Abstract
Simulations of cardiac bioelectric phenomena remain a significant challenge despite continual advancements in computational machinery. Spanning large temporal and spatial ranges demands millions of nodes to accurately depict geometry, and a comparable number of timesteps to capture dynamics. This study explores a new hardware computing paradigm, the graphics processing unit (GPU), to accelerate cardiac models, and analyzes results in the context of simulating a small mammalian heart in real time. The ODEs associated with membrane ionic flow were computed on traditional CPU and compared to GPU performance, for one to four parallel processing units. The scalability of solving the PDE responsible for tissue coupling was examined on a cluster using up to 128 cores. Results indicate that the GPU implementation was between 9 and 17 times faster than the CPU implementation and scaled similarly. Solving the PDE was still 160 times slower than real time.
I. INTRODUCTION
Computational modeling of the bioelectric activity of the heart has made major contributions to our understanding of cardiac bioelectric phenomena, such as the tissue response to stimulation [1], the formation of arrhythmias [2], and their termination by electrical shocks [3]. The large span of space and time scales involved, ranging from microseconds to minutes, and from micrometers (cell) to centimeters (organ), makes simulation an inherently expensive task from the computational standpoint. So far, virtually all studies have had to find a trade-off between level of detail and usage of resources keep simulations tractable. While some choose to simplify geometry, using strands, sheets, or slabs, others opt for coarser spatial discretization or simplified cellular dynamics.
Fueled by recent advances in mesh generation [4], the availability of parallel toolkits [5] that simplify the implementation of parallel solver strategies for PDEs [6], [7] and ODEs [8], and the increasing adoption of parallel hardware, current research aims to push the limits of multiscale in silico models of the heart. Researchers will be able to flexibly decide which simplifications are made, if any, based on the availability computing resources. Nonetheless, highly detailed simulations remain prohibitive. For instance, Plank has reported [9] that a monodomain simulation of an anatomically realistic rabbit ventricular model with state-of-the-art cellular dynamics [10] lags real-time by a factor of ~ 4200 when executed on a cluster with 128 processors. For a full-blown bidomain formulation, the numbers are even more sobering, with execution times lagging roughly 65,000 times behind real-time. Further, this problem cannot circumvented simply by increasing the number of processors, since some components of the solver schemes do not scale well, so a brute-force increase in the number of cores will not necessarily lead to a meaningful reduction in execution time.
With the advent of new PetaFLOPS machines, it can be expected that some restrictions will be lifted, but a dramatic change in hardware paradigms will take place. Although it is controversially debated which paradigm will dominate the near future of high performance computing or which one will prevail in the long run, it is widely accepted that increasing the number of computational cores will play a pivotal role. In addition to classical CPU computing, new paradigms that are currently being discussed include graphical processing units (GPUs), cell processors, and field-programmable gate arrays. In this study, we explore the use of GPUs to speed up cardiac monodomain and bidomain simulations. Unlike in a previous study where significant speedups for solving the elliptic portion of the bidomain equations on a GPU were reported [11], the focus here is applying GPUs to solve the set of ODEs that describes cellular. Recent models of cellular dynamics use increasingly detailed representations of sarcolemmal and subcellular mechanisms, which increase computational complexity due to an increased number of state variables and a more pronounced property of stiffness between equations. With such models, a substantial percentage of the overall computational expense can be attributed to solving the ODEs, which makes it worthwhile to investigate potential performance benefits from using GPUs.
The objective of this study is to demonstrate the feasibility of offloading the computational burden of solving the system of ODEs to the GPU. We compare a parallel high-performance implementation of the recent Mahajan and Shiferaw model [10] with a GPU implementation which was derived from the same code. Results suggest that a GPU implementation outperforms the CPU version by a factor of at least 10 to 15.
II. METHODS
A. Governing equations
The bidomain formulation (Plonsey et al. 1988), a continuum approximation that treats cardiac tissue as a syncytium, states that currents entering the intracellular space must leave the extracellular space by crossing the cell membrane (or vice versa). The equations can be written as:
| (1) |
| (2) |
| (3) |
| (4) |
where ϕe is the extracellular potential, Vm is the transmembrane voltage, and are the intracellular and extracellular conductivity tensors, β is the membrane surface to cell volume ratio, Im is the transmembrane current density, Itr is the transmembrane stimulus current density, Ie is the current density of the extracellular stimulus, Cm is the membrane capacitance per unit area, and Iion is the density of the total current flowing through the membrane ionic channels, pumps, and exchangers, which depends in turn on Vm and a set of state variables . At the tissue boundaries, electrical isolation is assumed, with the imposition of no-flux boundary conditions. Operator splitting was applied to (2) to decouple the non-linear parabolic PDE into a linear parabolic PDE and a non-linear set of ODEs. The Galerkin finite element method was employed for spatial discretization using tetrahedral elements with linear weighting functions.
