Abstract
Germline TP53 mutations predispose to multiple cancers defining Li-Fraumeni/Li-Fraumeni-like syndrome (LFS/LFL), a disease with large individual disparities in cancer profiles and age of onset. G-quadruplexes (G4s) are secondary structural motifs occurring in guanine tracks, with regulatory effects on DNA and RNA. We analyzed 85 polymorphisms within or near five predicted G4s in TP53 in search of modifiers of penetrance of LFS/LFL in Brazilian cancer families with (n = 35) or without (n = 110) TP53 mutations. Statistical analyses stratified on family structure showed that cancer tended to occur ~15 years later in mutation carriers who also carried the variant alleles of two polymorphisms within predicted G4-forming regions, rs17878362 (TP53 PIN3, 16 bp duplication in intron 3; P = 0.082) and rs17880560 (6 bp duplication in 3′ flanking region; P = 0.067). Haplotype analysis showed that this inverse association was driven by the polymorphic status of the remaining wild-type (WT) haplotype in mutation carriers: in carriers with a WT haplotype containing at least one variant allele of rs17878362 or rs17880560, cancer occurred ~15 years later than in carriers with other WT haplotypes (P = 0.019). No effect on age of cancer onset was observed in subjects without a TP53 mutation. The G4 in intron 3 has been shown to regulate alternative p53 messenger RNA splicing, whereas the biological roles of predicted G4s in the 3′ flanking region remain to be elucidated. In conclusion, this study demonstrates that G4 polymorphisms in haplotypes of the WT TP53 allele have an impact on LFS/LFL penetrance in germline TP53 mutation carriers.
Summary:
G-quadruplexes (G4) are regulatory secondary structures in guanine tracts in DNA or RNA. Polymorphisms in two G4-forming regions of TP53 modulate age at cancer onset in susceptible germline TP53 mutation carriers, suggesting that these G4 regulate p53 suppressor functions.
Introduction
Germline mutations in the TP53 tumor suppressor gene (17p13.1, OMIM #191170) predispose to a range of early-onset cancers that define the Li-Fraumeni syndrome (LFS) (1,2). The LFS tumor pattern is dominated by childhood adrenal cortical carcinoma, choroid plexus carcinoma, medulloblastoma and rhabdomyosarcoma, followed by soft tissue sarcoma, osteosarcoma, premenopausal breast cancer and brain tumors in adolescents and young adults. Several definitions of ‘Li-Fraumeni-like’ (LFL) syndrome have been proposed in families showing only partial LFS traits (3–5). Currently, the only recurrent germline alterations found in patients from families with LFS/LFL traits are TP53 mutations. Depending upon the clinical definition, TP53 mutations are identified in 20–70% of the cases (6–9). In current practice, probands with suspected LFS/LFL are referred for TP53 mutation testing based on a set of individual and familial criteria known as the ‘modified Chompret criteria’ (10). Mutations are detected in ~21–29% of probands matching these criteria (9,11). In TP53 mutation carriers, the penetrance of the disease is ~50% by age 30–35 years and reaches 90% over lifetime. In addition, TP53 mutation carriers show a tendency to develop a wide range of cancers at an earlier age than in the general population, although there are large individual and familial variations in the age at diagnosis. Thus, it is extremely difficult to develop a protocol for prediction and surveillance of the disease. The current practice is to enroll subjects who tested positive for TP53 mutations in complex surveillance programs (12) (NCCN guidelines version 2.2013).
The variable individual cancer patterns suggest that other genetic or epigenetic traits may act as modifiers. Studies by Bond et al. (13) observed that a single-nucleotide polymorphism (SNP) in MDM2, encoding a protein that regulates p53 protein stability, was associated with a reduction of 9 years in the age at diagnosis of first cancer. This effect was detected in several independent cohorts of LFS/LFL families (14–17). This SNP (rs2279744, SNP309) has been shown to alter the sequence-specific DNA binding of the transcription factor Sp1 to the MDM2 promoter, thus resulting in differences in levels of Mdm2 protein expression detected between individuals (13). There is evidence that the effect of rs2279744 might be modulated by another SNP in the MDM2 promoter (rs117039649, SNP285) (18). Borderline effects on age at diagnosis in TP53 mutation carriers have also been reported for a common non-synonymous SNP in exon 4 of TP53, R72P (rs1042522, TP53 PEX4) (15). In a previous analysis on a cohort of TP53 mutation carriers from Brazil, we have shown that a 16bp duplication polymorphism in intron 3 (rs17878362, TP53 PIN3) was associated with a large difference in the age of first cancer diagnosis, with carriers of two alleles without the duplication (A1 allele) developing their first cancer on average 20 years earlier than heterozygote carriers (A1/A2) (15). However, this Brazilian cohort included a large proportion of subjects from apparently unrelated families who carried the same germline TP53 mutation, R337H, which is common in South-Eastern Brazil due to a widespread founder effect detected in 0.3% of the general population (19). So far, the effect of rs17878362 has not been replicated in an independent LFS/LFL cohort. Thus, it is possible that the effect of the A1 allele of rs17878362 may be due, at least in part, to specific effects on the penetrance of R337H or to other genetic factors in this particular Brazilian background.
