Abstract
Breast cancer is the most common cancer and the second leading cause of cancer death among women of all races and Hispanic origin populations in the United States. In the present study, we reported that the survival time of the breast cancer patients is influenced by the expression level of mdig, a previously identified lung cancer-associated oncogene encoding a JmjC-domain protein. By checking the expression levels of mRNA and protein of mdig through both RT-PCR and immunohistochemistry in samples from 204 patients, we noticed that about 30% of breast cancer samples showed increased expression of mdig. Correlation of the mdig expression levels with the survival time of the breast cancer patients indicated a clear inverse relationship between mdig expression and patient survival, including poorer overall survival, distant metastasis free survival, relapse free survival, and post-progression survival. Taken together, these data suggest that an increased expression of mdig is an important prognostic factor for poorer survival time of the breast cancer patients.
Keywords: Breast cancer, Mdig, Prognosis, Patient survival
1. Introduction
Breast cancer is the most frequently diagnosed cancer among women in the US in the past several decades. In 2013, an estimation of 232,340 new cases of invasive breast cancer is expected (Siegel et al., 2013). A significant decrease in the incidence rate of breast cancer was noted since 2002, which was largely attributed to the reduced use of hormone replacement therapy (HRT) for the post-menopausal women and increased detection of early stage tumors through breast cancer screening. Based on the molecular features that represent distinct subtypes of the breast epithelial cells, breast cancers can be categorized into basal cell carcinoma, luminal carcinoma and Her2+ carcinoma. The current 5-year survival rate of breast cancer is around 90%. Many factors affect patient survival after diagnosis and curative tumor resection, such as overweight, cigarette smoke, alcoholism, and the molecular subtypes of the tumor.
Inherited germline mutations of DNA repair genes BRCA1 and BRCA2 predispose risks of developing breast cancer, which accounted about 5–10% of breast cancer patients with Caucasian origination (Maxwell and Domchek, 2012). A most recent retrospective study also revealed that women with breast cancer and are carriers of brca1 mutations have increased mortality compared with non-carriers (Boyle, 2012). In addition to BRCA1 and BRCA2, a number of studies have also identified other gene signatures that are associated with poorer prognosis or therapeutic responses of the breast cancer patients (Marchionni et al., 2013). However, scanty and contradictory data exist on the survival-associated gene signatures. Such an inconsistence is further complicated by the history of adjuvant therapy and differences of bioinformatics platforms used in each study.
The mineral dust-induced gene (mdig) was first identified from alveolar macrophages obtained from people with occupational lung diseases (Lu et al., 2009; Zhang et al., 2005). This gene was independently characterized in cell lines with an overexpression of c-myc oncogene and named as myc-induced nuclear antigen 53 (mina53) (Tsuneoka et al., 2002) or nucleolar protein 52 (NO52) (Eilbracht et al., 2005). The mdig protein contains a conserved JmjC domain and is indicated in cell growth regulation, possibly through its effect on tri-methylation of lysine 9 on histone H3 (H3K9me3) and hydroxylase activity on ribosomal proteins (Ge et al., 2012; Lu et al., 2009). Further studies demonstrated a strong association of mdig expression with human lung cancer (Lu et al., 2009), colon cancer (Teye et al., 2004), esophageal squamous cell carcinoma (Tsuneoka et al., 2004), neuroblastoma (Fukahori et al., 2007), renal cell carcinoma (Ishizaki et al., 2007), gastric carcinoma (Zhang et al., 2008), hepatocellular carcinoma (Ogasawara et al., 2010), and cholangiocarcinoma (Tan et al., 2012). It is unclear whether mdig is overexpressed in human breast cancer and if so, whether such an overexpression contributes to the prognostic outcomes of the breast cancer patients. In this report, we show that about 30% of breast cancers exhibited higher level of mdig expression and increased mdig expression predicts poorer survival of the breast cancer patients, including poorer overall survival (OS), distant metastasis free survival (DMFS), relapse free survival (RFS), and post-progression survival (PPS).