B. Reference implementations for the CPU
The LIMPET library is the ionic model backend of the Cardiac Arrhythmia Research Package (CARP) for solving the set of non-linear ODEs given by:
| (5) |
The implementation employs numerous optimizations including table lookups, temporal decoupling of different time scales and dynamic reformulation of equations [8]. When possible, ODEs are integrated using the Rush-Larsen technique; otherwise, other techniques, including forward Euler (FE) and Runge-Kutta methods, are used. The technique has been classified as a non-standard finite difference method with forward Euler (NSFD w/FE) [12]. LIMPET served as a starting point for the GPU implementation and as a reference for performance comparisons between GPU and CPU.
C. GPU implementation
The code was already optimized for performance under Message Passing Interface using PETSc, as well as for running under OpenMP. GPU code was derived from CPU code under the principle of perturbing the code as little as possible. Since the original code was written in C, and GPU code was compiled using the CUDA (copyright NVIDIA) compiler which uses C/C++, this was possible. Compiling for any of the particular hardware paradigms is controlled by preprocessor directives.
All tables and state variables were first computed on the CPU and then copied to the GPU. Each time step, Vm values for each node were copied to GPU memory, the total ionic current was computed and then transferred back to the CPU memory. The voltage was then updated based on Iion. State variables () were kept in GPU memory.
D. Performance Metrics
The computational load associated with solving the set of cellular ODEs is determined by the degrees of freedom in a tissue model, Nv, the complexity of the model in terms of number of state variables, Ns, and the stiffness of the system due to fast transients in the solution. Five different problem sizes, Nv = 0.05e6, 0.1e6, 0.5e6, 1.0e6, 5.0e6, were considered to reflect the range of vertices required to spatially discretize small mammalian hearts, ranging roughly from the mouse to rabbit scale, with an average element resolution of 250 μm or better. This range was estimated to lie between 50,000 (mouse heart at 250 μm) and 5 million (rabbit heart at 100 μm); The rabbit ventricular cell model published recently by Mahajan et al. [10] was chosen to describe cellular dynamics.
1) Numerical Setup
All benchmarks simulated electrical activity over one second using a time step of 25 μs, resulting in sufficiently large number of solver steps to average out systemic variations in performance. To benchmark ODE performance, all Nv cells were stimulated with a basic cycle length of 500 ms. Parabolic performance was measured by applying a transmembrane stimulus to the center of a tissue wedge of 0.25 cm transmural width, discretized at 100 μm, where the length in circumferential and apicobasal direction was chosen to arrive at the desired Nv. Temporal discretization of the parabolic PDE employed either the explicit FE method or the implicit Crank-Nicolson (CN) method using the same time step as for the ODEs.
2) Comparing performance between a single CPU and a single GPU
From a computational standpoint, the performance ratio between two different computing paradigms, in this case CPU versus GPU, and the scaling efficiency when increasing the number of computational cores, Np, is of primary interest. Benchmarks were run for all problem sizes on a single CPU and a single GPU. The minimum, average and maximum time required for advancing the solution by a single time step, Tdt, were recored as well as the time elapsed for simulating 1 second of activity, T1s.
3) Comparing performance between multiple CPUs and multiple GPUs
Although the solution of the system of ODEs is considered to be an embarrassingly parallel computing problem that scales linearly with the number of processors used, when using the GPU in a hybrid scenario, i.e. parts of the computation such as the parabolic PDE are solved on the host (CPU) and only the ODEs are solved on the device (GPU), additional communication arises between host and device, which may deteriorate scalability. To investigate whether saturation of the available bandwidth between host and device limits the scalability of hybrid computing schemes, the previous tests were repeated, varying numbers of CPUs and GPUs between 1 and 4, again for all problem sizes.
