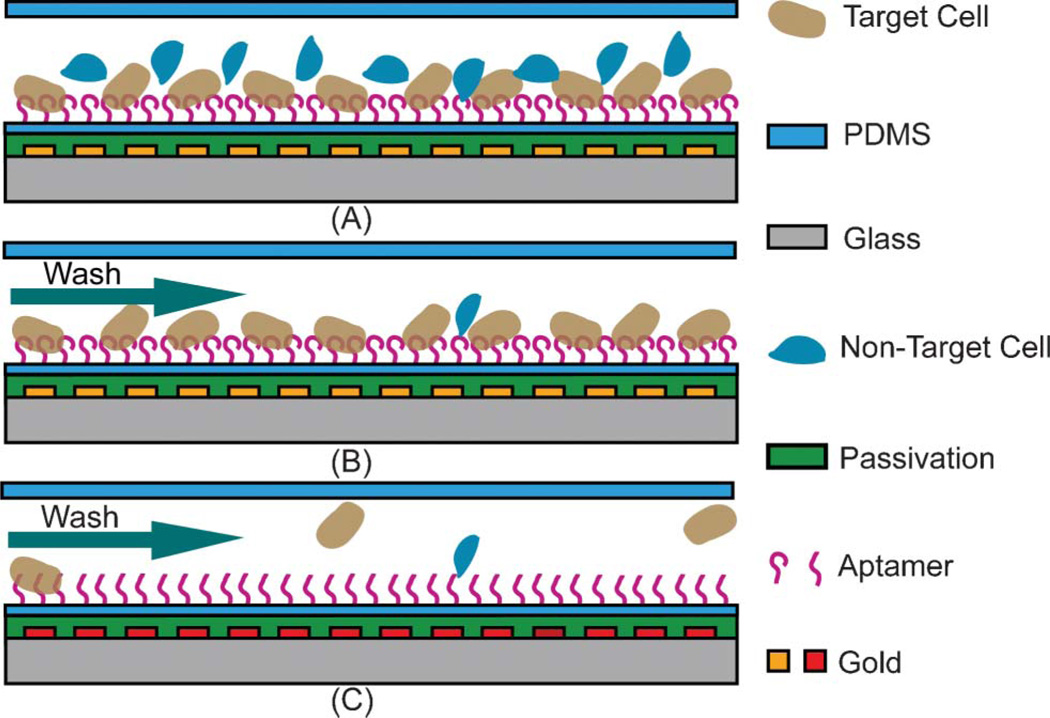
The principle of aptamer-based specific capture and temperature-mediated release of cells is as follows. Aptamer molecules specific to target cells are immobilized on solid surfaces within a microfluidic device. A cell suspension containing target and non-target cells is introduced into the device and incubated for an appropriate period of time. The target cells are specifically captured by the surface-immobilized aptamer molecules via affinity binding (Fig. 1A), while the non-target cells are removed by washing (Fig. 1B). Next, exploiting the strong temperature-dependence of affinity binding of aptamers and cells, the device temperature is increased to disrupt the binding and release the captured target cells from the surface-immobilized aptamer molecules (Fig. 1C). This temperature-mediated cell release can be accomplished in such a way that the temperature increase does not affect the viability of released cells.
Fig. 1.
Principle of specific cell capture and temperature-mediated release. (A) Cell capture at room temperature. (B) D-PBS wash after capture. (C) Cell release at a moderately higher temperature.
The principle of aptamer-based specific capture and temperature-mediated release of cellular targets will be demonstrated using CCRF-CEM cells, a human ALL cell line. ALL is the most common cancer for children from 0 to 14 years old, representing one third of all malignancies in that age group.44,45 CCRF-CEM cells are recognized by the DNA aptamer sgc8c.27,29,30 Toledo cells, a human diffuse large-cell lymphoma cell line not recognized by sgc8c, are used as a control target to study nonspecific binding.22,27
2.2. Design
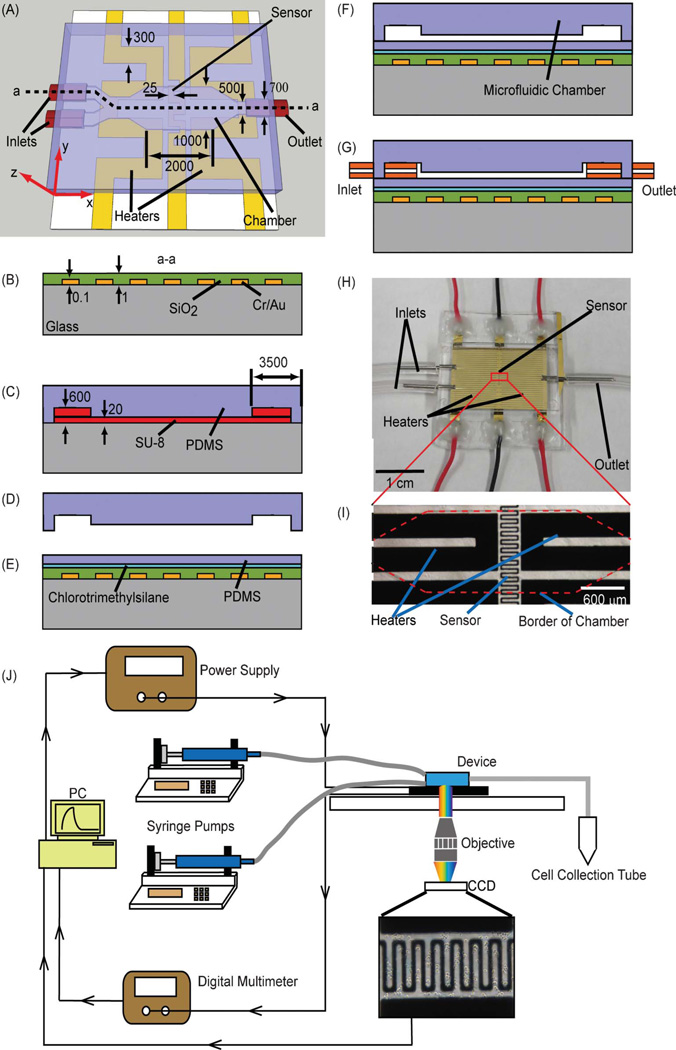
The microfluidic device used for cell capture and temperature-mediated cell release consists of a microchamber situated on a temperature control chip (Fig. 2A). The tapered chamber (2 mm in length, 1 mm in width and 20 µm in height), whose surfaces are functionalized with aptamers specific to a target cell type, is connected to two inlets (3.5 mm in length, 0.7 mm in width and 600 µm in height) respectively for introduction of sample and washing buffer, and one outlet for collection of released cells or waste fluids. The microfluidic channels connecting these fluidic ports and the chamber are 0.5 mm in width and 20 µm in height. Integrated on the temperature control chip are a serpentine-shaped temperature sensor (linewidth: 25 µm) beneath the center of the chamber, and two serpentine-shaped heaters (linewidth: 300 µm) on each side of the temperature sensor. The chamber temperature can be controlled in a closed loop using these integrated temperature sensor and heaters.