G-quadruplexes (G4s) consist of four-stranded structures occurring in guanine-rich sequences. Potential G4-forming structures are widespread in the genome and have been shown to have important regulatory effects on gene transcription (20), genomic stability (21–23) and DNA replication (24), messenger RNA (mRNA) splicing (25) and mRNA stability (25). Experimentally, drugs that modulate the stability of G4s exert significant effects on the expression (26) and splicing (27) of specific sets of genes. Two G4s have been experimentally demonstrated in the pre-mRNA of p53. A complex G4 structure is found in intron 3 in which the rs17878362 16 bp duplication is located, which regulates the splicing of p53’s pre-mRNA into either the fully spliced form (FSp53) generating the canonical p53 tumor suppressor protein, or into p53I2, a variant mRNA that retains intron 2 (28). This variant mRNA encodes a p53 protein isoform lacking the 39 N-terminal residues (29) that exerts regulatory effects on p53 suppressor activity (28,30–32). Another G4 has been identified downstream of the cleavage/polyadenylation site of the p53 pre-mRNA. This G4 contributes to p53 pre-mRNA 3′ end processing in response to DNA damage (33).
In this study, we have hypothesized that polymorphisms affecting G4 structures may exert modifier effects on the age at cancer onset in LFS/LFL subjects. We have identified, using bioinformatics approach, candidate G4 domains throughout the entire TP53 locus and analyzed the association between SNPs located within or in the close vicinity to these putative and demonstrated G4s and age at first cancer diagnosis in a series of 402 subjects from Brazilian families with LFS/LFL traits.
Materials and methods
LFS/LFL families
LFS/LFL Brazilian subjects were recruited from families attending the Cancer Risk Evaluation clinics in the Department of Oncogenetics of A.C. Camargo Cancer Center (São Paulo, Brazil) and Hospital de Clinicas, Universidade Federal do Rio Grande do Sul (Porto Alegre) (ethics statement number 0568/07). A total of 402 subjects from 145 families were included in the study (Supplementary Table 1, available at Carcinogenesis Online). Families matched at least one LFS/LFL criteria (classic LFS, Birch, Chompret, Eeles criteria), and their familial history was documented over several generations using the Progeny software (Progeny software, Wolfville, Nova Scotia, Canada). Of these families, 15 (10.3%) matched classic LFS criteria, whereas 66 (45.5%) matched Birch or Chompret criteria. Other families matched more relaxed criteria known as the Eeles criteria (8). The coding sequence (exons 2–11 including splice junctions) of TP53 was sequenced in DNAs extracted from blood samples (see below). Mutations were found in 35 families (24.1%), including 19 families with the ‘Brazilian founder’ R337H mutation and 16 families with previously reported DNA-binding domain mutations (34) (http://p53.iarc.fr/TP53GermlineMutations.aspx). The institutional ethics committees of participating institutions approved the study and all patients provided informed consent.
Controls
Four groups of control subjects were selected. Group 1 consisted of 487 cancer-free subjects from the general population of Southeast Brazil (age 40–69 years) (34). Groups 2–4 consisted of individuals included as reference samples in the HapMap project. Group 2 (Caucasians) included 30 trios (two parents and a child) of Northern or Western European ancestry. Group 3 (Asians) comprised 90 East Asian subjects (including two groups of 45 subjects from the Tokyo and Beijing areas, respectively). Group 4 (Africans) consisted of 30 trios (two parents, one child) from the Yoruba people in Nigeria (35).
