Abstract
Archaea display a variety of type IV pili on their surface and employ them in different physiological functions. In the crenarchaeon Sulfolobus acidocaldarius the most abundant surface structure is the aap pilus (archaeal adhesive pilus). The construction of in frame deletions of the aap genes revealed that all the five genes (aapA, aapX, aapE, aapF, aapB) are indispensible for assembly of the pilus and an impact on surface motility and biofilm formation was observed. Our analyses revealed that there exists a regulatory cross-talk between the expression of aap genes and archaella (formerly archaeal flagella) genes during different growth phases. The structure of the aap pilus is entirely different from the known bacterial type IV pili as well as other archaeal type IV pili. An aap pilus displayed 3 stranded helices where there is a rotation per subunit of ~ 138° and a rise per subunit of ~ 5.7 Å. The filaments have a diameter of ~ 110 Å and the resolution was judged to be ~ 9 Å. We concluded that small changes in sequence might be amplified by large changes in higher-order packing. Our finding of an extraordinary stability of aap-pili possibly represents an adaptation to harsh environments that S. acidocaldarius encounters.
Introduction
Attachment to different surfaces is a universal process of microbial cells among the three domains of life which is a prerequisite to form biofilms. Living in biofilms is thought to be the dominant mode of life for microorganisms as this cell community provides increased tolerance against antimicrobial agents and changing environmental conditions (Hall-Stoodley et al., 2004). Another beneficial feature for microorganisms attached to different surfaces is the accumulation of various nutrients such as proteins, polysaccharides and other molecules on these surfaces (Palmer et al., 2007). Initiation of attachment is determined by different factors like environmental conditions and signaling molecules (Petrova and Sauer, 2012). However, in many microorganisms the first physical contact with surfaces is mediated by different cell surface appendages. In Bacteria as well as in Archaea, it was shown that surface structures like flagella and type IV pili are promoting adhesion to different surfaces (Klausen et al., 2003; Jenkins et al., 2005; Shime-Hattori et al., 2006; Jarrell et al., 2011; Zolghadr et al., 2010). Various archaeal cell surface appendages were identified by electron microscopy (reviewed in Albers and Meyer, 2011). The assembly machinery of many of the newly identified structures, including the archaellum (former archaeal flagellum) (Jarrell and Albers, 2012), resemble the assembly system of bacterial type IV pili. However, others exhibit a unique morphology such as cannulae and hami (reviewed in Ng et al., 2008). Direct observations show that also Ignicoccus hospitalis cells adhere to surfaces (Müller et al., 2009; Yu et al., 2012). In the methanogenic euryarchaeon Methanococcus maripaludis, both pili and archaella are prerequisite structures for surface adhesion (Jarrell et al., 2011). In Haloferax volcanii, a deletion of the prepilin pepdidase PibD, that is essential for filament assembly, resulted in the loss of the ability to adhere to a glass surface. However, the lack of the archaella did not affect surface adhesion, emphasizing the role of other type IV pili-like structures for attachment (Tripepi et al., 2010). Archaella were identified to promote attachment to different surfaces in Pyrococcus furiosus and Methanocaldococcus villosus (Näther et al., 2006; Bellack et al., 2010), whereas in Sulfolobus solfataricus, adhesion to surfaces was also strongly influenced by the UV-inducible pili (ups) (Koerdt et al., 2010; Zolghadr et al., 2010).
Thus far, all identified archaeal filaments that are key components for surface adhesion share an assembly system which is homologous to the assembly machinery of bacterial type IV pili. In bacteria, type IV pili are involved in several unique functions like a flagella-independent movement called twitching motility (Mattick, 2002), DNA uptake during transformation (Averhoff, 2004) and phage transduction (Budzik et al., 2004). Furthermore, type IV pili play a distinct role in adhesion and also biofilm formation (Klausen et al., 2003), which has a huge impact on the virulence of many pathogenic bacteria (Craig et al., 2004). Archaeal type IV pili-like surface appendages share an evolutionary origin with bacterial type IV pili and therefore their assembly process is similar (Pohlschröder et al., 2011).
The thermoacidophilic crenarchaeon Sulfolobus acidocaldarius possesses three distinct surface appendages which share homology with bacterial type IV pili. The best studied structure so far is the archaellum, which is a functional analogue to the bacterial flagellum (Jarrell and Albers, 2012) and is responsible for motility (Lassak et al., 2012) but was also identified to be involved in surface adhesion (Henche et al., 2012). The assembly of this filament is predominantly induced in late growth stages which is caused by nutrient starvation (Lassak et al., 2012). The UV-induced pili (ups) are only assembled after UV-irradiation and other stress signals which promote DNA double-strand breaks (Fröls et al., 2008). This structure enables a species-specific exchange of genetic material to repair DNA damages (Ajon et al., 2011). The third structure is the archaeal adhesive pilus (aap) which is the most abundant structure on the cell surface of S. acidocaldarius. Whereas the archaellum and the ups pilus are conserved among the Sulfolobales, the aap pilus is unique to S. acidocaldarius. In a previous study it was shown that the aap pili promote the attachment to surfaces, whereas ups pili and archaella only display a minor role in surface adhesion in S. acidocaldarius (Henche et al., 2012). This is in contrast to S. solfataricus, where the archaellum and ups pili are indispensable for surface attachment (Zolghadr et al., 2010). Biofilm formation in S. acidocaldarius is strongly influenced by the presence of aap pili which were shown to important to build up tower-like structures (Henche et al., 2012).
In this study we wanted to gain insights into the assembly of the archaeal adhesive pilus of S. acidocaldarius. Therefore, we constructed systematic in-frame deletion mutants of the putative aap genes and analyzed the respective strains by transmission electron microscopy, by their ability to swarm on a semi-solid surface and by their ability to form biofilms. In addition, the aap filaments were isolated and analyzed by mass spectrometry as well as by electron microscopic methods.
Results
Identification of the aap pili
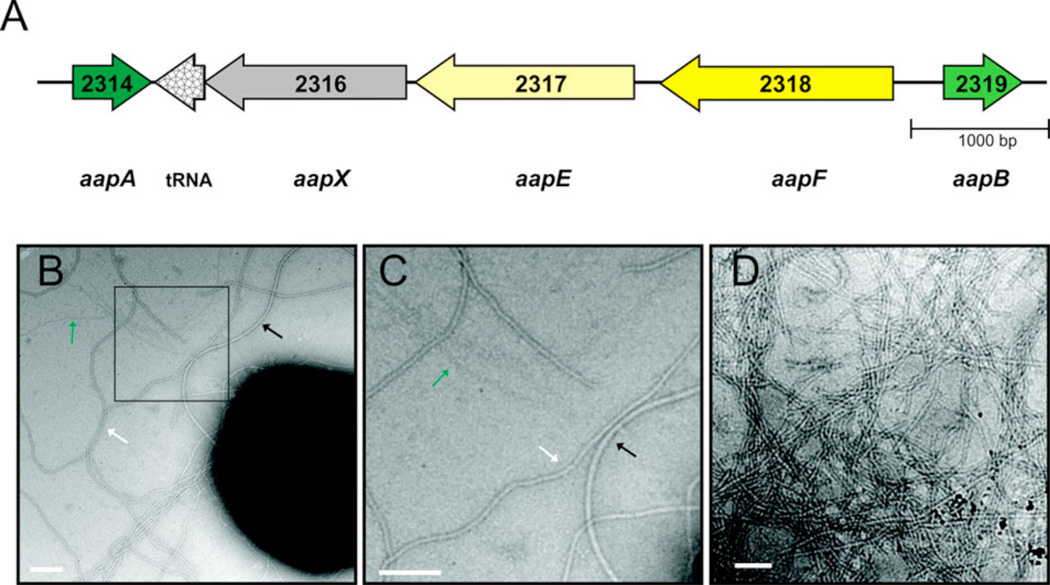
The cell surface of S. acidocaldarius cells is under normal growth conditions covered with three kinds of surface structures: archaella, very thin filaments (‘threads’; 5 nm in width) and abundant aap pili (Fig 1B). After UV exposure, ups pili (UV inducible pili) are assembled on the surface of S. acidocaldarius. The operons encoding the archaellum and the ups pili assembly components have been described recently (Lassak et al., 2012; Ajon et al., 2011). In the genome of S. acidocaldarius only one other gene locus is present that encompasses two putative pilin subunits in close proximity to a PilB (TadB) and PilC (TadC) homologs, which are the assembly ATPase and the central membrane protein in bacterial type IV pili assembly systems, respectively (Pohlschröder et al., 2011). The genes encoding for a PilB homolog, aapE (Saci2317), and the PilC homolog aapF (Saci2318), are present in a gene locus with a gene aapX (Saci2316) coding for a putative iron-sulfur oxidoreductase (Fig. 1A). The putative pilin genes, aapA (Saci2314) and aapB (Saci2319) are transcribed in opposite directions of the main operon (Fig. 1). Spatial proximity of the prepilin genes to the other aap genes suggested that the encoded products of these genes are the pilin subunits of the aap filament. However, to unequivocally verify this assumption, pili were isolated from a S. acidocaldarius strain lacking the archaellum and ups pili, ΔupsEΔflaJ (Henche et al., 2012). The isolated fraction was analyzed by TEM which only contained long filamentous structures (Fig 1B). However, subsequent SDS-PAGE and glycostain could not identify distinct bands at the expected sizes of ~ 14–15 kDa. In all analyzed samples, a large amount of protein did not enter the separating gel of the SDS-PAGE. Mass spectrometry analysis of this fraction identified aapB (Saci2319) which encodes the pilin AapB, indicating that the aap pilus is a very stable filament. This stability might stem from strong hydrophobic interactions of the pilin subunits and also posttranslational modification by glycosylation of the aap pilus which can also be the cause for the difficulties to identify the pilins by mass spectrometry. Harsh treatment of the isolated pili with phenol and detergents like Triton X-100 did not lead to the disassociation of the pilin subunits. As only AapB was identified in the isolated fraction it is assumed that this protein is the major pilin of the aap filament.