Fig. 2.
(A) Schematic of the microfluidic device for the specific capture and temperature-mediated release of CCRF-CEM cells. (B) Deposition, patterning and passivation of gold sensor and heaters. (C) Fabrication of SU-8 mold. (D) Demolding of PDMS microchamber. (E) Treatment with chlorotrimethylsilane and PDMS spin-coating. (F) Bonding of PDMS microchamber to PDMS-coated temperature control chip. (G) Insertion of inlet and outlet capillary tubes. (H) Photograph of a fabricated device. (I) Micrograph of the temperature sensor and heaters in the device. (J) Experimental setup for specific capture and temperature-mediated release of cells. Dimensions are given in micrometers.
3. Experimental
3.1. Materials
Chlorotrimethylsilane, (3-mercaptopropyl) trimethoxysilane (3-MPTS), 4-maleimidobutyric acid N-hydroxysuccinimide ester (GMBS), streptavidin and bovine serum albumin (BSA) were obtained from Sigma-Aldrich (St. Louis, MO). 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1), propidium iodide (PI), RPMI-1640 media, fetal bovine serum (FBS), penicillin-streptomycin (P/S, penicillin 10 000 unit mL−1, streptomycin 10 000 µg mL−1), Dulbecco’s phosphate-buffered saline (D-PBS) and the Vybrant® multicolor cell-labeling kit (DiI, DiO and DiD) were purchased from Invitrogen (Carlsbad, CA). CCRF-CEM and Toledo cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The biotinylated sgc8c aptamer with a polyT(9) spacer at the 5′ end of the sequence (biotin-5′-TT TTT TTT TAT CTA ACT GCT GCG CCG CCG GGA AAA TAC TGT ACG GTT AGA-3′, Kd = 0.78 nM) was synthesized and purified with high-performance liquid chromatography (HPLC) by Integrated DNA Technologies (Coralville, IA).
3.2. Microfluidic device fabrication
The temperature control chip was fabricated using standard microfabrication techniques. A glass slide (Fisher HealthCare, Houston, TX) was cleaned by piranha. Chrome (10 nm) and gold (100 nm) thin films were deposited by thermal evaporation and patterned by wet etching to generate the temperature sensor and heaters which were then passivated by 1 µm of silicon dioxide that was deposited using plasma-enhanced chemical vapor deposition (PECVD). Finally, contact regions for electrical connections to the sensor and heaters were opened by etching the oxide layer using hydrofluoric acid (Fig. 2B).
In parallel, the microchamber was fabricated from polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning Inc., Midland, MI) using soft lithography techniques. Layers of SU-8 photoresist (MicroChem Corp., Newton, MA) were spin-coated on a silicon wafer (Silicon Quest International, Inc., San Jose, CA), exposed to ultraviolet light through photomasks, baked and developed to form a mold defining the microfluidic features. Next, a PDMS prepolymer solution (base and curing agent mixed in a 10 : 1 ratio) was cast onto the mold and cured on a hotplate at 72 °C for 1 h (Fig. 2C). The resulting sheet bearing the microfluidic features was then peeled off the mold (Fig. 2D).
Subsequently, the surface of the temperature control chip was treated with chlorotrimethylsilane, and a PDMS layer (approximately 100 µm) was spin-coated onto the chip (Fig. 2E). Then, the PDMS sheet was bonded to the PDMS membrane on the chip after treatment of the bonding interfaces with oxygen plasma for 15 s (Fig. 2F). Finally, capillary tubes (O.D. = 813 µm and I.D. = 495 µm) were inserted into the inlet and outlet ports (Fig. 2G), resulting in a packaged device. Following each experiment, the PDMS film and chamber were easily removed from the temperature control chip, allowing the chip to be reused for the next experiment. A fabricated and packaged device is shown in Fig. 2H and I.