4) Estimations of performance of large-scale hybrid parallel simulations
Based on performance metrics determined for previous benchmarks in a desktop environment, additional tests were run in a cluster environment to obtain an approximate measure of the absolute performance one could expect when building a hybrid cluster where each blade is equipped with 4 GPUs as coprocessors. Such a cluster was not available for running these tests, but could easily be built with off-the-shelf components.
To measure strong scaling of the ODE solver and parabolic PDE, benchmarks were conducted on a Linux cluster, The problem size was kept fixed at Nv =1e6 , and the number of processors Np was increased from 4 to 128 where Np was incremented by doubling. Execution times spent on solving the ODEs were recorded as well as time dedicated to solving the parabolic PDE for both FE and CN method. Theoretical performance of a hybrid CPU-GPU cluster was estimated then by combining the measured performance data with the GPU performance measured in the desktop environment for the respective local problem sizes. That is, to estimate the performance for Np processors, the local problem size was determined by Nl = Nv/Np and the corresponding performance data where taken from the desktop benchmark.
E. Hardware
Desktop benchmarks were run on a single quad-core computer (AMD Phenom(tm) 9950 Quad-Core Processor, clocked at 2.6 GHz) and equipped with 4 GPUs (GTX 280, 1GB memory each, clocked at 1.4 GHz). Benchmarks for Np > 4 were conducted on a Linux cluster equipped with an Infiniband low-latency network interconnect. Each compute node was equipped with four cores (AMD Opteron(tm) 2216 Processor).
III. RESULTS
A. Comparing performance between a single CPU and a single GPU
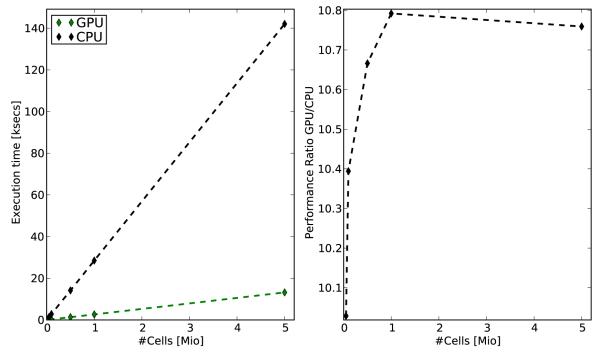
For all problem sizes, the GPU implementation executed between ~ 10.0 and ~ 10.8 times faster than the CPU analog. Although minor variations in performance were observed between runs, the speedup factor was robust for all problem sizes under study. As expected, execution time T1s increased linearly with Nv. Fig. 1 shows single core execution times T1s for and performance ratios between GPU and CPU as a function of problem size Nv.
Fig. 1.
Left panel: Execution times for solving the Mahajan model over 1 second of activity was measured for varying Nv for both the CPU and the GPU where a single computational core was employed for both paradigms. Right panel: Performance ratio between GPU and CPU.
B. Comparing performance between multiple CPUs and multiple GPUs
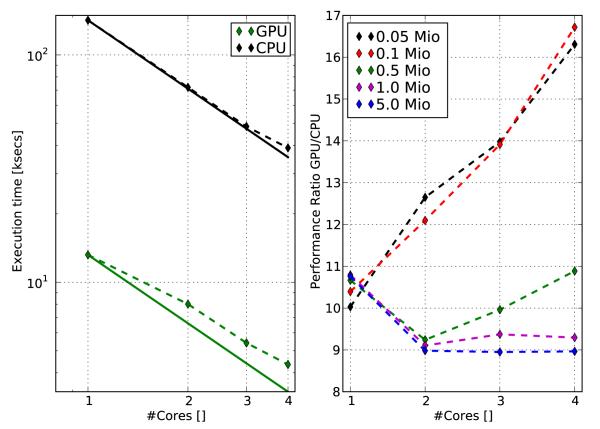
To compare scalability between CPU and GPU implementations, the same benchmarks were conducted as before, but the number of computational cores Np was varied from 1 to 4. Results of the scalability benchmark are summarized in Fig. 2. Scalability was slightly better for the CPU code, but, again, overall performance was substantially better for the GPU code. On the CPU, a speedup of 3.65 was achieved for the largest test case with Nv = 5 million when going from 1 to 4 computational cores whereas this factor was slightly lower for the GPU (3.1). The GPU implementation outperformed the CPU implementation by at least a factor of 9, but for smaller problems with Nv up to 100,000 a speedup factor of ~ 17 were achieved.