Figure 1.
Aap pili and organization of gene cluster encoding the aap pili. (A) Organization of the aap pili gene cluster. The S. acidocaldarius ORF numbers are indicated in the arrows and given names below the arrows. A tRNA gene for glycine is located inside the aap gene cluster (S. acidocaldarius DSM639 complete genome GenBank accession number: CP000077.1). (B) Electron micrograph showing S. acidocaldarius DSM639 on a carbon-coated nickel grid. The cells were negatively stained with uranyl acetate. Aap pili are marked with a white arrow, whereas archaella are depicted by a black arrow. Threads are indicated by a green arrow. The square marks the magnified area shown in (C). (D) Electron micrograph showing isolated aap pili filaments. Scale bars are 100 nm.
AapA and AapB are type IV pilins
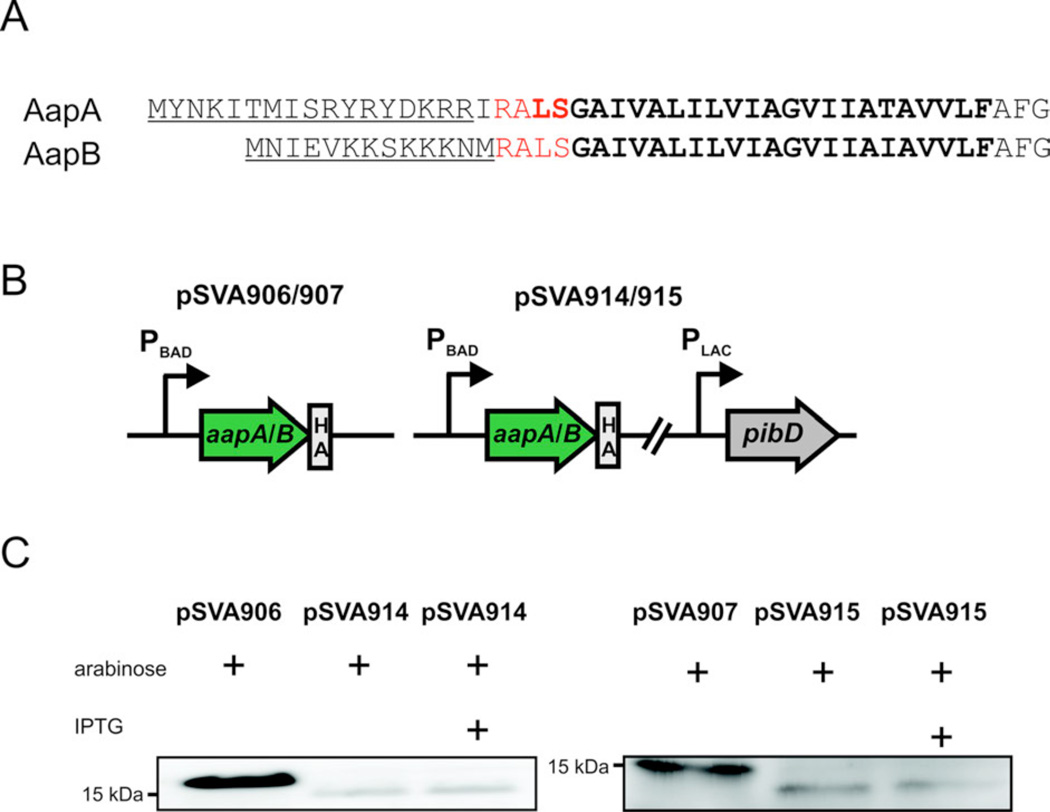
The two pilins AapA and B contain the conserved class III signal peptides that can be predicted by the FlaFind program (Fig 2) (Szabo et al., 2007). The class III signal peptidase PibD from S. solfataricus has been characterized in detail (Albers et al., 2003; Szabo et al., 2006). The PibD (Saci_0139) from S. acidocaldarius was cloned under the control of an IPTG inducible promoter and either the AapA or AapB were cloned with an HA-tag under the control of an arabinose inducible promoter on the same plasmid (Fig. 2B). Using these plasmids an in vivo cleavage assay in E.coli was performed in which the expression of the pilins was induced by the addition of arabinose two hours before the induction of PibD by the addition of IPTG (Szabo et al., 2007). Membrane extracts of the expressing E. coli strains were separated on SDS-PAGE and AapA/B were detected by immunoblotting using a HA-tag specific antibody. AapA and AapB were clearly processed by the S. acidocaldarius PibD confirming their identity as class III pilins (type IV pilins). As described before cleavage of the pilins was already observed before the expression of PibD was induced as the IPTG inducible promoter was leaky (Szabo et al., 2007).
Figure 2.
In vivo processing of AapA and AapB in E.coli. (A) Signal peptides of AapA/B. The predicted cleavage site is indicated by red letters and the hydrophobic domain is marked by bold letters. (B) Schematic representation of the used plasmids. The plasmids pSVA906 and pSVA914 encode the full length putative prepilin aapA, whereas pSVA907 and pSVA915 encode the full length putative prepilin aapB. pSVA914 and 915 encode additionally to the respective pilin gene also for the type IV prepilin peptidase PibD. (C) Detection of the cleavage of AapA/B by immunoblot analysis using HA-specific antibodies. The fulllength AapA has a size of 17.6 kDa. After pibD expression, AapA displays band at 15 kDa. The fulllength and the processed AapB have a size of 15.9 kDa and 13.9 kDa, respectively.