3.3. Surface modification
The biotinylated sgc8c aptamer was functionalized in a freshly fabricated device using an established protocol with slight modifications.1 The microchamber was first treated with 4% (v/v) 3-MTPS in ethanol for 30 min at room temperature, followed by an ethanol wash. Then, 2 mM GMBS in ethanol was introduced and incubated for 20 min at room temperature, followed by an ethanol wash and drying by nitrogen. Next, the chamber was incubated overnight with 100 µg mL−1 streptavidin in D-PBS at 4 °C, followed by a D-PBS wash. Finally, 10 µM of biotinylated sgc8c aptamer in D-PBS was introduced into the chamber and incubated at room temperature for 20 min. A D-PBS wash was used to remove free aptamer molecules, leaving immobilized aptamer molecules on the surface. Just prior to cell introduction, the chamber was incubated with 1 mg mL−1 BSA solution in D-PBS at room temperature for at least 30 min to minimize nonspecific adsorption of cells.
3.4. Cell Culture and preparation
Both CCRF-CEM and Toledo cells were incubated with RPMI-1640 media supplemented with 10% FBS and 1% P/S, and were kept at 37 °C in a humidified incubator containing 5% CO2. Before microfluidic experiments, each cell type was collected through centrifugation, resuspended at 1 × 108 cells mL−1 in D-PBS supplemented with 1 mg mL−1 BSA, and then kept on ice. Cells were mixed or diluted to different concentrations prior to introduction into the microfluidic device.
3.5. Experimental setup
Closed-loop temperature control of the chamber was achieved using the integrated temperature sensor and heaters with a proportional-integral-derivative (PID) algorithm implemented in a LabVIEW (National Instruments Corp., TX) program on a personal computer. The resistance of the sensor was measured by a digital multimeter (34420A, Agilent Technologies Inc., CA), and the two heaters were connected to a DC power supply (E3631, Agilent Technologies Inc., CA). The microfluidic device’s two inlets were connected to syringes that respectively contained cell mixture and D-PBS, and were each driven by a syringe pump (KD210P, KD Scientific Inc., MA). The outlet was connected to a microcentrifuge tube for collection of released cells or experimental waste. Unless indicated otherwise, all phase contrast images and fluorescent images of the chamber were taken using an inverted epifluorescence microscope (Diaphot 300, Nikon Instruments Inc., NY) with a CCD camera (Model 190CU, Micrometrics, NH) (Fig. 2J).
3.6. Testing procedure
During cell capture experiments, a batch of CCRF-CEM cells was introduced into the chamber and incubated without any fluid flow for 1 min. This process was repeated several times, followed by a wash with D-PBS at 5 µL min−1 for approximately 1 min. An image of the cell-laden chamber was taken and used to manually count the number of captured cells, which was used to compute the captured cell density on the surface. To test the specificity of cell capture, CCRF-CEM and Toledo cells were labeled with the fluorescent dyes DiO and DiI, respectively, and fluorescent images were taken after the first introduction of the cell mixture as well as after D-PBS washing.
In temperature-mediated cell release experiments, the chamber was heated using the integrated heaters via closed loop temperature control to a desired temperature for 2 min, and flows of D-PBS at various rates were used to rinse the chamber. Images of the chamber were taken every 2 s, and used to manually count the cells that remained on the aptamerimmobilized surface.
To test cell viability, the retrieved cells were kept in D-PBS with 10% FBS containing PI (2 µM) and JC-1 (10 µg mL−1) at 37 °C for 1 h, and then phase contrast and fluorescent images were taken with an inverted microscope (DMI6000B, Leica Microsystems Inc., IL) equipped with a digital camera (Retiga 2000R, Qimaging, Canada) and commercial image acquisition software (InVitro, Media Cybernetics Inc., MD). Moreover, a batch of cells was treated in a water bath at 48 °C for 2 min and then cultured for 4 days. The concentration of cultured cells was determined each day using a hemacytometer (Chang Bioscience Inc., CA).
4. Results and discussion
This section presents and analyzes experimental results from the aptamer-based microfluidic device. We first characterize the temperature control chip and evaluate the uniformity of the temperature field in the chamber (Supplementary Information†). Then, specific capture and temperature-mediated release of cells are performed using CCRF-CEM and Toledo cell lines to investigate factors that influence the efficiency of cell capture and release. Finally, the viability of retrieved cells is evaluated to confirm the non-destructive nature of the cell release mechanism.