2. Materials and methods
2.1. Breast cancer tissue samples
A panel of cDNAs (TSCE101) derived from total RNAs of the individual breast cancer tissues was purchased from Origene (Rockville, MD). A tissue microarray containing 192 breast cancer tissue samples was purchased from US Biomax, Inc. (Rockville, MD).
2.2. PCR
The expression level of mdig and actin was determined by PCR using One-step RT-PCR kit (Promega, Madison, WI) by omitting the reverse transcription reaction. The PCR primers for mdig are: 5′-TCA TGT CGG GCC TAA GAG AC-3′ and mdig-b: 5′-GGC ATT TGA TTC TGC AAA GG-3′, which amplifies a 1510 bp DNA fragment covering the whole coding region of the mdig gene. PCR primers for β-actin are: 5′-TTC TAC AAT GAG CTG CGT GTG-3′ and 5′-GGG GTG TTG AAG GTC TCA AA-3′.
2.3. Immunohistochemistry
The tissue microarray slide BR2086 was purchased from US Biomax, Inc. (Rockville, MD) and deparaffinized with Xylene and hydrated in series of alcohol. To quench the endogenous peroxidase activity, slides were incubated with 1.5% H2O2 for 20 min at room temperature. Heat-mediated antigen retrieval was performed by boiling the slides in Citrate buffer, pH 6 for 20 min. To block the non-specific binding of immunoglobulin, slides were incubated with a solution consisting of 5% goat serum and 0.2% triton-X 100 in PBS for 2 h at room temperature, followed by incubation with monoclonal antibody against Mdig/Mina 53 (mouse anti-Mina 53, Invitrogen) in 1:100 dilution overnight at 4 °C. Next day goat anti-mouse biotinylated secondary antibody was applied at 1:200 dilution and incubated for 2 h at room temperature. Slides were then incubated with an ABC reagent (Vectastain Elite ABC kit) for 45 min at room temperature and the chromogen was developed with diaminobenzidine (DAB). The slides were counterstained with hematoxylin and mounted with entellan. All incubation steps were carried out in a humidified chamber and all washing steps were performed with 1× PBS. The images were captured under the bright field optics of the Nikon Eclipse Ti-S Inverted microscope (Mager Scientific, Dexter MI, USA).
2.4. Kaplan–Meier survival analysis
A Kaplan–Meier survival database that contains survival information of 2880 breast cancer patients and gene expression data obtained by using three different versions of Affymetrix HG-U133 microarrays was used (Gyorffy et al., 2010). Two different mdig probe sets, probes 213188_s_at and 213189_at, are presented in this database. Probe set 213188_s_at was excluded because it detects the far end of the 3′-UTR of mdig mRNA and the antisense of 3′-UTR of the β-γ-crystallin domain containing 3 (CRYBG) mRNA. The probe set 213189_at that detects the open-reading frame (ORF) of mdig mRNA was used in this survival analysis. Survival curves resulting in p values of <0.05 between mdig higher (mdighigh) and mdig lower (mdiglow) groups were considered significantly different.