Fig. 2.
Left panel: Scaling results for CPU and GPU implementations for varying numbers of computational cores and Nv = 5e6 when executing the T1s benchmark. Solid lines represent linear scaling. Right panel: Performance ratio between GPU and CPU as a function of computational cores employed for varying Nv.
C. Estimations of performance of large-scale hybrid parallel simulations
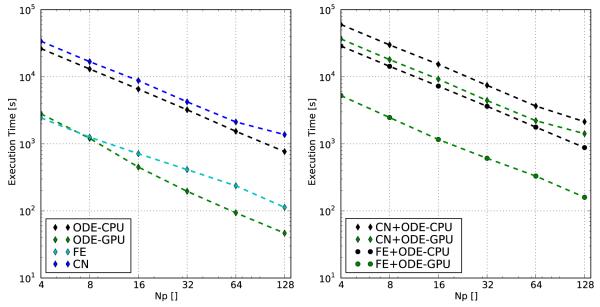
Strong scalability was tested for Nv = 1 million vertices with 4–128 processors. The hypothetical hybrid performance was estimated by dividing the execution time dedicated to solving the ODEs by the performance gain factor measured for a particular local problem size. Results are summarized in Fig. 3. Since the time step was common for all subproblems, the explicit FE scheme outperformed the implicit CN scheme roughly by a factor of 10. Time spent on solving the set of ODEs on the CPU was comparable to that required for solving the parabolic PDE with the CN approach. The predicted GPU performance on a hybrid cluster is within the range of execution times required for the solution of the parabolic PDE when using the FE scheme. When running on a cluster with equipped with 128 GPUs the performance for the ODE solver step is only 46 times slower than realtime which is indeed sufficiently close to real-time for most applications. The total execution time with Np =128 for both components of the computing scheme, the parabolic PDE and the set of ODEs, lags by a factor of 160 and 1420 behind real-time for the FE and the CN scheme, respectively.
Fig. 3.
Left panel: Strong scaling benchmark with Nv = 1e6 vertices for the individual sub-problems, the parabolic PDE solved with FE and CN scheme and the ODE solver running on CPU and GPU. Right panel: Strong scaling benchmark for the overall monodomain problem for different combinations of parabolic PDE solver scheme and ODE hardware implementation.
IV. DISCUSSION
In this study, a high-performance implementation of a recent rabbit ventricular model of cellular dynamics, designed to perform well on CPUs, was ported to the GPU following the principle of minimal perturbation. Monodomain simulations were conducted and execution times spent on the two main computational tasks, the solution of the parabolic PDE and the set of ODEs, were measured. Two different temporal discretization schemes, explicit FE and implicit CN, were used to solve the parabolic PDE with varying numbers of CPUs. The solution of the set of ODEs was computed either on a CPU or on a GPU where the number of computational cores involved matched those used for the solution of the parabolic PDE. The performance of CPU and GPU implementation was benchmarked for single and multiple core scenarios. Benchmark results suggest that implementations for the GPU offer significant performance gains over traditional CPU codes. The scalability between the implementations was comparable with a stable performance advantage for the GPU in the range between 9.3 and 17. Benchmarks were also executed on a cluster with up to 128 cores. Hypothetical performance for the same cluster that is equipped with additional GPU blades to arrive at a 1:1 CPU:GPU ratio were obtained by extrapolating GPU performance data obtained at a desktop computer. These estimations predict that the solution of the set of ODEs in isolation can be solved with a near-realtime performance at a rate roughly 50 times slower than real-time. Depending on the solver strategy applied to solve the parabolic PDE, execution times lagged a factor of 160 and 1420 behind realtime for FE and CN, respectively.
V. CONCLUSION AND FUTURE WORK
Results suggest that the GPU implementation was 9 to 17 times faster than the CPU implementation and scaled similarly. Using clusters equipped with GPU co-processors indeed allowed to scale down execution times for the ODE portion to a near-realtime performance. Due to the embarrassingly parallel nature of the ODE problem, further substantial gains can be achieved by using thousands of processors.