4.1. Specific cell capture
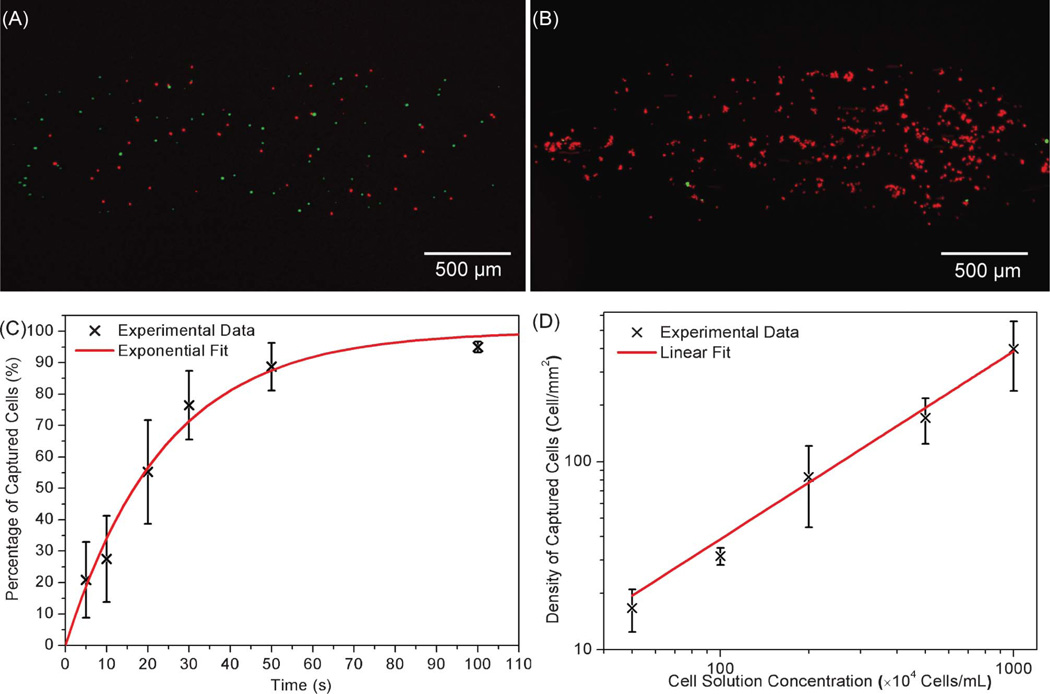
To verify specific cell capture at room temperature, a mixture of CCRF-CEM cells (target cell type, 3.5 × 106 cells mL−1) and Toledo cells (non-target cell type, 5.0 × 106 cells mL−1) was introduced into the sgc8c aptamer-modified microchamber and incubated for 1 min. The total number of CCRF-CEM cells observed on the surface, 51 in total, was less than that of Toledo cells, 78 in total (Fig. 3A). However after washing, all nonspecifically adsorbed Toledo cells were removed, leaving only specifically captured CCRF-CEM cells on the surface. Moreover, after 10 cell samples were introduced (each followed by rinsing with D-PBS), the target cells dominated the chamber surface, with only 8 non-target Toledo cells visible amongst a few hundred CCRF-CEM cells (Fig. 3B). This demonstrates the specific and effective capture of CCRF-CEM cells using the surface-immobilized aptamers, and the capability of the device to enrich target cells from a heterogeneous mixture.
Fig. 3.
(A) Image of the chamber after the introduction of a cell sample. (B) Image of the chamber after the introduction of 10 cell samples and D-PBS washing. (C) Time response of cell capture: percentage of cells captured (η) versus incubation duration (t). The solid line represents an exponential fit to the experimental data with a regression equation: η = 1 − e−t/24 (R2 = 0.982, n = 3). (D) Concentration response of cell capture: density of captured cells (ρcapture) as a function of the cell suspension concentration (ccell). The solid line represents a linear fit to the experimental data with a regression equation: ρcapture = 0.3874ccell (R2 = 0.995, n = 3).
To test the transient behavior of the cell capture process, CCRF-CEM cell suspensions with concentrations of 5.0 × 106 cells mL−1 were introduced into the aptamer-functionalized chamber and allowed to incubate for varying lengths of time. After incubation, D-PBS was used to remove unbound cells. The fraction of captured cells in each introduction was calculated by η ≈ Na/Nb, where Na is the number of captured cells, i.e., cells that remained on the microfluidic aptamer-functionalized surface after washing, and Nb is the maximum number of cells that can be captured due to geometric limitations. Because of the height of the chamber (20 µm) and the low cell density of the introduced cell suspension, we assumed that only a single monolayer of cells could be arranged on the lower surface of the chamber. Therefore, Nb is also equal to the number of cells observed in the chamber before washing. As shown in Fig. 3C, increasing incubation time resulted in an increase in cell surface density. The captured cell percentage (calculated from three repeated experiments, n = 3) revealed an approximately exponential dependence on incubation duration η = 1 − e−t/τ, where τ is a constant, and t is the incubation duration. According to this relationship, cell loss during washing could be almost completely eliminated via incubation by setting t (incubation time) to a value such that η approximates to 1. The constant τ indicates the rate at which the surface density of captured cells approaches its maximum value, and can be used to calculate the time needed to isolate a number of target cells from the heterogeneous cell suspension. An exponential fit to the experimental data indicated an accurate relationship (coefficient of determination R2 = 0.982), and yielded a value of τ equal to 24 s. Based on this first-order exponential fit, it was estimated that approximately 92% of introduced cells exposed to the aptamer-functionalized surface were captured after incubating for 1 min. These results, which were similarly obtained at other cell concentrations ranging from 0.5 × 106 to 10 × 106 cells mL−1, could be further improved by optimizing the channel design,1 surface topography,34 and operation conditions such as flow rates.46
We next investigated the effects of the cell suspension concentration on the surface density of captured cells. Cell capture experiments were conducted using samples with varying cell concentrations (0.5 to 10 × 106 cells mL−1). In each experiment, 5 aliquots of cells were introduced into the chamber, each followed by a 1-min incubation. Each test was performed in triplicate simultaneously on identical devices (n = 3). All of the devices were fabricated at the same time to guarantee chamber surfaces were generated with nominally identical aptamer densities to ensure consistent experimental data. Experiments with the most dilute cell suspension (0.5 × 106 cells mL−1) yielded captured cells with a surface density of 17 ± 4 cells mm−2 (n = 3), while those with the most concentrated cell suspension (10 × 106 cells mL−1) resulted in a captured cell density of approximately 399 ± 160 cells mm−2 (n = 3), as shown in Fig. 3D. It can be seen that in this range of cell concentrations, the captured cell density was approximately proportional to the cell concentration ρcapture = A ccell, where ccell is the cell suspension concentration (cells mL−1), and A is a proportionality constant that depends on device characteristics such as the surface density of immobilized aptamer molecules and equilibrium cell-aptamer affinity association, and testing parameters such as the number of samples introduced to the chamber. The linear equation fitted the experimental data well (R2 > 0.99), resulting in a value of A equal to 0.3874 mL mm−2. These results indicate that there is a large dynamic range of cell suspension concentrations over which the device can capture a predictable surface concentration of cells for downstream analysis.