3. Results
3.1. Increased expression of mdig in breast cancer
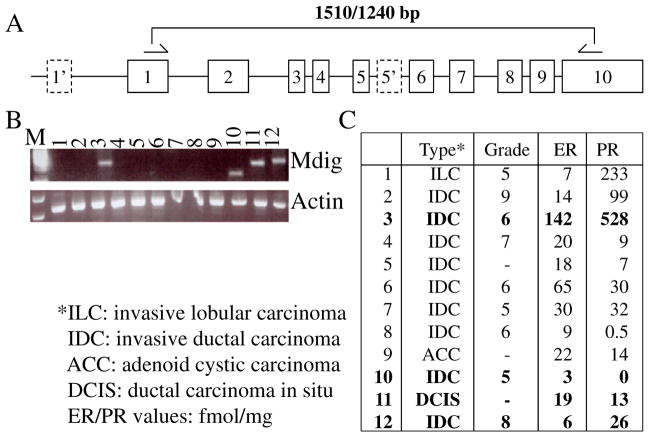
An overexpression of mdig has been observed in a number of human cancers, implying its important role in the pathogenesis of human malignancies. To investigate whether mdig is also overexpressed in human breast cancer, the level of mdig expression was determined using a panel of cDNA derived from human breast cancer tissues by PCR. In previous studies using human lung cancer cell lines, we had detected multiple alternatively spliced isoforms of mdig mRNA, which rendered difficulties for quantitative real-time PCR. Accordingly, we used a traditional PCR procedure and primers that amplify a 1510 bp fragment covering the entire coding region of the mdig mRNA (Fig. 1A). As exhibited in Fig. 1B, mdig expression was detected in 4 out of 12 breast cancer samples, among which sample 10 showed a size-reduced fragment due to alternative splicing. Further analyses of the mdig expression and the pathological features of these samples indicated no correlation of mdig expression with the types, stages and levels of blood estrogen (ER)/progesterone (PR) in the cancer patients (Fig. 1C).
Fig. 1.
Detection of mdig mRNA in breast cancer samples. A. Schematic indication of the mdig gene structure. Dashed boxes indicate alternative exons identified in some alternatively spliced mdig mRNAs. Half arrows denote the corresponding regions of the PCR primers. B. Expression of mdig in the indicated breast cancer samples. The expression levels of mdig were calibrated by β-actin. C. Tumor and patient information of B.
3.2. Mdig protein can be detected in breast cancer samples but not the normal breast tissues
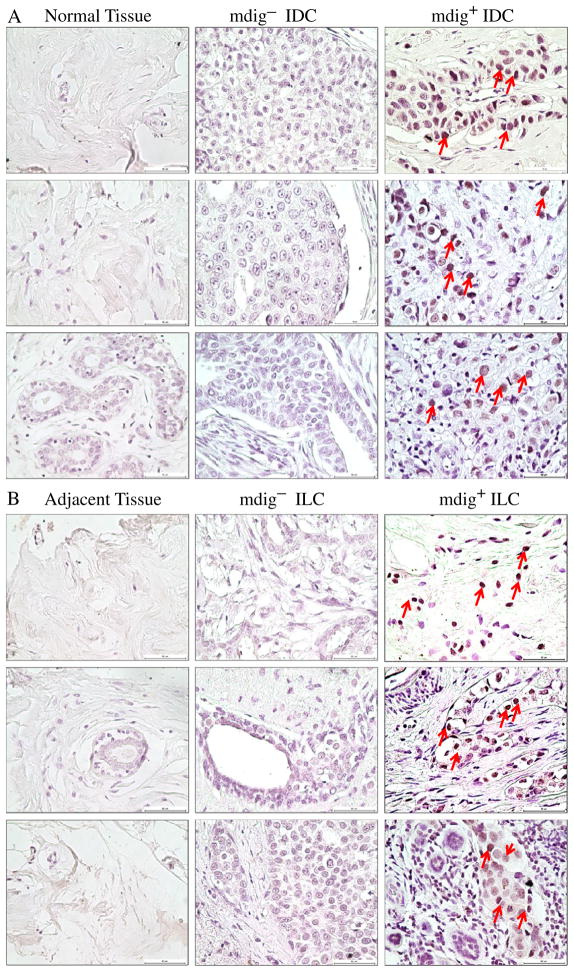
To determine whether the protein levels of Mdig is altered in the breast cancer tissues, we next evaluated mdig protein expression by immunohistochemistry on a breast cancer tissue microarray slide that contains 3 intraductal carcinomas (IntraDC), 140 invasive ductal carcinomas (IDC), 45 invasive lobular carcinomas (ILC), 1 invasive papillary carcinoma (IPC), 2 mucinous carcinomas (MuC), 1 medullary carcinoma (MedC), 7 normal breast tissues, and 9 adjacent normal tissues of breast cancer. No mdig expression was detected in all the normal breast tissues (Table 1 and Fig. 2A). Among the adjacent non-cancerous tissues of the breast cancer, 6 out of 9 samples showed extremely weak mdig expression, mostly in only one or few cells/per microscopic field. No mdig expression was detected in the remaining 3 adjacent tissues. Higher levels of mdig expression were detected in 30% of IDC and 33% of ILC (Table 1 and Figs. 2A and B). Table 1 shows percentage of mdig+ samples among the breast tumors at different stages. It appears that stage IIa of breast cancer has the highest percentage of mdig+ cancer tissues.