Structure of the aap pili
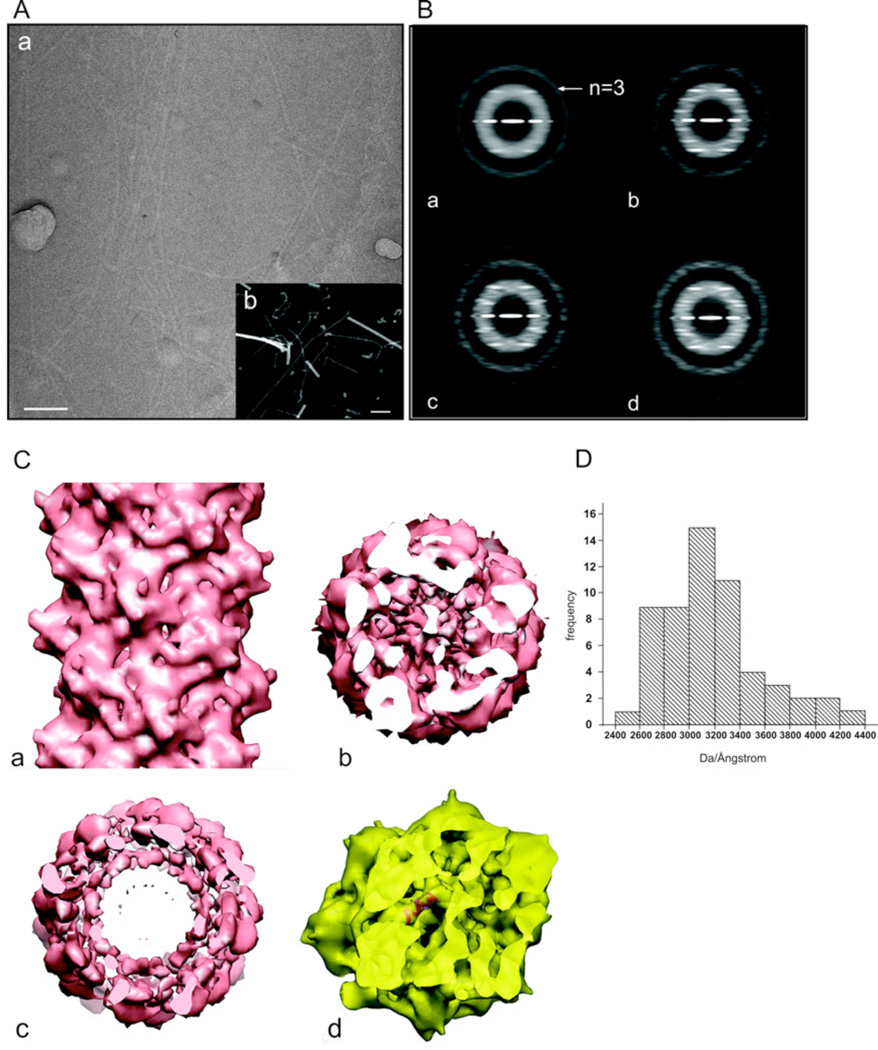
The pili were examined by using electron cryo-microscopy (cryo-EM) of unstained frozen-hydrated filaments (Fig. 3A(a)). Since a preliminary analysis of such images suggested that there might be ambiguities in determining the correct helical symmetry (Egelman, 2010), we also used scanning transmission electron microscopy (STEM) of freeze-dried filaments (Fig. 3A(b)) to measure the mass per unit length (Engel et al., 1982; Wall and Hainfeld, 1986). The mass per unit length histogram (Fig. 3D) shows a rather broad distribution, with a mode at ~ 3.1 kDa/Å. Given a predicted molecular weight of the subunit (in the absence of any glycosylation) of 14.8 kDa, this would predict an axial rise per subunit of ~ 4.8 Å. Using the Iterative Helical Real Space Reconstruction method (Egelman, 2000) with ~ 21,000 segments extracted from the cryo-EM images, the only solution that was found that was consistent with the STEM measurements had ~ 2.6 subunits per turn of a ~ 15 Å pitch helix. This would correspond to a mass per subunit of 17.4 kDa. We expect that the difference from the STEM value is due to the known glycosylation.
Figure 3.
Electron microscopic analysis of the aap pili. (A) Electron microscopic images of the pili by cryo-EM of unstained frozen-hydrated samples (a), and by scanning transmission electron microscopy (STEM) of unstained freeze-dried samples (b). The filaments in (a) are imaged in a hole present in a carbon film, while the filaments in (b) are on a solid support. The larger objects in (b) are tobacco mosaic virus particles, used as an internal mass standard. The scale bars are 100 nm. (B) Power spectra from cryo-EM images of the pili. (a) A power spectrum from the entire set of segments (n=21,369) shows only a single layer line, identified as |n|=3, that is at a spacing of ~ 1/(41 Å). Sorting the population by twist and axial rise yields subsets that show three layer lines at relatively low resolution, Three examples are shown: (b) 136.9°, 5.7 Å. This is the subset that was used for subsequent reconstructions, and contained 4,613 segments. The near equatorial layer line would therefore be n=−8, and the layer line below the n=−3 would be n=+5; (c) 138.9°, 6.7 Å; (d) 137.9°, 6.2 Å. To boost the signal-to-noise ratio in the images and to correct for phase reversals introduced by the defocus of the microscope, all electron micrographs were multiplied by the contrast transfer function determined for that micrograph. This produces the very visible Thon rings in the power spectra. (C) A side (a) and top (b) view of the three-dimensional reconstruction of the pili. A single α-helix fits very nicely into the tubular densities seen in the inner core of the reconstructed volume (c), consistent with the notion that the hydrophobic N-terminal α-helices are forming this core, with the globular heads on the outside of the filament. (D) A histogram of mass per unit length measurements of the pili determined by STEM.
Further analysis showed great variability in these helical parameters, explaining why the averaged power spectrum from all segments (Fig. 3B) was so poor, and only displayed a single layer line which arose from the 3-stranded helices. Significant improvements in power spectra were obtained by sorting the segments into subsets corresponding to different values of twist and axial rise per subunit. The subset that was most heavily populated (136.9° twist, 5.7 Å axial rise) was used for the final reconstruction (Fig. 3C). The filaments have a diameter of ~ 110 Å, with a weaker density in the core (Fig. 3C (c)). The resolution was judged (FSC=0.5) to be ~ 9 Å, and the volumes were filtered to this resolution. At this resolution the central core is seen to contain rod-like densities, and this would be consistent with the packing of the highly hydrophobic N-terminal α-helices in the core of these pili, as determined for the Neisseria gonorrhoeae Type IV pili (Craig et al., 2006). However, the packing of these α-helices is very different between Neisseria gonorrhoeae and Sulfolobus acidocaldarius. In N. gonorrhoeae there is a rise per subunit of 10.5 Å and a rotation per subunit of ~ 100°, and these parameters appeared to be rather fixed. The Klebsiella oxytoca PulG Type IV pili have a rotation of ~ 85° and an axial rise of 10.4 Å (Campos et al., 2010), while the adhesion filament from I. hospitalis that is built from subunits having homology in the α-helical N-terminal region to bacterial Type IV pilin have a rotation of ~ 107° and an axial rise of 5.3 Å (Yu et al., 2012). In contrast, in S. acidocaldarius, there is a rotation per subunit of ~ 138°, a rise per subunit of ~ 5.7 Å, and these parameters are quite variable. Examples are shown in Fig. 3B of power spectra from subsets classified as having axial rises of 6.2 and 6.7 Å. The fact that these subsets, classified using models, display the expected power spectra shows that the classification works. An alternative scenario is that due to a poor signal to noise ratio filament segments can be found to have the best cross-correlation with models having a different symmetry than they actually have, but the averaged power spectra from such subsets would display the same features as the global average. We can therefore exclude this possibility.
The hand of this reconstruction has not been determined, and the available resolution (~ 9 Å) does not allow us to directly determine the hand of the α-helices. The reconstruction is shown assuming a left-handed ~ 15 Å pitch one-start helix, which means that the prominent three-start helices would also be left-handed.
Expression pattern of the aap pili
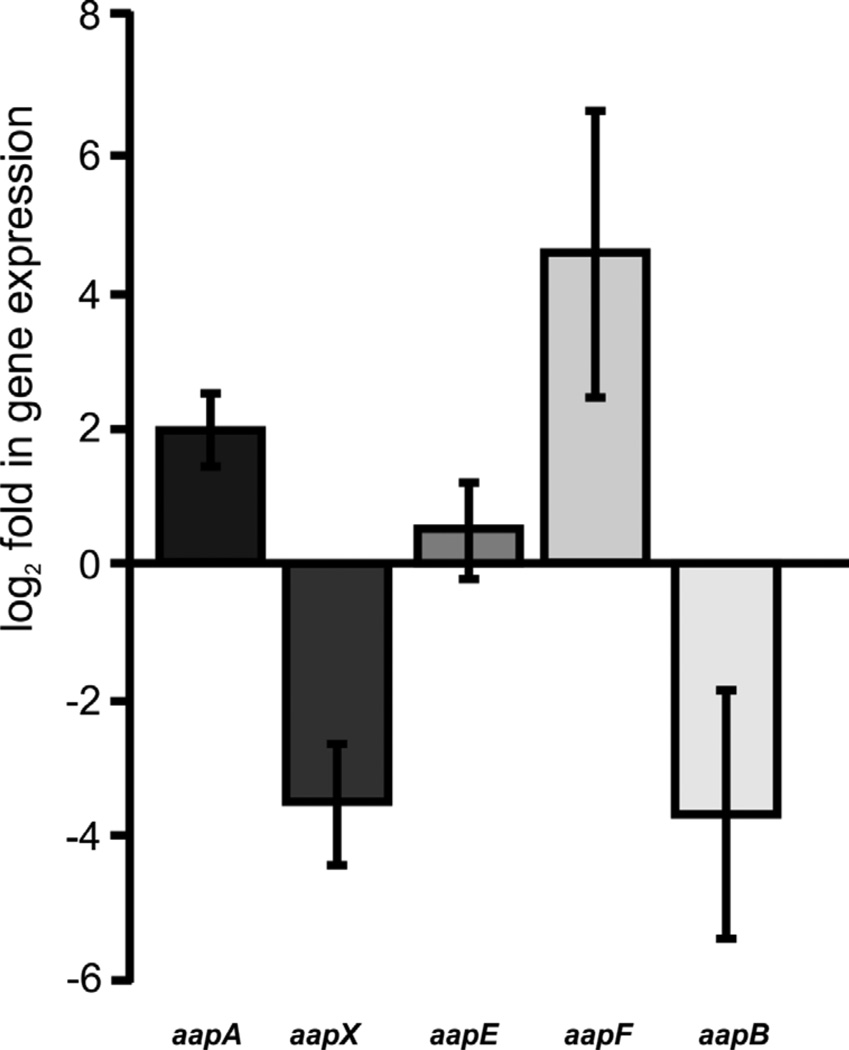
In an earlier study, it was observed that in an aapF deletion mutant archaella expression and production was highly induced (Lassak et al., 2012; Henche et al., 2012). As archaella were mainly expressed during the stationary phase in wild type cells, the expression of the different aap genes was analyzed comparing exponential phase against stationary phase grown cells. Quantitative RT-PCR on total RNAs isolated from the cells from the two different growth phases using primers for each aap gene, showed that transcript levels for aapX and aapB were strongly reduced in the stationary phase which corresponds to the lower abundance of aap pili in the stationary growth phase (Fig 4). Whereas transcript levels of aapE were not significantly changed, aapA transcripts were slightly and aapF transcripts strongly stimulated in the stationary phase. In RNA-seq data obtained from S. solfataricus in the aapF homologue SSO2386 two antisense RNAs were described (Wurtzel et al., 2010). If that is the case for S. acidocaldarius too, the qRT-PCR data might just be the result of a high expression and accumulation of the antisense RNA. Although primers were chosen in such a way that they should bind outside of the possible transcribed antisense RNAs, the transcription of these might not be perfectly terminated leading to a wrong signal in the qRT-PCR suggesting higher expression levels.