4.2. Temperature-mediated cell release
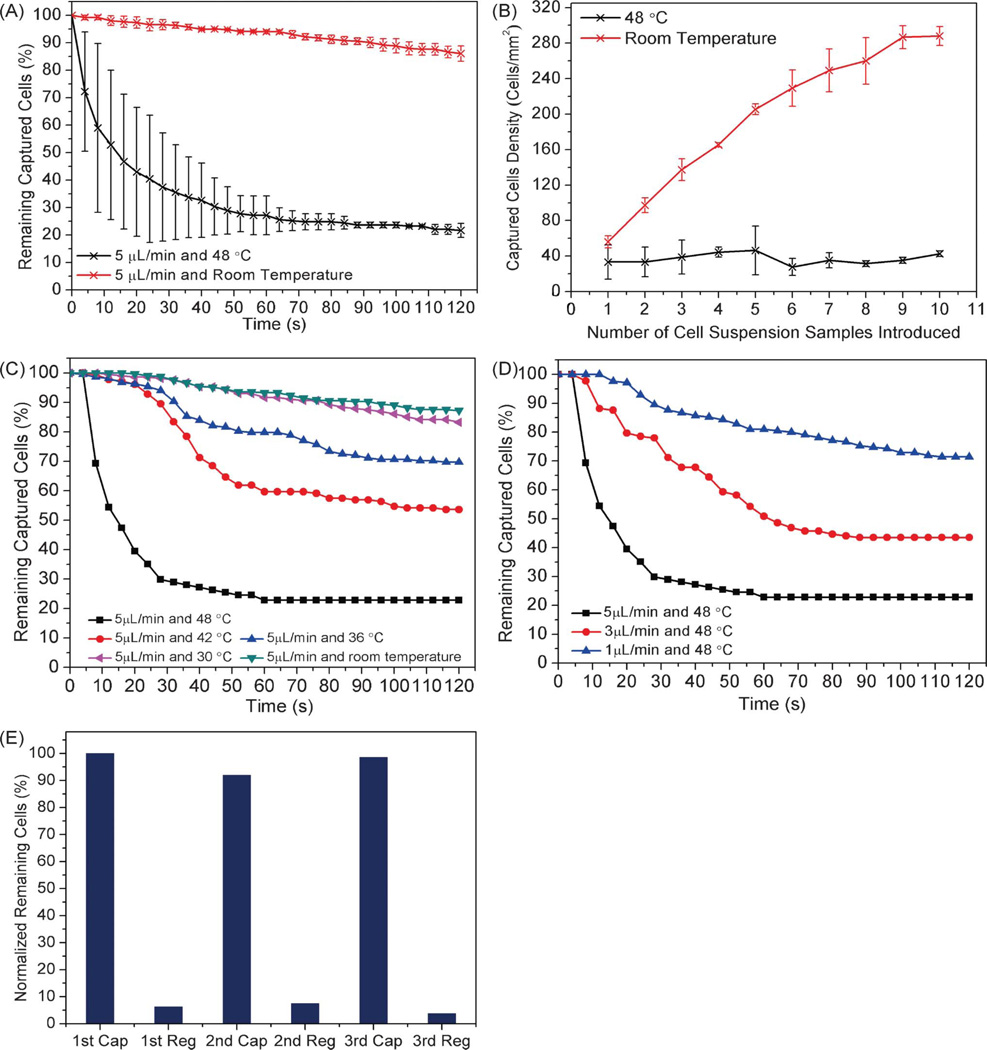
We next tested the thermally induced release of captured cells from the aptamer-functionalized chamber surfaces. Prior to the experiment, CCRF-CEM cells were captured by the surface-immobilized sgc8c aptamer, and unspecific bound cells were removed by D-PBS washing. Then, the cell-laden chamber was rinsed at either room temperature or 48 °C (Fig. 4A). It was seen that approximately 80% of cells were released from the surfaces after rinsing with D-PBS at 5 µL min−1 and 48 °C for 2 min, whereas negligible cell release was observed when rinsing at room temperature with an identical buffer solution and flow rate. These results suggest that the release of CCRF-CEM cells may have been caused by the conformational changes in the aptamer structure at the elevated temperature.
Fig. 4.
(A) Percentage of captured cells remaining on the substrate as a function of time while rinsing at constant temperature (48 °C and room temperature) and flow rate (5 µL min−1). (B) Captured cell density versus the number of cell suspension samples introduced while the temperature was maintained at either 48 °C or room temperature. (C) Effect of temperature on cell release efficiency while rinsing at 5 µL min−1. (D) Effect of flow rate on cell release efficiency while the chamber temperature was maintained at 48 °C. (E) Cell capture and re-capture on the regenerated aptamer-functionalized surface: the normalized percentage of remaining cells after the first, second and third capture and regeneration cycle.