Acknowledgments
This research is supported by a grant of the Austrian Science Fund grant (SFB F3210-N18) to GP, a grant from the Natural Sciences and Engineering Research Council of Canada to JL, a grant from the Mathematics of Information technology and Complex Systems to EV and JL, and funding from the Alberta Ingenuity Fund for PB
Contributor Information
Edward J. Vigmond, Department of Electrical and Computer Engineering, University of Calgary, Calgary, AB T2N 1N4, Canada, vigmond@ucalgary.ca
Patrick M. Boyle, Department of Electrical and Computer Engineering, University of Calgary, Calgary, AB T2N 1N4, Canada, pmjboyle@ucalgary.ca
L. Joshua Leon, Faculty of Engineering, Dalhousie University, Halifax, B3H 4R2, Canada, Joshua.Leon@dal.ca.
Gernot Plank, Institute of Physiology, Medical University of Graz, Graz, A-8010, Austria and the Computational Biology Group, University of Oxford, Oxford, OX1 3QG, UK. gernot.plank@medunigraz.at or gernot.plank@comlab.ox.ac.uk.
REFERENCES
- [1].Sepulveda NG, Roth BJ, Wikswo JP., Jr Current injection into a two-dimensional anistropic bidomain. Biophysical Journal. 1989;55:987–999. doi: 10.1016/S0006-3495(89)82897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Beaumont J, Davidenko N, Davidenko JM, Jalife J. Spiral waves in two-dimensional models of ventricular muscle: formation of a stationary core. Biophys J. 1998;75(1):1–14. doi: 10.1016/S0006-3495(98)77490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ashihara T, Constantino J, Trayanova NA. Tunnel propagation of postshock activations as a hypothesis for fibrillation induction and isoelectric window. Circ Res. 2008;102(6):737–45. doi: 10.1161/CIRCRESAHA.107.168112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prassl A, Kickinger F, Ahammer H, Grau V, Schneider J, Hofer E, Vigmond E, Trayanova N, Plank G. Automatically Generated, Anatomically Accurate Meshes for Cardiac Electrophysiology Problems. IEEE Trans Biomed Eng. 2009 doi: 10.1109/TBME.2009.2014243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Balay S, Buschelman K, Gropp WD, Kaushik D, Knepley M, McInnes LC, Smith BF, Zhang H. PETSc users manual. Argonne National Laboratory, Tech. Rep. ANL-95/11 - Revision 2.1.5. 2002 [Google Scholar]
- [6].Weber dos Santos R, Plank G, Bauer S, Vigmond EJ. Parallel multigrid preconditioner for the cardiac bidomain model. IEEE Trans Biomed Eng. 2004;51(11):1960–8. doi: 10.1109/TBME.2004.834275. [DOI] [PubMed] [Google Scholar]
- [7].Plank G, Liebmann M, Weber dos Santos R, Vigmond EJ, Haase G. Algebraic multigrid preconditioner for the cardiac bidomain model. IEEE Trans Biomed Eng. 2007;54(4):585–96. doi: 10.1109/TBME.2006.889181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Plank G, Zhou L, Greenstein JL, Cortassa S, Winslow RL, O’Rourke B, Trayanova NA. From mitochondrial ion channels to arrhythmias in the heart: computational techniques to bridge the spatio-temporal scales. Philos Trans A Math Phys Eng Sci. 2008;366(1879):3381–409. doi: 10.1098/rsta.2008.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Plank G, Burton RAB, Hales P, et al. Generation of histoanatomically representative models of the individual heart: tools and application. Philos Trans A Math Phys Eng Sci. 2009;367:2257–2292. doi: 10.1098/rsta.2009.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mahajan A, Shiferaw Y, Sato D, Baher A, Olcese R, Xie LH, Yang MJ, Chen PS, Restrepo JG, Karma A, Garfinkel A, Qu Z, Weiss JN. A rabbit ventricular action potential model replicating cardiac dynamics at rapid heart rates. Biophys J. 2008;94(2):392–410. doi: 10.1529/biophysj.106.98160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liebmann M, Plank G, Prassl A, Haase G. GPU accelerated algebraic multigrid preconditioner for the virutal heart model; nVision Conference; San Jose, USA. 25-27 Aug. 2008. [Google Scholar]
- [12].Maclachlan MC, Sundnes J, Spiteri RJ. A comparison of non-standard solvers for ODEs describing cellular reactions in the heart. Comput Methods Biomech Biomed Engin. 2007;10(5):317–26. doi: 10.1080/10255840701259301. [DOI] [PubMed] [Google Scholar]