Figure 4.
qRT-PCR analysis of aap transcript levels in MW001 during stationary phase in comparison to the exponential growth phase. The relative gene expression was normalized against an internal control gene secY. The bars represent the mean of the biological replicates of the log2 fold expression of each aap gene in the stationary compared to the exponential growth phase. Standard deviations are indicated.
Deletion mutant analysis
To dissect which subunits of the aap pilus operon are essential for assembly, single in frame deletion mutants were obtained of all five aap subunit genes. Using a pop-in/pop-out method as described recently by Wagner et al (Wagner et al., 2012) in frame deletion mutants were generated in an uracil-auxotrophic background strain of S. acidocaldarius MW001. Additionally, a double mutant lacking both potential pilin genes was constructed. All mutants were analyzed in terms of their growth kinetics, however no significant difference compared to the wild-type growth behavior was observed.
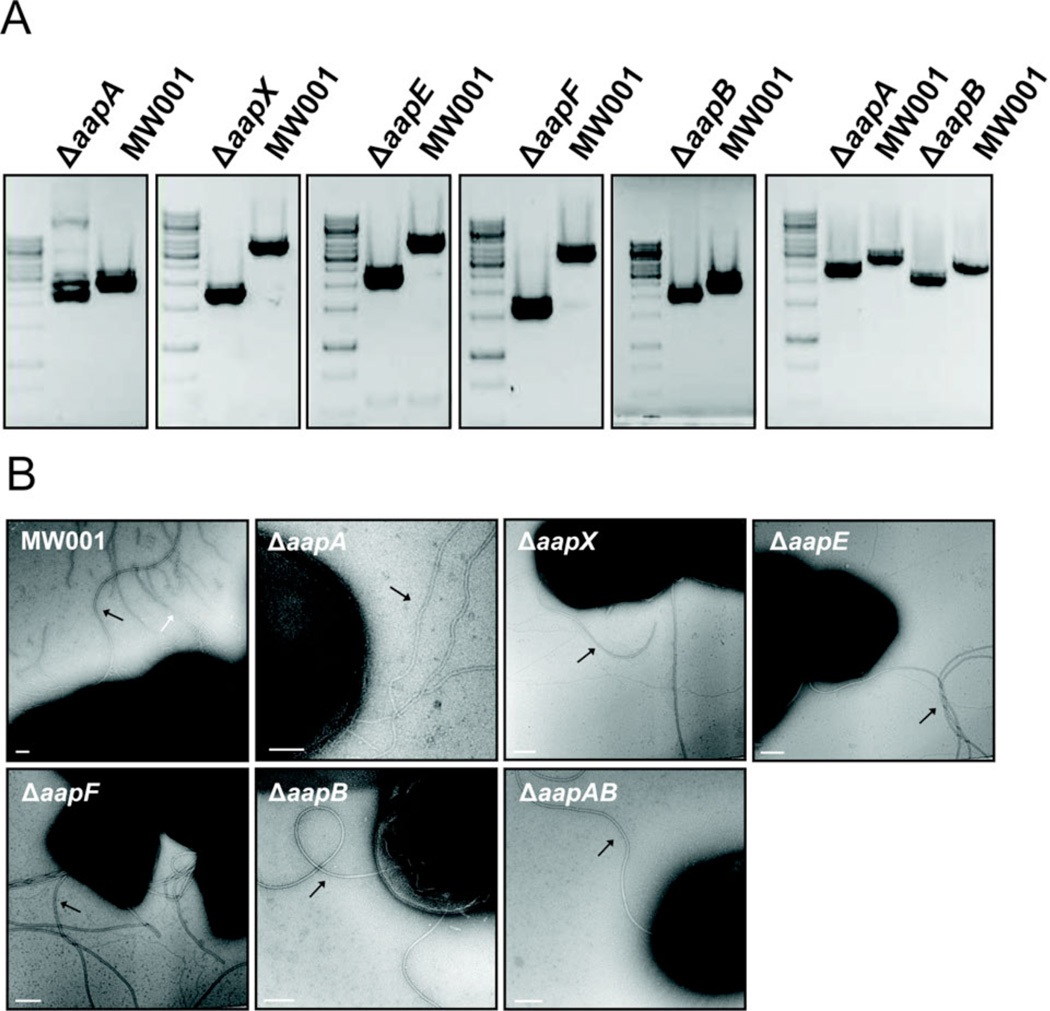
Transmission electron microscopic images of all aap deletion mutants were compared to the wild-type cells, which exhibits archaella, aap pili and threads (Fig 5B). In none of the aap deletion strains aap pili could be detected indicating that all five genes are indispensible for pilus assembly. However, in all deletion strains the expression of archaella was increased and especially in the ΔaapF strain. The deletion of aapF led to a hyperarchaellated phenotype as observed previously (Henche et al., 2012). This phenotype was confirmed by swarming assays on soft gelrite plates (Fig 6). The wild type cells MW001 and all aap deletion mutants were tryptone starved and inoculated on a soft gelrite plate which induces the expression of archaella leading to swarming motility. Especially the ΔaapX, E and F strains exhibited very strong swarming motility, whereas ΔaapA/B swarmed only slightly faster than the wild type. This confirms that the aap pili are regulatorily interlinked to the expression of the archaella operon.
Figure 5.
Construction and characterization of aap deletion mutants. (A) PCRs were performed on wild type and the respective aap deletion mutant genomic DNA revealing a downshift of the corresponding DNA band of the mutant DNA. (B) Electron microscopy images of the MW001 and aap deletion mutants. White arrow indicates aap pilus, black arrows indicate archaella. Scale bars are 100 nm.
Figure 6.
Swarming motility assay of aap deletion mutants. The cells were grown to an OD600nm of 1 in Brock medium containing 0.1 % dextrin and 0.1% tryptone and spotted on a Brock plate containing only 0.15% gelrite. The tryptone concentration in the plates was reduced to 0.005% to induce the production of archaella and incubated at 75°C in special metal boxes to prevent evaporation of medium. After seven days, the swarming of the different strains was analysed by scanning the plates.