We then tested this hypothesis of cell release due to heat-induced conformational changes to aptamer structure, seeking to exclude denaturation of cell membrane proteins as the cause of cell release. To do so, we conducted additional tests in which cells were heated prior to capture in the device, and compared the results to those from heating the device itself during cell capture. The cell suspension, diluted to 5 × 106 cells mL−1, was heated at 48 °C for 2 min, followed by introduction to the chamber at room temperature. In parallel, an unheated cell solution of 5 × 106 cells mL−1 was introduced into a chamber with the chamber temperature set to 48 °C. In both tests, 10 aliquots of cells were introduced into the chamber, followed by 1 min of incubation after each cell introduction. Heat treated cells were captured at room temperature up to a concentration of 288 ± 10 cells mm−2 (n = 3), as shown in Fig. 4B. Unheated cells in a 48 °C chamber achieved a surface density of only 43 ± 3 cells mm−2 (n = 3), and the presence of these remaining surface-bound cells was attributed to non-specific adsorption. These results suggest that the conformational changes in the aptamer structure, rather than the denaturation of the target cell membrane protein PTK733 at the increased temperature, caused the release of the specifically captured cells.
Having eliminated denaturation of the cell surface protein as the cause of cell release, we then investigated the relative impact on cell release of the chamber temperature compared to the hydrodynamic shear stress applied by the buffer flow. Cell detachment from aptamer-functionalized substrates is governed by the balance between the hydrodynamic shear stress applied on cell surfaces and the temperature-dependent binding strength of aptamers and their target cells. Therefore, changes in either the chamber temperature or the buffer flow rate would result in different cell release efficiencies. We first tested the effects of temperature on cell release by varying the chamber temperature from 30 °C to 48 °C while rinsing with D-PBS (Fig. 4C). It was clearly seen that with the elevated temperature, an increasing number of cells were detached from the substrate. Moreover, as the local temperature increased from 30 to 48 °C, the viscosity of the aqueous washing buffer decreased by approximately 35%,47 which led to a 35% lower shear stress at the cell membranes. This indicates that at higher temperatures there is a greater loss of binding between the aptamers and the cells, most likely due to temperature-dependent changes in conformational structure of aptamers. Next, we further evaluated the effect of shear stress on cell release, by performing identical experiments while varying the flow rate through the chamber. As shown in Fig. 4D, a higher flow rate caused more cells to detach from the substrate, as a result of increased shear stress disrupting the cell-aptamer binding. As either a higher temperature or a larger shear stress poses a greater risk of cell damage, the trade-off between them is an important design consideration.
As conformational changes in aptamer structures are reversible, this indicates that we can create a regenerable cell-capture surface, which would reduce the average cost of assays using the microfluidic device. To verify the reusability of the aptamer-functionalized surface, three experimental cycles were performed in the same device, with each cycle consisting of first introducing a dilute cell solution to the microchamber at room temperature, then releasing cells at 48 °C and 5 µL min−1 for 2 min, and finally regenerating the aptamer-functionalized surface (releasing all remaining cells) via washing with D-PBS at 60 °C and 50 µL min−1 for 2 min, and then at room temperature and 50 µL min−1 for 2 min. Following the first cycle, similar densities of captured cells were observed for succeeding cycles, with a maximum difference of captured cell density of only 8% between the first and the second capture (Fig. 4E). Given these results, we conclude that the regeneration of cell capture function of the microfluidic device can be both effective and consistent. Although some residual cells remained on the surfaces after each regeneration, this could be addressed by using a higher temperature and flow rate.
4.3. Cell viability assay
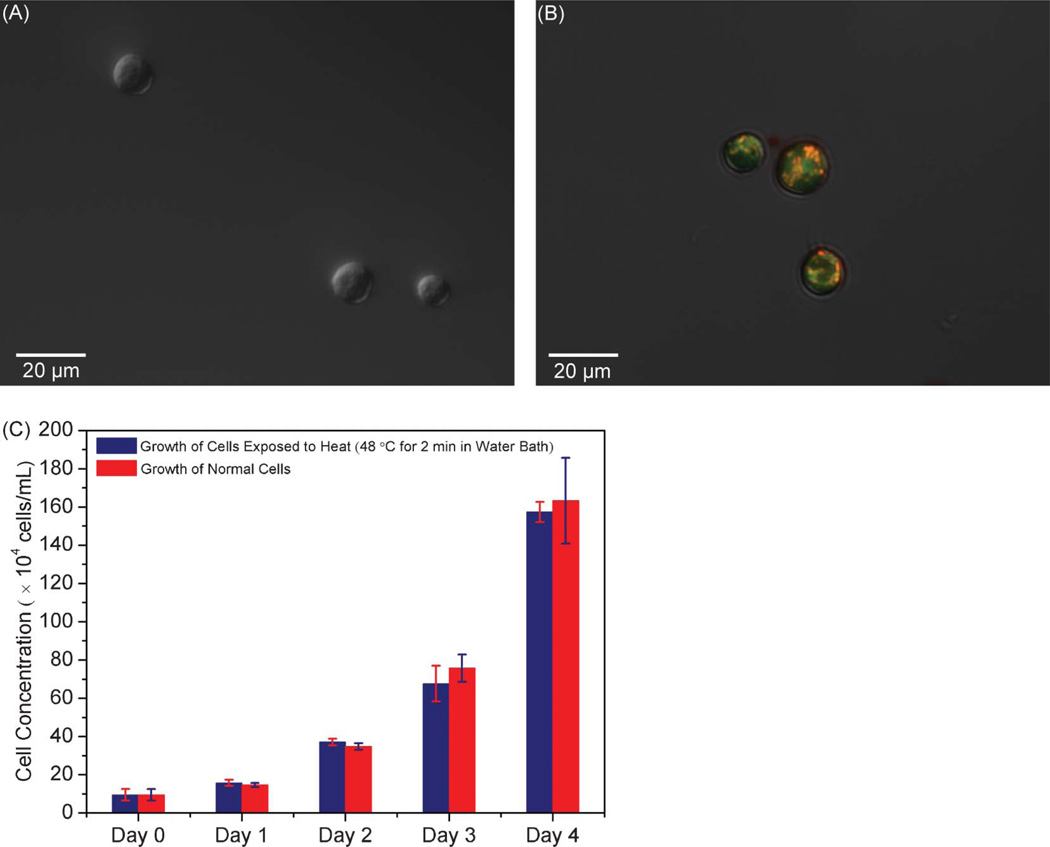
Cell viability is important for downstream applications such as tissue engineering and cell-based therapeutics.3,4 To evaluate cell viability, released cells were collected after rinsing at 5 µL min−1 and 48 °C for 2 min, at which point PI48 and JC-149,50 were used to stain cells. PI is a red-fluorescent nuclear stain that is not permeant to live cells. JC-1 accumulates in healthy mitochondria as indicated by red fluorescence, the intensity of which decreases along with mitochondrial depolarization occurring in the early stage of apoptosis. Our experimental results showed that the PI stained cells did not emit any red fluorescence (Fig. 5A), and the JC-1 stained cells exhibited bright red fluorescence (Fig. 5B), indicating that the collected cells were viable.