Prediction of G4 motifs and selection of polymorphisms
The TP53 sequence NC_000017.10 (ENSG00000141510) containing the entire TP53 gene sequence plus 500bp upstream and downstream of the first and last exons, respectively, was used for in silico prediction of G-rich regions likely to form G4 structures using the web-based prediction softwares Quadfinder (http://miracle.igib.res.in/quadfinder/) and QGRS Mapper (http://bioinformatics.ramapo.edu/QGRS/index.php), each set at its own default parameters (Quadfinder: G stretch = 3–5; loop size = 1–7; maximum length = 45; QGRS mapper: G stretch = minimum 2; loop size = 0–30; maximum length = 30) (36,37). A total of five G4 regions were identified by the two softwares. Polymorphisms located in these regions or within 100bp upstream or downstream of these regions were selected in dbSNP (http://www.ncbi.nlm.nih.gov/snp/). To this set, were added the well-defined TP53 SNPs rs1642785 (G/C, intron 2) and rs1042522 (G/C, exon 4). Overall, a total of 87 SNPs were selected (Supplementary Table 2, available at Carcinogenesis Online and Figure 1).
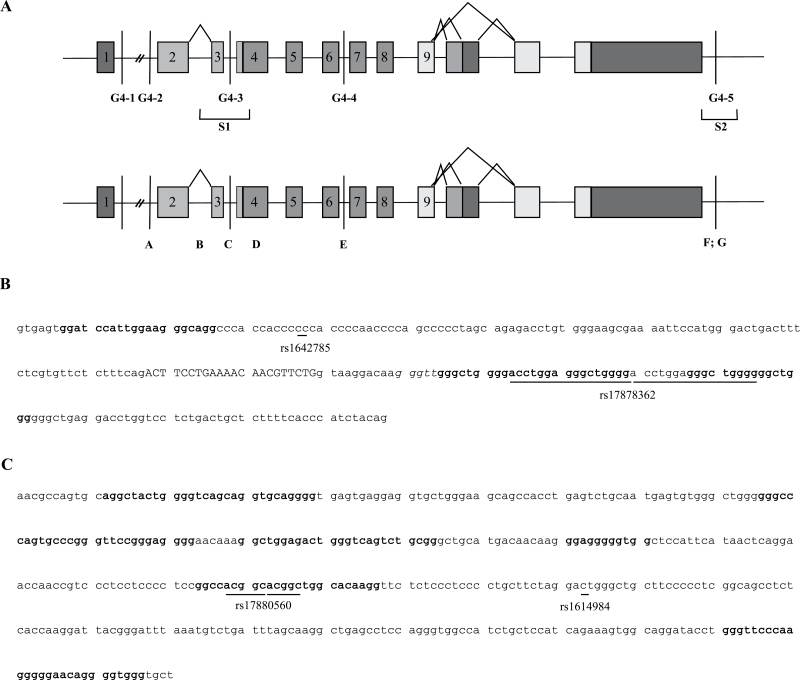
Fig. 1.
Predicted localization of G4s and associated polymorphisms in TP53. (A) Schematic representation of G4 localization and polymorphisms at the TP53 locus. G1 to G5: position of G4 motifs predicted using the QGRS mapper software set at its default parameters (see Materials and methods). A to G: position of polymorphisms within or near these G4. S1: sequence 1 encompassing G3; S2: sequence 2 encompassing G5. (B) Sequence of S1 region, from intron 2 to intron 3 (small letters), encompassing exon 3 (capital letters). Two G-tract domains forming putative G4 are shown in bold. The position of polymorphisms B in intron 2 (G/C, rs1642785) and C in intron 3 (16bp duplication, rs17878362) are underlined. (C) Sequence of S2 region encompassing the 3′ flanking region of TP53 after cleavage site. Bold letters: G-tracts forming putative G4. The position of polymorphisms F (6bp duplication, rs17880560) and G (C/T, rs1614984) are underlined.