Table 1.
Percentage of mdig expression among normal breast tissues and breast cancer samples with different subtypes and stages.
| # of samples | Mdig+ | +% | |
|---|---|---|---|
| Tissue types | |||
| Normal | 7 | 0 | 0 |
| Adjacent | 9 | (6) | (67)a |
| IntraDC | 3 | 0 | 0 |
| IDC | 140 | 42 | 30 |
| ILC | 45 | 15 | 33 |
| IPC | 1 | 1 | 100 |
| MuC | 2 | 1 | 50 |
| MedC | 1 | 1 | 100 |
| Stages | |||
| 0 | 3 | 0 | 0 |
| I | 25 | 8 | 32 |
| IIa | 109 | 39 | 35.8 |
| IIb | 40 | 8 | 20 |
| IIIb | 12 | 3 | 25 |
| IV | 3 | 9 | 0 |
Exhibited extremely weak staining of mdig protein.
Fig. 2.
Immunohistochemistry of breast cancer tissue microarray. Immunostaining was performed as described in “Materials and methods”. A. Mdig expression was detected in 30% of IDC breast cancer samples (mdig+, right column), but not in normal breast tissues (left column). Panels in middle column show mdig negative IDC samples (mdig−). Red arrows denote cells positive for the mdig staining. B. Mdig expression was detected in 33% of ILC breast cancer samples (mdig+, right column). The adjacent non-cancerous tissues showed very weak or no expression of mdig (left column). Panels in middle column show mdig negative ILC samples (mdig−). Red arrows denote cells positive for the mdig staining in the cancer tissues. Magnification 40× and scale bar = 50 μm.
3.3. Mdig is a potential new prognostic factor for breast cancer patients
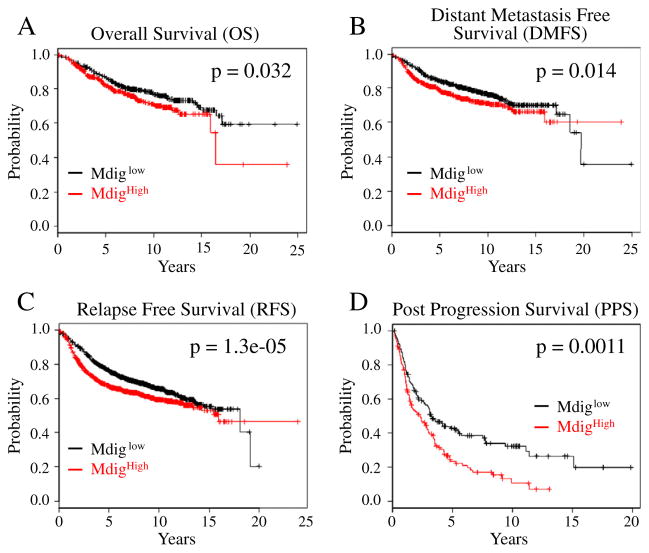
The possible contribution of mdig to the prognosis of the cancer patients remains to be elucidated. As indicated above, neither PCR nor tissue microarray studies showed significant correlation of mdig expression with the tumor stages or molecular subtypes, possibly due to the limited number of samples that was analyzed. To explore the potential association of mdig expression with the prognostic outcomes, such as tumor progression and overall survival, of the breast cancer patients, we examined contribution of mdig expression to the survival of breast cancer patients in a clinical microarray database (Gyorffy et al., 2010). The database collected gene expression data that were obtained by using three different versions of Affymetrix HG-U133 microarrays and the survival information of 2880 breast cancer patients. There are two different mdig probe sets, probes 213188_s_at and 213189_at, in this database. The probe set 213188_s_at was excluded from the patient survival assay because it detects the far end of the 3′-UTR of mdig mRNA and the sequence region corresponding