Fig. 5.
Image of PI stained cells (A) and JC-1 stained cells (B) following cell capture and release experiment, generated by a combination of phase contrast and fluorescent micrographs. (C) Concentrations of normal cells and heat-treated cells as a function of culture duration.
To obtain further confirmation of cell viability, we performed cell culture testing. Because of practical difficulties51 in conducting cell culture using cells directly retrieved from the microfluidic device without an on-chip cell culture component, we performed off-chip cell proliferation assays, in which cells from a well-mixed suspension were treated in a water bath at 48 °C for 2 min and then cultured for several days. Meanwhile, cells from the same suspension were also cultured without any treatment for the same period to serve as a control. The growth curves of normal and heat-treated cells are shown in Fig. 5C, in which heat-treated cells are seen to have a similar proliferation rate as normal cells. This indicates that the brief period of modestly elevated temperature used in the cell release experiment would not induce detectable cell damage, allowing the thermally released cells to remain viable.
5. Conclusion
The ability to isolate and retrieve live cells from biological samples is critical in both basic cell biology studies and clinical diagnostics. In this work, we have developed an aptamer-based microfluidic device for specific capture and temperature-mediated release of cells. The device consists of a microchamber situated on a temperature control chip that includes an integrated temperature sensor and heaters. The chamber surface is functionalized with aptamers to capture target cells with high specificity. The integrated temperature sensor and heaters allow closed-loop control of the chamber temperature, enabling thermally induced release of the captured cells without causing any physical or chemical damage.
The temperature control chip was first characterized experimentally and numerically to ensure temporal and spatial accuracy of the temperature field, respectively. Further experiments demonstrated highly specific capture and enrichment of CCRF-CEM cells using the sgc8c aptamer, followed by measurements of the sensitivity of cell capture to the concentration of cell suspension and to the incubation time within the chamber. Subsequently, efficient temperature-mediated release of cells was achieved, and the effects of the chamber temperature and shear stress on the aptamer-cell interaction were investigated. It was found that both higher temperatures and higher shear stresses facilitate cell release, and the trade-off between these two factors is an important design consideration, due to the greater risk of cell damage when using either a higher temperature or a larger shear stress. Finally, the released cells were shown to be viable, and the aptamer-functionalized surfaces were successfully regenerated and shown to be reuseable. These results demonstrate that our approach can potentially be used in basic biological research and clinical diagnostics for the isolation of viable pure cell subpopulations.
Supplementary Material
Acknowledgements
We gratefully acknowledge financial support from the National Science Foundation (Award No. CBET-0854030) and the National Institutes of Health (Award Nos. RR025816-02 and CA147925-01).