Influence of the aap genes biofilm formation
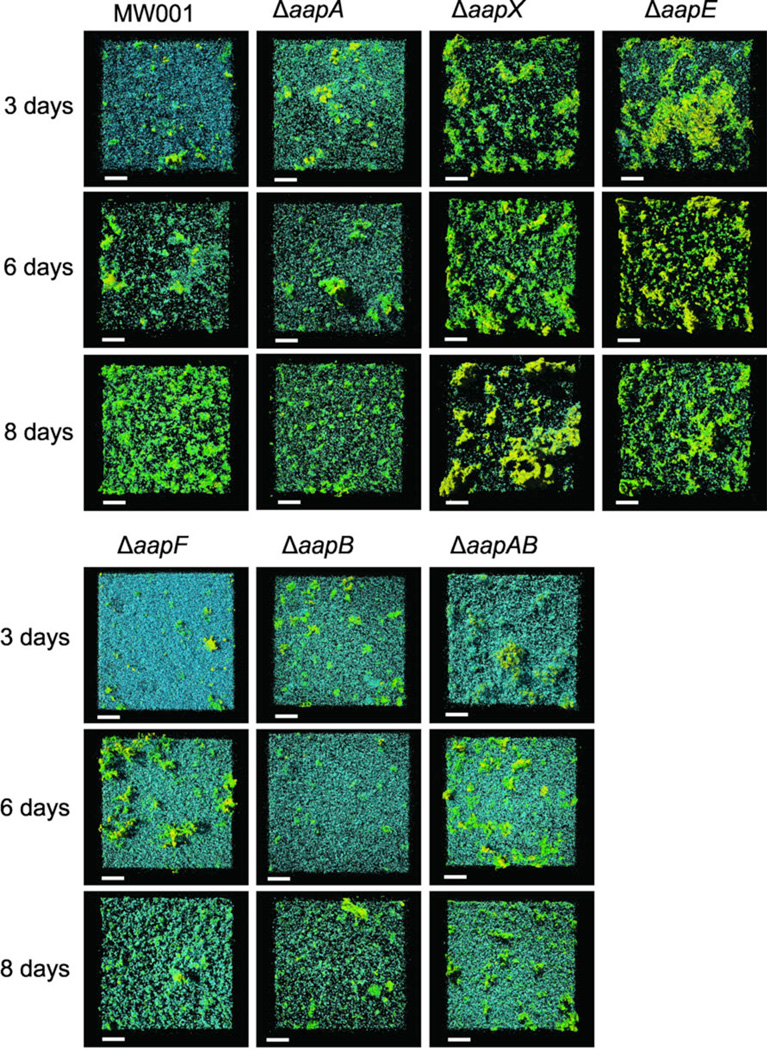
In a previous study, it was already shown that the aap pilus is strongly involved in shaping the S. acidocaldarius biofilm (Henche et al., 2012). Strains lacking the aap pilus displayed a carpet-like flat biofilm with an increased cell density at the bottom layer and strongly reduced tower-like structure formation. However, as different aap deletion strains already displayed a different behavior in distribution of cell surface appendages we wanted to test how the different deletion mutants behave during biofilm formation. Biofilms of the wild type MW001 and all aap deletion mutants were grown for 3, 6 and 8 days and analyzed by confocal laser scanning microscopy (CLSM). As previously shown, the ΔaapF deletion mutant showed a flat biofilm with a high density of cells possibly caused by the overexpression of the archaella. The double pili deletion strain ΔaapAΔaapB and the ΔaapB were most similar to this phenotype, whereas the ΔaapA mutant was comparable to the wild type. Surprisingly, the ΔaapX and ΔaapE strains showed a strong increase in EPS production already at day 3. Secretion of galactosyl containing EPS was already present for the ΔaapE strain at day 3 and continued through day 6, whereas in ΔaapX mainly mannose/glucose containing EPS was produced during the first 7 days whereas at the 8th day galactosyl residues containing EPS was secreted.
Discussion
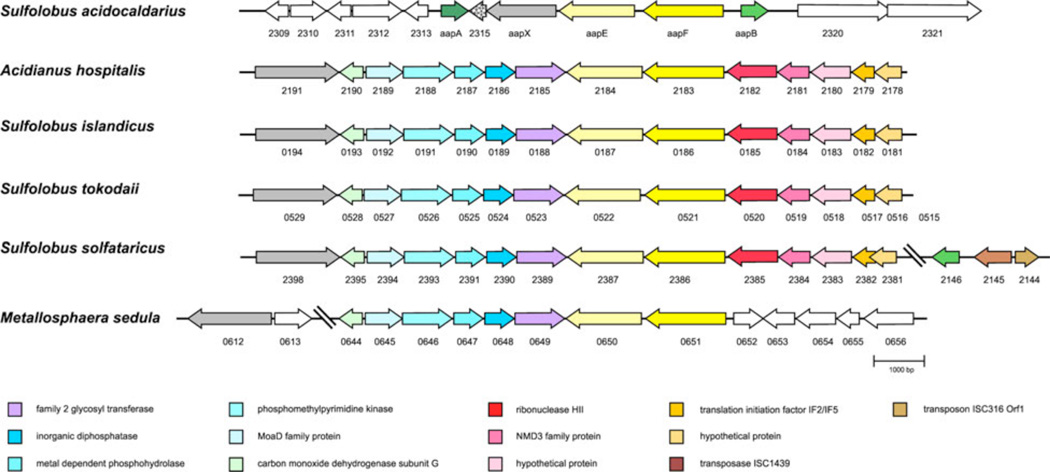
Archaeal type IV pili have been shown to be involved in numerous functions such as adhesion, biofilm formation, motility, cell aggregation and DNA exchange. The aap pilus, which is unique for S. acidocaldarius in contrast to other Sulfolobales, has been shown to be essential for surface attachment and shaping the S. acidocaldarius biofilm (Henche et al., 2012). During this study, we could show that the pilus is very stable against different denaturing conditions including hot phenol and detergent treatment. Considering the fact that S. acidocaldarius thrives at temperatures ranging from 70–90°C and low pH in its natural habitat (Brock et al., 1972), the high rigidity of the filament seemed to be an adaptation to this environment. For bacteria, a high stability of type IV pili was observed which was linked to their biological functions in these organisms. In Neisseria gonorrhoeae and Myxococcus xanthus, it was demonstrated that the type IV pilus requires a high stability to withstand mechanical forces when retracting the pilus to perform twitching motility (Merz et al., 2000; Clausen et al., 2009). Recently, thermal and proteolytic stability could also be linked to the GC pilus of N. gonorrhoeae which was suggested to stem from aromatic residues present in the pilin PilE (Li et al., 2012). The C-terminal region of the two pilins AapA and AapB of the aap pilus contains phenylalanine and tyrosine residues which could then contribute to a more firm binding of the pilins in the filament. This would suggest that not only hydrophobic interactions of the N-terminal α-helices of the pilins are forming the filament, but also the C-terminus of the pilins might be involved in forming a rigid structure as well. Additionally, numerous asparagines are present in the pilins which are possible target residues of the N-glycosylation machinery. The high stability observed for the aap structure is reflected in the biological role of this pilus which is predominantly to promote adhesion to a surface (Henche et al., 2012). Especially, the establishment of sufficient binding in an environment characterized by extreme acidity, heat and consequently high Brownian motion, demands a stable connection of the cell with a surface. In nature, S. acidocaldarius is predominantly found in the crusty edges of geothermal springs, indicating that this organism prefers the biofilm lifestyle. In contrast to this, other Sulfolobaceae do not form this filament and show a rather simple biofilm development (Koerdt et al., 2010). Nevertheless, homologs of most the aap genes are also found in the other Sulfolobaceae (Fig.8). However, the genetic organization of the aap homologues differs from S. acidocaldarius, as the aapX homologue is not in the direct neighborhood of aapE and aapF, but is located up to five genes upstream. Additionally, the direction of the transcription start site is in the opposite direction. Homologs of aapA and aapB are completely missing in the other species except for aapB for which one homolog was identified in S. solfataricus. In close vicinity to aapB in S. solfataricus, a transposase is present, suggesting that previously the aap pilus might have been present in all Sulfolobaceae. Due to transposition events that occur in species other than S. acidocaldarius more frequently (Mao and Grogan, 2012), the aap gene cluster was split up. It is intriguing to correlate these findings with the natural habitat for different species of Sulfolobales. As already mentioned earlier, it is known that S. acidocaldarius is more abundant at the periphery of the hot volcanic mud pool, where as S. solfataricus and S. islandicus are found to be abundant inside the mud pool ecosystem in the geothermal springs. Recent evidences have suggested that microbial population dynamics are largely dependent on different biological/ecological parameters such as mutation, selection, recombination, DNA exchange and transposition. Such events ultimately result into species differentiation which finally leads to the speciation of different species under same genus in a given ecological niche (Cadillo-Quiroz et al., 2012). As S. acidocaldarius prefers to grow in the periphery of the volcanic mud pools the aap pili may enable the organism to stay confined in this niche. In contrary, surface attachment is possibly not the pivotal feature for other species, e.g., S. solfataricus and S. islandicus, and therefore aap pili have been negatively selected in these organisms. The aap homologs in these species have been selected for so far unknown purposes as it is evident for S. solfataricus. Although the function of AapX has not been identified so far, bioinformatic analysis suggest a role in sugar- or iron-binding activities which might not only be important for S. acidocaldarius but for other Sulfolobales members as well. As AapX is essential to assemble the aap pilus as an aapX-deficient mutant is lacking this filament, the homologs of the polytopic membrane protein AapF might also be important in the other Sulfolobus species to correctly anchor AapX to the membrane. However, experimental analysis is needed to be performed to support these ideas.The ATPase homolog (SSO_2387) from S. solfataricus was shown to autophosphorylate (Albers and Driessen, 2005; Lower and Kennelly, 2003). However, such an autophosphorylation event is absent in AapE of S. acidocaldarius (data not shown).