to the anti-sense strand of 3′-UTR of the β-γ-crystallin domain containing 3 (CRYBG) mRNA, which may not reflect the authentic expression levels of mdig mRNA in tumors. In contrast, the probe set 213189_at targets the open-reading frame (ORF) of mdig mRNA, which may detect the true expression level of the mdig mRNA. An inverse correlation of mdig expression with the patient survival was noted. All four different survival assays of the breast cancer patients, including overall survival (OS), relapse free survival (RFS), distant metastasis free survival (DMFS), and post-progression survival (PPS), revealed that higher expression of mdig predicts poorer survival of the breast cancer patients (Fig. 3). The most prominent predictive power of mdig is for the poorer RFS, which showed the smallest p value of the statistic difference of the survival time between mdig higher (mdighigh) and mdig lower (mdiglow) patient groups.
Fig. 3.
Increased expression of mdig predicates poorer overall survival (OS, A), distant metastasis free survival (DMFS, B), relapse free survival (RFS, C), and post-progression survival (PPS, D) of the breast cancer patients.
3.4. Increased expression of mdig predicts poorer survival of the patients with luminal A subtype of the breast cancer
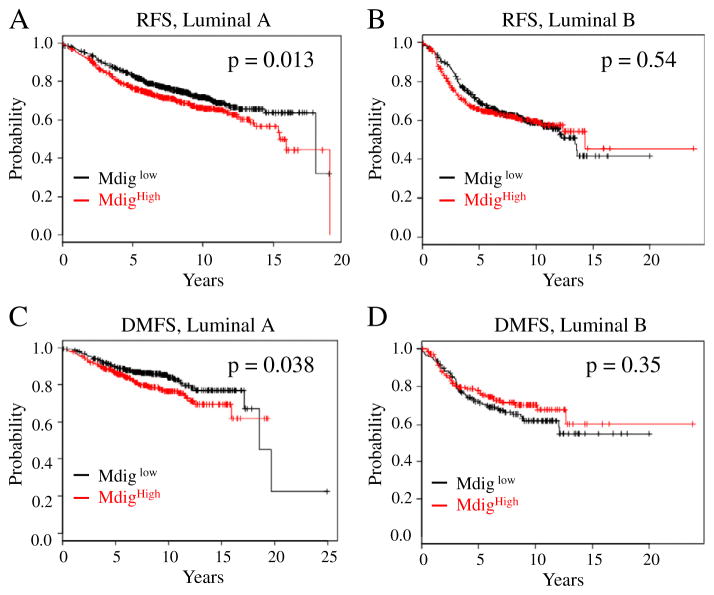
Accumulating evidence suggested that breast cancer is a highly heterogeneous disease. Based on well-characterized molecular features, breast cancer can be categorized into four major subtypes, including basal-like, luminal A, luminal B, and Her2+ tumors, which exhibit distinct oncogenic activation pathways, tumor progression pattern and prognostic outcomes (Tran and Bedard, 2011). To determine the potential linkage of mdig expression and the survival times of the patients with different tumor subtypes, we next analyzed survival data of each tumor subtypes by stratifying the patients based on the mdig expression levels. These analyses indicated that mdig expression was not statistically associated with the survival times of the patients who have basal-like, luminal B (RFS: p = 0.54; DMFS: p = 0.35) or Her2+ breast cancer (Fig. 4 and data are not shown). However, we noted that there was a statistically significant effect of higher mdig expression for the poorer survival of the patients who have luminal A breast cancer (RFS: p = 0.013; DMFS: p = 0.038, Figs. 4A and C). These data suggest that increased expression of mdig is an unfavorable factor for the prognosis of luminal A subtype of the breast cancer.