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2lc40411g
References
- 1.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murthy SK, Sin A, Tompkins RG, Toner M. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- 3.Bianco P, Robey PG. Nature. 2001;414:118–121. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Barrena E, Rosset P, Muller I, Giordano R, Bunu C, Layrolle P, Konttinen YT, Luyten FP. J. Cell. Mol. Med. 2011 doi: 10.1111/j.1582-4934.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Recktenwald D, Radbruch A. Cell separation methods and applications. New York: M. Dekker; 1998. [Google Scholar]
- 6.Kompala DS, Todd P. Cell separation science and technology. Washington, DC: American Chemical Society; 1991. [Google Scholar]
- 7.Kumar A, Bhardwaj A. Biomed. Mater. 2008;3:034008. doi: 10.1088/1748-6041/3/3/034008. [DOI] [PubMed] [Google Scholar]
- 8.Hunt SV. Cell Separation Techniques Used in Immunology. John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 9.Adams JD, Kim U, Soh HT. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhagat AAS, Bow H, Hou HW, Tan SJ, Han J, Lim CT. Med. Biol. Eng. Comput. 2010;48:999–1014. doi: 10.1007/s11517-010-0611-4. [DOI] [PubMed] [Google Scholar]
- 11.Chmela E, Tijssen R, Blom MT, Gardeniers HJ, van den Berg A. Anal. Chem. 2002;74:3470–3475. doi: 10.1021/ac0256078. [DOI] [PubMed] [Google Scholar]
- 12.Petersson F, Nilsson A, Holm C, Jonsson H, Laurell T. Lab Chip. 2005;5:20–22. doi: 10.1039/b405748c. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Joo S, Duhon M, Heller M, Wallace B, Xu X. Anal. Chem. 2002;74:3362–3371. doi: 10.1021/ac011273v. [DOI] [PubMed] [Google Scholar]
- 14.Du Z, Cheng KH, Vaughn MW, Collie NL, Gollahon LS. Biomed. Microdevices. 2007;9:35–42. doi: 10.1007/s10544-006-9010-x. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen TH, Pei R, Stojanovic M, Lin Q. Sens. Actuators, B. 2011;155:58–66. doi: 10.1016/j.snb.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenberg M, Nygren H. J. Immunol. Methods. 1988;113:3–15. doi: 10.1016/0022-1759(88)90376-6. [DOI] [PubMed] [Google Scholar]
- 18.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 19.Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP. Biochemistry. 1997;36:9726–9734. doi: 10.1021/bi9700633. [DOI] [PubMed] [Google Scholar]
- 20.Nieuwlandt D, Wecker M, Gold L. Biochemistry. 1995;34:5651–5659. doi: 10.1021/bi00016a041. [DOI] [PubMed] [Google Scholar]
- 21.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 22.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Anal. Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 24.Guo KT, Schafer R, Paul A, Ziemer G, Wendel HP. Mini-Rev. Med. Chem. 2007;7:701–705. doi: 10.2174/138955707781024481. [DOI] [PubMed] [Google Scholar]
- 25.Jayasena SD. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 26.Shangguan D, Cao ZC, Li Y, Tan W. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 27.Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. ChemBioChem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 28.Bouchard PR, Hutabarat RM, Thompson KM. Annu. Rev. Pharmacol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 29.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan WH. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Anal. Chem. 2009;81:7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, Kiselyuk A, Teply B, Eng G, Langer R. Anal. Chem. 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 32.Dharmasiri U, Balamurugan S, Adams AA, Okagbare PI, Obubuafo A, Soper SA. Electrophoresis. 2009;30:3289–3300. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S. Adv. Mater. 2011;23:4376–4380. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 35.Baumann H, Doyle D. J. Biol. Chem. 1979;254:3935–3946. [PubMed] [Google Scholar]
- 36.Jung K, Hampel G, Scholz M, Henke W. Cell. Physiol. Biochem. 1995;5:353–360. [Google Scholar]
- 37.Fujioka N, Morimoto Y, Takeuchi K, Yoshioka M, Kikuchi M. Appl. Spectrosc. 2003;57:241–243. doi: 10.1366/000370203321535187. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen T, Pei R, Stojanovic M, Lin Q. Microfluid. Nanofluid. 2009;6:479–487. [Google Scholar]
- 39.Nguyen T, Pei RJ, Landry DW, Stojanovic MN, Lin Q. Sens. Actuators, B. 2011;154:59–66. doi: 10.1016/j.snb.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilton JP, Nguyen TH, Pei RJ, Stojanovic M, Lin Q. Sens. Actuators, A. 2011;166:241–246. [Google Scholar]
- 41.Nguyen TH, Pei RJ, Qiu CM, Ju JY, Stojanovic M, Lin Q. J. Microelectromech. Syst. 2009;18:1198–1207. [Google Scholar]
- 42.Zhu J, Nguyen T, Pei R, Stojanovic M, Lin Q. presented in part at the IEEE Int. Conf. Solid-State Sensors, Actuators and Microsystems (Transducers ’11); Beijing, China. 2011. [Google Scholar]
- 43.Zhu J, Nguyen T, Pei R, Stojanovic M, Lin Q. presented in part at the Int. Conf. on Miniaturized Chemical and Biochemical Analysis Systems (MicroTAS ’10); Groningen; the Netherlands. 2010. [Google Scholar]
- 44.Shah A, Coleman MP. Br. J. Cancer. 2007;97:1009–1012. doi: 10.1038/sj.bjc.6603946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkin D, Kramárová E, Draper G, Masuyer E, Michaelis J, Neglia J, Qureshi S, CA S, editors. International Incidence of Childhood Cancer, Vol. II (IARC Scientific Publication, No 144) Lyon: International Agency for Research on Cancer; 1998. [Google Scholar]
- 46.Dickson MN, Tsinberg P, Tang ZL, Bischoff FZ, Wilson T, Leonard EF. Biomicrofluidics. 2011;5 doi: 10.1063/1.3623748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry RH, Green DW. Knovel (Firm) 8th edn. New York: McGraw-Hill; 2008. [Google Scholar]
- 48.Hoernlein RF, Orlikowsky T, Zehrer C, Niethammer D, Sailer ER, Simmet T, Dannecker GE, Ammon HP. J. Pharmacol. Exp. Ther. 1999;288:613–619. [PubMed] [Google Scholar]
- 49.Avci CB, Gunduz C, Baran Y, Sahin F, Yilmaz S, Dogan ZO, Saydam G. J. Cancer Res. Clin. Oncol. 2011;137:41–47. doi: 10.1007/s00432-010-0857-0. [DOI] [PubMed] [Google Scholar]
- 50.Lossi L, Alasia S, Salio C, Merighi A. Prog. Neurobiol. 2009;88:221–245. doi: 10.1016/j.pneurobio.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Mather JP, Roberts PE. Introduction to cell and tissue culture : theory and technique. New York: Plenum Press; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.