Figure 8.
Synteny of aap gene cluster in different Sulfolobaceae as obtained by Absynte (Despalins et al., 2011). Conserved genes are depicted in the same color. Numbers below genes represent ORF numbers in the respective organism (S. acidocaldarius DSM639 complete genome GenBank accession number: CP000077.1; Acidianus hospitalis W1 complete genome GenBank accession number: CP002535.1; S. islandicus L.D.8.5 complete genome GenBank accession number: NC_013769.1; S. tokodaii str. 7 complete genome GenBank accession number: BA000023.2; S. solfataricus P2 complete genome GenBank accession number: AE006641.1; Metallosphaera sedula DSM5348 complete genome GenBank accession number: CP000682.1).
Analysis of the expression patters of the aap genes showed that they seem to be regulated individually. Also for the archaellum, it was demonstrated that the archaellin was differently regulated than the remaining genes of the archaellum assembly system (Lassak et al., 2012). Intriguingly, the transcript levels of aapF were up-regulated in stationary phase. A RNA-seq analysis already revealed that the aapF homolog SSO_2386 in S. solfataricus contains two non-coding RNAs on its antisense strand (Wurtzel et al., 2010). The presence of the antisense RNA might interfere with the detection of the actual aapF transcripts. However, the presence of the antisense RNAs indicates that the regulation of the aap pilus might involve also non-coding RNAs. In contrast to Bacteria, non-coding RNAs seem to play a more important role in Archaea, as archaeal genomes comprise a high number of non-coding RNAs (Wurtzel et al., 2010).
Analysis of the different aap mutants by TEM, swimming motility and biofilm formation demonstrated that the deletion of the individual aap genes did not result in a uniform phenotype and once more outlined that the regulatory network of the aap pilus is more complex than previously expected and seems to influence different processes. Whereas the EM analysis revealed that only the aapF-deficient strain induces visible hyperarchaellation of the cell surface, the swarming motility assay showed that also the strains ΔaapX and ΔaapE are hypermotile on semi-solid plates. Consequently, elevated swarming motility cannot be solely linked to the higher abundance of archaella. The analysis of the static biofilm formation demonstrated that the deletion of aapE and aapX is causing an increased level of EPS secretion, which was already visible after day 3. The huge amount of extracellular polysaccharides might have facilitated the swarming motility by increasing the medium viscosity. This effect was recently observed for Pseudomonas aeruginosa. By increasing the mucin concentration on swarming plates, the surface motility of P. aeruginosa was positively influenced (Yeung et al., 2012). Additionally, type IV pili-dependent motility in M. xanthus was also shown to be linked to the presence of EPS (Lu et al., 2005).
Conserved building blocks, such as actin-like proteins (Derman et al., 2009), can be found widely across evolution. While early expectations were that these actin-like proteins would polymerize into filaments similar to F-actin (Van Den Ent et al., 2002), it has now been established that this is not the case, and different bacterial actin-like proteins polymerize into different filaments (Galkin et al., 2009; Orlova et al., 2007; Popp et al., 2010a; Popp et al., 2010b; Polka et al., 2009). Similarly, the expectation existed that bacterial flagellins, all sharing highly conserved coiled-coil domains (Beatson et al., 2006), would pack together the same way as done in Salmonella (Yonekura et al., 2003). However, we have shown that this is not the case (Galkin et al., 2008), and the Campylobacter flagellins form a different filament with seven protofilaments, rather than 11, even though they are built from a conserved flagellin subunit.
We have now shown that although the Sulfolobus pili are built from a homolog of bacterial type IV pilin, they have a different architecture from the best studied bacterial type IV pilus, that of Neisseria gonorrhoeae (Craig et al., 2006), that modeled for the PulG pilus from Klebsiella oxytoca (Campos et al., 2010), or that formed by the Ignicoccus hospitalis adhesion filament (Yu et al., 2012). This supports our argument (Galkin et al., 2008) that quaternary structure may be quite labile, and that small changes in sequence may be amplified by large changes in higher-order packing.
Based on the results we gained so far, we concluded that the aap pilus is a unique cell surface appendage of S. acidocaldarius, which influences the mode of life by attaching the cells to different surfaces. Due to the high stability of this filament, adherence can be successfully promoted also in a harsh environment. Transcriptional and deletion mutant analysis, suggested that the regulation of the aap gene cluster appears to be quite complex as the single aap genes are regulated separately and non-coding RNAs might be involved in the regulatory process as well. Future studies on the regulatory network of the aap pilus may shed more light into the interplay of the different surface structures in S. acidocaldarius.
Materials and Methods
Strains and growth conditions
Sulfolobus acidocaldarius MW001 and all constructed aap deletion mutants were aerobically grown in Brock media (Brock et al., 1972) with a pH of 3 at 76°C. The media were supplemented with 0.1% (w/v) tryptone or with 0.1% (w/v) N-Z-Amine and 10 µg ml−1 uracil. The growth of the cells was monitored by measurement of the optical density at 600 nm.
Isolation of the aap pili
The strain ΔupsEΔflaJ expressing only aap pili was grown for two days (OD600nm: 0.7) in 3 l Brock media supplemented with 0.1% tryptone, 0,1% dextrine and 10 µg ml−1 uracil. The cells were collected (5000 × g, 30 min) and resuspended in 10 ml Brock media. The filaments were sheared off by processing the cell suspension in a blender for 4 × 15 sec. After removal of the cells by a low spin centrifugation at 4500 × g for 20 min, the filaments were pelleted from the supernatant by ultracentrifugation at 340.000 × g for 90 min and resuspended in 100 µl Brock media. This fraction was further purified by a CsCl density gradient (0,5 g ml−1) at 250.000 × g for 18 h. The resulting whitish band was isolated, diluted in Brock medium to maintain optimal salt concentrations, subsequently concentrated by ultracentrifugation at 250.000 × g for 30 min and resuspended in 100 µl Brock media. The isolated fraction was analysed by TEM analysis, SDS-PAGE and glycostained using the Pro-Q ®Emerald 300 Glycoprotein Gel Stain Kit (Invitrogen).
Construction of deletion mutant plasmids and deletion strains
All S. acidocaldarius deletion strains generated in this study derived from the uracil-auxotrophic strain MW001 using two consecutive homologous recombination events (Wagner et al., 2012). For the construction of deletion mutant plasmids, 800 bp of the respective up-and downstream flanking region of the genes aapA (Saci2314), aapX (Saci2316), aapE (Saci2317), and aapB (Saci2319) were amplified by PCR using S. acidocaldarius genomic DNA as a template and the corresponding primer pairs listed in table 3. By overlap extension PCR, the up- and downstream flanking regions were joined and amplified using the outward bound primers. The PCR products were cloned in the suicide vector pSVA406 yielding the deletion mutant plasmids pSVA218 (aapA), pSVA222 (aapX), pSVA220 (aapE) and pSVA219 (aapB). The plasmid for the aapF in-frame deletion was already constructed before (Henche et al., 2012). All plasmid constructs were confirmed by sequence analysis. Before transformation in MW001, the plasmids were methylated in E. coli ER1821 containing pM.EsaBC4I to be protected against degradation by endogenous nucleases of S. acidocaldarius. Via electroporation (Berkner et al, 2007), methylated plasmids were transformed into electro-competent S. acidocaldarius MW001 cells. After regeneration in 2× recovery solution (1% sucrose, 20 mM β-alanine, 1.5 mM malate buffer, pH 4.5, 10 mM MgSO4) for 30 min at 75°C, the cells were streaked on gel-rite plates lacking uracil and incubated at 75°C for 5–7 days. Integrants were confirmed by colony PCR and plated on counter selection plates supplemented with uracil and 5-FOA to promote a second homologous recombination event. After 5–7 days, uracil-auxotrophic colonies containing the deletion construct were identified by colony PCR and confirmed by sequence analysis using the respective sequencing primers.