Fig. 4.
Increased expression of mdig predicates poorer RFS and DMFS of the luminal A breast cancer patients (A and C), but not luminal B breast cancer patients (B and D).
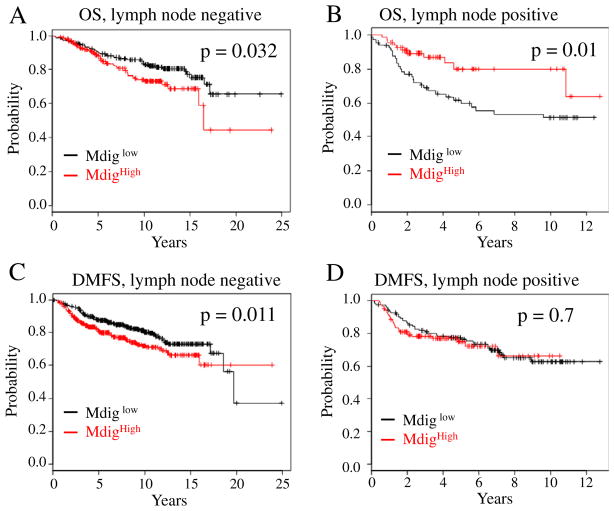
3.5. Increased expression of mdig predicts opposite outcomes of the breast cancer patients with different lymph node status
Lymphatic spread, esp. to the axillary lymph nodes, is an earlier sign of invasion and metastasis of the breast cancer (Lee et al., 2012). Analyses of the patient survival data had clearly indicated an unfavorable effect of mdig expression on OS, RFS, DMFS, and PPS (Fig. 3). A compelling question to be asked is whether higher expression of mdig predicts poorer survival of the breast cancer patients with different lymph node statuses. Unexpectedly, we observed a statistically significant effect of higher mdig expression on the poorer survival of the breast cancer patients who were lymph node negative (OS: p = 0.032; DMFS: p = 0.011), but not the patients who were lymph node positive (Fig. 5). In fact, an opposite effect of mdig on the survival of lymph node positive patients was observed (Fig. 5B). Among the breast cancer patients who were lymph node positive, increased expression of mdig appears to be a favorable factor for the prolonged overall survival (OS: p = 0.01). No statistically significant effect of the mdig expression statuses was observed on the RFS, DMFS and PPS of the patients who were lymph node positive (Fig. 5D and data are not shown). It is very likely, thus, that the effect of mdig expression on the survival of the patients may be context dependent, esp., the metastatic status of the breast cancer. For the breast cancer patients who were lymph node negative, increased expression of mdig predicts poorer survival of the patients. In contrast, increased expression of mdig may predict better survival of the patients whose tumor cells had been metastasized to lymph nodes and other organs of the body.
Fig. 5.
The predicative power of mdig expression on patient survival is determined by the lymph node status. A. Increased expression of mdig predicates poorer OS of the breast cancer patients who were lymph node negative. B. Increased expression of mdig predicates better OS of the breast cancer patients who were lymph node positive. C. Increased expression of mdig predicates poorer DMFS of the breast cancer patients who were lymph node negative. D. Increased expression of mdig has no predicative power for DMFS of the breast cancer patients who were lymph node positive.
4. Discussion
Breast cancer is by far the most common malignant disease in women worldwide. The incidence has increased in most countries across the world in the last decades, esp. in many of the developing countries (Ferlay et al., 2010). Several risk factors, including reproductive behavior, the use of exogenous hormones, obesity, and alcohol consumption, have been linked to the occurrence of breast cancer. An estimated nearly half million women died from breast cancer annually. In these breast cancer patients, it is believed that the death is not caused by the primary tumor, but the metastasized tumors at distant sites. Thus, improving our understanding of the molecular mechanisms of the progression of breast cancer is essential for achieving effective clinical management of the disease and favorable prognosis of the patients.