Construction of in vitro cleavage assay plasmids
To study the processing of the two putative aap pilus subunits by Saci0139, the PibD homolog in S. acidocaldarius, several plasmids were constructed containing Saci0139 and the two class III signal peptide-harbouring substrates under differentially regulated promoters. The genes encoding for the putative pilins aapA (saci_2314) and aapB (saci_2319) were amplified by PCR by using S. acidocaldarius genomic DNA as a template, primers with appropriate restriction sites and devoid of the native stop codon. The PCR products were cloned into pSVA133 via the restriction enzymes NcoI and BamHI which adds a C-terminal hemagglutinin (HA)-tag to the introduced gene resulting in the plasmids pSVA901 (aapA)and pSVA903 (aapB). The pilin genes including the C-terminal epitope-tag were transferred to pSVA134 using the restriction enzymes NcoI and HindIII yielding pSVA906 (aapA+HA) and pSVA907 (aapB+HA). Consequently, the expression of the genes was regulated by an arabinose-inducible promoter. To establish plasmids for co-expression of the prepilin peptidase pibD and the the pilin genes, the S. acidocaldarius pibD homolog Saci0139 was first cloned into the vector pUC18-pibD as described previously (Szabo et al., 2007). Using SphI restriction enzyme, a cassette containing a T7 promoter, the pibD open reading frame, a C-terminal six-histidine tag, and a T7 terminator was transferred into the plasmids pSVA906 and 907 resulting in pSVA914 (aapA+HA, pibD+6xHis) and pSVA915 (aapB+HA, pibD+6xHis).
In vitro cleavage assay
The plasmids were transformed into E. coli BL21 (DE3) RIL (Agilent Technologies) cells. Expression of the pili genes was induced by the addition of 0.2% L-arabinose, whereas PibD expression was initiated by the addition of 0.1 mM IPTG. After 4 hours of protein production, the cells were centrifuged, resuspended in 8 ml of HEPES buffer (50 mM HEPES pH 7.5, 150 mM KCl) and lysed by sonication. After a lowspin (4500 × g, 20 min) and an ultracentrifugation step (250.000 × g, 45 min) the membrane fraction was resuspended in 500 µl buffer. Additionally, SDS was added up to a final concentration of 1% (v/v) in order to solubilize membrane proteins. The samples were analysed by SDS-PAGE and immune-detected using monoclonal anti-hemagglutinin antibodies (Sigma-Aldrich).
RNA isolation, cDNA generation and quantitative real-time PCR
Total RNA was isolated from S. acidocaldarius MW001 cells grown up to exponential and stationary growth phase by using the TRIzol® Reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions. Contaminating genomic DNA was removed by incubating the RNA with DNaseI at 37°C for 2 h. Before cDNA synthesis, the quality of the RNA was analyzed with gel-electrophoresis, nano-photometry and PCR using the qRT-primer pair for aapB. 2 µg DNA-free total RNA was reverse-transcribed into cDNA using the First Strand cDNA Synthesis Kit. The quantitative PCR analysis was carried out according to the protocol and materials provided by Applied Biosystems. For each gene of interest, a triplicate set-up of 25 µl PCR mixture was prepared from 12.5 µl Maxima® SYBR Green/ROX qPCR Master Mix (2×), 2 µl of 0.3 µM primer pair stock solution, 1 µl cDNA and 9.5 µl nucleotide-free water. The negative control assays were done with RNA mixtures that were used for cDNA synthesis. Primer efficiencies were calculated from the average slope of the linear regression curves according to the calculation model advised by Applied Biosystems. The fluorescence quantities of the reactions were measured with ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA).
Electron microscopy
The different S. acidocaldarius strains were grown to an OD600nm 1 and fixed with 1.25% glutaraldehyde. After 15 minutes, the cells were collected applied to hydrophobilized carbon-coated nickel-grids and negatively stained with 2% uranyl acetate. Images were obtained on a JEOL 3010, 300 kV transmission electron microscope (JEOL, Eching, Germany). Cryo-EM was performed on filaments isolated from cells as described above and were applied to freshly glow-discharged C-flat grids, blotted, and plunged into liquid ethane using a home-built cryo-plunger. Grids were imaged using a Tecnai F20 microscope (FEI) at an accelerating voltage of 200 keV and a nominal magnification of 50,000×, and recorded on film. Micrographs were scanned with a Nikon Coolscan 8000 at a raster of 1.25 Å per pixel. The helixboxer routine in EMAN (Ludtke et al., 1999) was used for cutting filaments from micrographs. The SPIDER software package (Frank et al., 1996) was used for most of the subsequent processing. Samples for STEM were prepared as previously described (Wall, Simon, Lin, & Vinogradov, 2008). Digital dark-field micrographs of the freeze-dried samples were recorded in 512×512 frames at either 1.0 or 2.0 nm/pixel at the Brookhaven STEM Facility. Images were analyzed using PCMass 3.0.
Biofilm
Biofilms of the Sulfolobus strains were grown in small Petri dishes (µ-dishes, 35 mm, Ibidi, Martinsried) in 4ml Brock media with 0,1% NZ-amine as a standing culture. Two biological replicates were performed for each of the eight strains and grown for 3, 6 and 8 days. The Petri dishes were put in a specially designed metal box (25 cm L × 20 cm W × 20 cm D) with ~ 500 ml of water in the bottom to minimize evaporation of the media, as described by Koerdt and colleagues (2010). To visualize the biofilms by CLSM different fluorescent probes were employed and was essentially performed as described in Koerdt and colleagues (2010). In brief, DAPI (4′,6-Diamidino-2-phenylindol) was used to stain the cells of the biofilm whereas fluorescently labelled lectins (ConA, specific for mannose/glucose and GS-IB4 specific for galactosyl residues) were employed to visualize the EPS (extracellular polymeric secretions) of the biofilms. The lectin biofilm mixtures were incubated at room temperature for 20–30 min, in the absence of light. After incubation, the biofilm was washed with Brock media (pH 5) to remove excess label and images were taken with CSLM. Image data obtained were processed by using the IMARIS software package (Bitplane AG, Zurich, Switzerland).
Supplementary Material
Figure 7.
Static biofilm formation of all aap deletion mutants in comparison to the wild type MW001 analyzed by confocal laser scanning microscopy. Cells were incubated for three, six and eight days in small petri dishes at 75°C in special metal boxes to prevent evaporation of medium. The cells were visualized using DAPI (blue channel) and EPS was stained using the two fluorescently labeled lectins ConA and IB4. The green channel represents ConA which binds to glucose and mannose residues, whereas IB4 binds to α-galactosyl residues (yellow channel).. Overlay of all three channels are shown. Scale bars are 40 µm.
Table 1.
List of archaeal and bacterial strains used in this study
| Strain no. | Genotype | Source/reference |
|---|---|---|
| MW001 | Deletion of pyrEpyrF in S. acidocaldarius | Michaela Wagner |
| MW155 | Deletion of aapF in strain MW001 | (Henche et al., 2012) |
| MW153 | Deletion of aapA in strain MW001 | This work |
| MW154 | Deletion of aapB in strain MW001 | This work |
| MW159 | Deletion of aapX in strain MW001 | This work |
| MW160 | Deletion of aapE in strain MW001 | This work |
| MW161 | Deletion of aapA and aapB in strain MW001 | This work |
| MW158 | Deletion of upsE and flaJ in strain MW001 | (Henche et al., 2012) |
| DH5α | Escherichia coli K-12 cloning strain | Gibco |
| ER1821 | F- glnV44 e14-(McrA-) rfbD1 relA1 endA1 spoT1 | New England Biolabs |
| BL21 (DE3) RIL | thi-1 Δ(mcrC-mrr)114::IS10E. coli B F-ompT hsdS(rB-mB-) dcm+ Tetr gal λ endA Hte [argU ileY leuW Camr] | Agilent Technologies |
Table 2.