The regulation of mdig gene and the functional characteristics of the mdig gene product remain to be fully elucidated. Several studies indicated increased expression of mdig in a number of human cancers (Lu et al., 2009; Tsuneoka et al., 2004). However, there are no reports showing altered expression of mdig in human breast cancer. Accordingly, the present report is the first to reveal increased expression of mdig and its prognostic value on the survival of the breast cancer patients. The encoded protein of mdig gene contains a conserved JmjC domain, a domain that occurred in the majority of the histone demethylases (Kooistra and Helin, 2012). It is unknown whether mdig is also a histone demethylase. Overexpression of mdig enhances cell proliferation and reduces signals of histone H3 lysine 9 trimethylation (H3K9me3) in some human immortalized cell lines, which indicates possible contribution of mdig to the regulation of the histone methylation states (Lu et al., 2009). In A549 lung cancer cells, our recent studies suggested that although mdig showed limited effect on the global level of H3K9me3, the role of mdig is pronounced on the reduction of H3K9me3 in gene loci of rRNA, H19 and Oct4 (Chen et al., 2013). In gene knockout studies, we found that heterozygotic knockout of mdig blunted fibrotic response of the lung and inhibited FasL expression induced by silica particle (Thakur et al., unpublished observation). In addition, by ectopic expression of mdig in lung epithelial cells or lung cancer cell lines, we noted that mdig enhanced cell proliferation but repressed cell motility and invasiveness in an in vitro cell migration and invasion assay (Yu et al., under review), which may support the notion that higher level of mdig predicts poorer survival of the breast cancer patients who were lymph node negative but better survival of the patients who were lymph node positive.
The most important hallmark of cancers, such as breast cancer, is the uncontrolled proliferation of the cancer cells, which can be assessed by immunohistochemical staining of Ki67 nuclear antigen (Inwald et al., 2013). Considerable challenging remains in using Ki67 staining as a prognostic marker to predict the survival of the breast cancer patients, molecular subtypes of the tumors, the responses to chemotherapy or adjuvant therapy, and the risk of recurrence. Even for the highly proliferative breast cancers, inconsistence of Ki67 staining exists due to differences of tumor regions that were analyzed, such as the invasive tumor edge vs the core area of the tumor section (Dowsett et al., 2011). Nevertheless, some studies showed that the levels of Ki67 were linearly related to tumor grades, patient survival and lymphatic invasion (Inwald et al., 2013; Jacquemier et al., 2009). Since mdig had been shown to be associated with cell proliferation (Tsuneoka et al., 2002; Yu et al., under review; Zhang et al., 2005), it will be important to determine whether mdig expression is correlated with the levels of Ki67 in breast cancer tissues. However, notable distinctions between mdig and Ki67 can be found, for example, mdig predicts poorer survival of the breast cancer patients who were lymph node negative, but not the lymph node positive (Fig. 5), and mdig is an unfavorable factor for the survival of patients with luminal A breast cancer that is less proliferative (Fig. 4). These differences suggest that the function of mdig may be beyond the known property in promoting cell proliferation. One possibility is the contribution of mdig to the epigenetic regulation of the self-renewal or pluripotent genes for the cancer stem cells. We are currently expanding our studies on the role of mdig in proliferation, invasion, migration, and possibly the expression of stemness genes of the breast cancer cells.
Acknowledgments
This research project is partially supported by the NIH R01 ES017217 and R01 ES020137 to F.C.