List of plasmids used in this study
| Plasmid No. | Description | Source/reference |
|---|---|---|
| Δ2pyrEF | Plasmid used for in-frame deletion in S. acidocaldarius, contains pyrEF of S. solfataricus, Ampr | (Wagner et al., 2009) |
| pSVA906 | Overexpression plasmid carrying C-terminal HA-tagged AapA, Ampr | This work |
| pSVA907 | Overexpression plasmid carrying C-terminal HA-tagged AapB, Ampr | This work |
| pSVA914 | Overexpression plasmid carrying C-terminal HA-tagged AapA and C-terminal 6xHis-tagged PibD, Ampr | This work |
| pSVA915 | Overexpression plasmid carrying C-terminal HA-tagged AapB and C-terminal 6xHis-tagged PibD, Ampr | This work |
| pSVA406 | Plasmid used for inframe deletion in S. acidocaldarius, contains pyrEF of S. solfataricus, Ampr | Michaela Wagner pBluescript |
| pSVA180 | Deletion plasmid for ΔaapF (Δsaci2318), Ampr | Δ2pyrEF, (Henche et al., 2012) |
| pSVA218 | Deletion plasmid for ΔaapA (Δsaci1174), Ampr | pSVA406, this work |
| pSVA219 | Deletion plasmid for ΔaapB (Δsaci1179), Ampr | pSVA406, this work |
| pSVA220 | Deletion plasmid for ΔaapE(Δsaci2317), Ampr | pSVA406, this work |
| pSVA222 | Deletion plasmid for ΔaapX (Δsaci2316), Ampr | pSVA406, this work |
References
- Ajon M, Fröls S, van Wolferen M, Stoecker D, Teichmann D, Driessen AJ, Grogan DW, Albers SV, Schleper C. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol Microbiol. 2011;82:807–817. doi: 10.1111/j.1365-2958.2011.07861.x. [DOI] [PubMed] [Google Scholar]
- Albers SV, Szabó Z, Driessen AJ. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers SV, Driessen AJ. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology. 2005;151:763–773. doi: 10.1099/mic.0.27699-0. [DOI] [PubMed] [Google Scholar]
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- Averhoff B. DNA transport and natural transformation in mesophilic and thermophilic bacteria. J Bioenerg Biomembr. 2004;36:25–33. doi: 10.1023/b:jobb.0000019595.66733.fa. [DOI] [PubMed] [Google Scholar]
- Beatson SA, Minamino T, Pallen MJ. Variation in bacterial flagellins: from sequence to structure. Trends Microbiology. 2006;14:151–155. doi: 10.1016/j.tim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Bellack A, Huber H, Rachel R, Wanner G, Wirth R. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon adhering to surfaces and forming cell-cell contacts. Int J Syst Evol Microbiol. 2010;61:1239–1245. doi: 10.1099/ijs.0.023663-0. [DOI] [PubMed] [Google Scholar]
- Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Rosche WA, Rietsch A, O'Toole GA. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J Bacteriol. 2004;186:3270–3273. doi: 10.1128/JB.186.10.3270-3273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML, Krause DJ, Whitaker RJ. Patterns of Gene Flow Define Species of Thermophilic Archaea. PLoS Biol. 2012;10:e1001265. doi: 10.1371/journal.pbio.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA, Francetic O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc Natl Acad Sci U S A. 2010;107:13081–13086. doi: 10.1073/pnas.1001703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. High-Force Generation Is a Conserved Property of Type IV Pilus Systems. J Bacteriol. 2009;191:4633–4638. doi: 10.1128/JB.00396-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions. Mol Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Van Den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despalins A, Marsit S, Oberto J. Absynte: a web tool to analyze the evolution of orthologous archaeal and bacterial gene clusters. Bioinformatics. 2011;27:2905–2906. doi: 10.1093/bioinformatics/btr473. [DOI] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Baumeister W, Saxton WO. Mass mapping of a protein complex with the scanning transmission electron microscope. Proc Natl Acad Sci U S A. 1982;79:4050–4054. doi: 10.1073/pnas.79.13.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Rademacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fröls S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, Boekema EJ, Driessen AJ, Schleper C, Albers SV. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol. 2008;70:938–952. doi: 10.1111/j.1365-2958.2008.06459.x. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH. Divergence of quaternary structures among bacterial flagellar filaments. Science. 2008;320:382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17:1253–1264. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Henche AL, Koerdt A, Ghosh A, Albers SV. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ Microbiol. 2012;14:779–793. doi: 10.1111/j.1462-2920.2011.02638.x. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Stark M, Nair DB, Chong JP. Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol Lett. 2011;319:44–50. doi: 10.1111/j.1574-6968.2011.02264.x. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends Microbiol. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Jenkins AT, Buckling A, McGhee M, Ffrench-Constant RH. Surface plasmon resonance shows that type IV pili are important in surface attachment by Pseudomonas aeruginosa. J R Soc Interface. 2005;2:255–259. doi: 10.1098/rsif.2005.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- Koerdt A, Gödeke J, Berger J, Thormann KM, Albers SV. Crenarchaeal biofilm formation under extreme conditions. PLoS One. 2010;5:e14104. doi: 10.1371/journal.pone.0014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak K, Neiner T, Ghosh A, Klingl A, Wirth R, Albers SV. Molecular analysis of the crenarchaeal flagellum. Mol Microbiol. 2012;83:110–124. doi: 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and Stability Comparison with the Neisseria gonorrhoeae Type IVa Pilus. J Mol Biol. 2012;418:47–64. doi: 10.1016/j.jmb.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower BH, Kennelly PJ. Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J Bacteriol. 2003;185:3436–3445. doi: 10.1128/JB.185.11.3436-3445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Cho K, Black WP, Duan XY, Lux R, Yang Z, Kaplan HB, Zusman DR, Shi W. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol Microbiol. 2005;55:206–220. doi: 10.1111/j.1365-2958.2004.04369.x. [DOI] [PubMed] [Google Scholar]
- udtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mao D, Grogan D. Genomic evidence of rapid, global-scale gene flow in a Sulfolobus species. ISME J. 2012;6:1613–1616. doi: 10.1038/ismej.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Müller DW, Meyer C, Gurster S, Kuper U, Huber H, Rachel R, Wanner G, Wirth R, Bellack A. The Iho670 fibers of Ignicoccus hospitalis: a new type of archaeal cell surface appendage. J Bacteriol. 2009;191:6465–6468. doi: 10.1128/JB.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Zolghadr B, Driessen AJ, Albers SV, Jarrell KF. Cell surface structures of archaea. J Bacteriol. 2008;190:6039–6047. doi: 10.1128/JB.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther DJ, Rachel R, Wanner G, Wirth R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. J Bacteriol. 2006;188:6915–6923. doi: 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova A, Garner EC, Galkin VE, Heuser J, Mullins RD, Egelman EH. The structure of bacterial ParM filaments. Nat Struct Mol Biol. 2007;14:921–926. doi: 10.1038/nsmb1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J, Flint S, Brooks J. Bacterial cell attachment, the beginning of a biofilm. J Ind Microb Biotechnol. 2007;34:577–588. doi: 10.1007/s10295-007-0234-4. [DOI] [PubMed] [Google Scholar]
- Petrova OE, Sauer K. Sticky situations - Key components that control bacterial surface attachment. J Bacteriol. 2012;194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschröder M, Ghosh A, Tripepi M, Albers SV. Archaeal type IV pilus-like structures-evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol. 2011;14:357–363. doi: 10.1016/j.mib.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Polka JK, Kollman JM, Agard DA, Mullins RD. The Structure and Assembly Dynamics of Plasmid Actin AlfA Imply a Novel Mechanism of DNA Segregation. J Bacteriol. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Narita A, Iwasa M, Maéda Y, Robinson RC. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem Biophys Res Commun. 2010a;391:1598–1603. doi: 10.1016/j.bbrc.2009.12.078. [DOI] [PubMed] [Google Scholar]
- Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, Goshdastider U, Maéda Y, Robinson RC, Schumacher MA. Structure and Filament Dynamics of the pSK41 Actin-like ParM Protein. J Biol Chem. 2010b;285:10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shime-Hattori A, Iida T, Arita M, Park KS, Kodama T. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol Lett. 2006;264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- Szabó Z, Albers SV, Driessen AJ. Active-site residues in the type IV prepilin peptidase homologue PibD from the archaeon Sulfolobus solfataricus. J Bacteriol. 2006;188:1437–1443. doi: 10.1128/JB.188.4.1437-1443.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschröder M. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol. 2007;189:772–778. doi: 10.1128/JB.01547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripepi M, Imam S, Pohlschröder M. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J Bacteriol. 2010;192:3093–3102. doi: 10.1128/JB.00133-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Berkner S, Ajon M, Driessen AJ, Lipps G, Albers SV. Expanding and understanding the genetic toolbox of the hyperthermophilic genus Sulfolobus. Biochem Soc Trans. 2009;37:97–101. doi: 10.1042/BST0370097. [DOI] [PubMed] [Google Scholar]
- Wagner M, van Wolferen M, Wagner A, Lassak K, Meyer BH, Reimann J, Albers SV. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front Microbiol. 2012;3:214. doi: 10.3389/fmicb.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JS, Hainfeld JF. Mass mapping with the scanning transmission electron microscope. Annu Rev Biophys Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- Wall JS, Simon MN, Lin BY, Vinogradov SN. Mass mapping of large globin complexes by scanning transmission electron microscopy. Methods Enzymol. 2008;436:487–501. doi: 10.1016/S0076-6879(08)36027-3. [DOI] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ATY, Parayno A, Hancock RE. Mucin Promotes Rapid Surface Motility in Pseudomonas aeruginosa. mBio. 2012;3 doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Yu X, Goforth C, Meyer C, Rachel R, Wirth R, Schröder GF, Egelman EH. Filaments from Ignicoccus hospitalis Show Diversity of Packing in Proteins Containing N-terminal Type IV Pilin Helices. J Mol Biol. 2012;422:274–281. doi: 10.1016/j.jmb.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolghadr B, Klingl A, Koerdt A, Driessen AJ, Rachel R, Albers SV. Appendage-mediated surface adherence of Sulfolobus solfataricus. J Bacteriol. 2010;192:104–110. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.