Abbreviations
- ACC
adenoid cystic carcinoma
- BRCA1
breast cancer 1 gene
- DCIS
ductal carcinoma in situ
- DMFS
distant metastasis free survival
- HRT
hormone replacement therapy
- IDC
invasive ductal carcinoma
- ILC
invasive lobular carcinoma
- Mdig
mineral dust-induced gene
- OS
overall survival
- PPS
post-progression survival
- RFS
relapse free survival
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–vi12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- Chen B, et al. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget. 2013;4:1427–1437. doi: 10.18632/oncotarget.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbracht J, Kneissel S, Hofmann A, Schmidt-Zachmann MS. Protein NO52—A constitutive nucleolar component sharing high sequence homologies to protein NO66. Eur J Cell Biol. 2005;84:279–294. doi: 10.1016/j.ejcb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Fukahori S, et al. Immunohistochemical expressions of Cap43 and Mina53 proteins in neuroblastoma. J Pediatr Surg. 2007;42:1831–1840. doi: 10.1016/j.jpedsurg.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8:960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Inwald EC, et al. Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res Treat. 2013;139:539–552. doi: 10.1007/s10549-013-2560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki H, et al. Overexpression of the myc target gene Mina53 in advanced renal cell carcinoma. Pathol Int. 2007;57:672–680. doi: 10.1111/j.1440-1827.2007.02156.x. [DOI] [PubMed] [Google Scholar]
- Jacquemier J, et al. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 2009;11:R23. doi: 10.1186/bcr2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Lee MC, Joh JE, Chau A. Axillary staging prior to neoadjuvant chemotherapy: the roles of sentinel lymph node biopsy and axillary ultrasonography. Cancer Control. 2012;19:277–285. doi: 10.1177/107327481201900404. [DOI] [PubMed] [Google Scholar]
- Lu Y, et al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle. 2009;8:2101–2109. doi: 10.4161/cc.8.13.8927. [DOI] [PubMed] [Google Scholar]
- Marchionni L, Afsari B, Geman D, Leek JT. A simple and reproducible breast cancer prognostic test. BMC Genomics. 2013;14:336. doi: 10.1186/1471-2164-14-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Domchek SM. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol. 2012;9:520–528. doi: 10.1038/nrclinonc.2012.123. [DOI] [PubMed] [Google Scholar]
- Ogasawara S, Komuta M, Nakashima O, Akiba J, Tsuneoka M, Yano H. Accelerated expression of a Myc target gene Mina53 in aggressive hepatocellular carcinoma. Hepatol Res. 2010;40:330–336. doi: 10.1111/j.1872-034X.2009.00604.x. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Tan XP, Zhang Q, Dong WG, Lei XW, Yang ZR. Upregulated expression of Mina53 in cholangiocarcinoma and its clinical significance. Oncol Lett. 2012;3:1037–1041. doi: 10.3892/ol.2012.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teye K, et al. Increased expression of a Myc target gene Mina53 in human colon cancer. Am J Pathol. 2004;164:205–216. doi: 10.1016/S0002-9440(10)63111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran B, Bedard PL. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011;13:221. doi: 10.1186/bcr2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka M, Koda Y, Soejima M, Teye K, Kimura H. A novel myc target gene, mina53, that is involved in cell proliferation. J Biol Chem. 2002;277:35450–35459. doi: 10.1074/jbc.M204458200. [DOI] [PubMed] [Google Scholar]
- Tsuneoka M, et al. Mina53 as a potential prognostic factor for esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:7347–7356. doi: 10.1158/1078-0432.CCR-03-0543. [DOI] [PubMed] [Google Scholar]
- Yu M, Sun J, Chen B, Lu Y, Zhao H, Chen F. Paradoxical roles of mineral dust induced gene expression on cell proliferation and migration/invasion. Cell Cycle. 2013w doi: 10.1371/journal.pone.0087998. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene. 2005;24:4873–4882. doi: 10.1038/sj.onc.1208668. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Hu CM, Yuan YS, He CH, Zhao Q, Liu NZ. Expression of Mina53 and its significance in gastric carcinoma. Int J Biol Markers. 2008;23:83–88. doi: 10.1177/172460080802300204. [DOI] [PubMed] [Google Scholar]