Analysis of mutations and polymorphisms
DNA was extracted from peripheral white blood cells obtained by venipuncture with the Qiagen DNA Extraction kit according to the manufacturer’s instructions (QIAamp DNA blood Maxi kit; Qiagen, Hilden, Germany). Purified DNA from HapMap subjects were obtained from the Coriell Institute Biorepository as described earlier (35). The TP53 coding sequence (exon 2–11 including flanking intronic regions containing splice sites) was analyzed according to the protocols of the International Agency for Research on Cancer (http://p53.iarc.fr/Download/TP53_DirectSequencing_IARC.pdf). The genotyping of TP53 polymorphisms that fell outside these regions was performed by direct sequencing using primers and PCR conditions described in Supplementary Table 2, available at Carcinogenesis Online. Genotyping for MDM2 rs2279744 was performed using a 5′ exonuclease SNP genotyping assay (Proligo, St Louis, MO) (probes FAM 5′-cccgcgccgcagc-3′ and Hex 5′-cccgcgccgaagc-3′; primers 5′-ttcagggtaaaggtcacggg-3′ and 5′-tcaacctgcccactgaacc-3′) or by restriction fragment length polymorphism (RFLP) after PCR with primers and condition as described in Supplementary Table 2, available at Carcinogenesis Online. RFLP was performed using 5 μl of PCR product, digested with 1 unit of MspAI restriction enzyme (New England Biolabs, Ipswich, UK) during 4h at 37°C. The results were analyzed on 3% agarose electrophoresis gels. The T allele of rs2279744 generated two RFLP fragments (53 and 158bp), whereas the G allele generated three fragments (46, 53 and 112bp). Comparison of the results obtained by the 5′ exonuclease and RFLP assays showed complete concordance.
Haplotype reconstruction
The haplotypes carried by Brazilian subjects were reconstructed using pooled sequence variant data of individuals from the LFS/LFL families and the Brazilian control group (Group 1). Rare variants (minor allele frequency < 0.04) were excluded from the analysis. Haplotypes were constructed based on five SNPs (rs1642785, rs17878362, rs1042522, rs17880560 and rs1614984) and the TP53 germline mutation status, using R software haplo.stats package (http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm).
Statistical analysis
Hardy–Weinberg equilibrium was assessed for each study group using a web-based calculator (http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls). Statistical analyses were performed using R software (weights package; http://cran.r-project.org/web/packages/weights/weights.pdf) and running a hierarchical model that takes into account family size to compensate for family bias caused by multiple subjects belonging to the same large pedigree. Briefly, analyses were adjusted for the number of subjects in the family who were sequenced for TP53 mutation and for polymorphisms, thus taking into account possible bias due to specific characteristics of large families in the dataset. Comparisons between average ages at first cancer diagnostic were performed by t-test weighted by family size, or by taking into account family sizes either for the whole cohort (‘All’) or for a particular subgroup analyzed (e.g. only families with defined wild-type (WT) or mutant TP53 status; ‘Group’). Unadjusted Kaplan–Meier disease-free probability estimates were calculated using tools available at http://vassarstats.net/. A ‘disease’ event was assigned as the age at first cancer diagnosis (subsequent diagnoses in the same subjects were not taken into account). For each year of age, subjects with no diagnosis were censored at their date of last follow-up.
Results
Distribution of G4s and polymorphisms in TP53
The presence of at least three consecutive guanines along a DNA or RNA strand is (as a first approximation) the classical minimal requirement for intramolecular G4 formation. Longer DNA/RNA sequences containing multiples of four G-tracts can, in principle, accommodate higher order structures defined by multiple G4 blocks, with wide topological and structural variations. The human genome has been predicted to contain up to 376 000 G4s (38). Using in silico prediction programs, we identified five regions predicted to contain G4 structures in a region of 20.2 kb encompassing the entire TP53 sequence (Figure 1A). The sequences and positions of these predicted G4s are given in Figure 1B and C and Supplementary Figure 1, available at Carcinogenesis Online. They are located in the proximal and distal part of intron 1, in intron 3, intron 6 and the 3′ flanking region, respectively (Figure 1B and C). Of these predicted G4s, the one located in intron 3 (28) and another one (among several predicted domains) in the 3′ flank (33), have been previously identified by structural and molecular techniques. A total of 85 polymorphisms reported in dbSNP were located within or near the five predicted G4s and their distributions were analyzed in LFS/LFL subjects, Brazilian controls (Group 1) and in the HapMap series (Groups 2–4). Only 11 of these polymorphisms showed allelic variation in these series. Four had a minor allele frequency <4% including rs78378222 that has been associated with basal cell carcinoma (prostate cancer, glioma and colorectal adenoma) in an Iceland population (39). The allele distributions of other polymorphisms (polymorphisms A, B, C, E, F and G) and rs1042522 (TP53 PEX4, codon R72P) in LFS/LFL families and in the four control groups are given in Supplementary Table 3, available at Carcinogenesis Online. The distribution of these polymorphisms was in Hardy–Weinberg equilibrium except for SNPs D and E in Group 2 (HapMap Caucasian, P = 0.04 for both SNPs), SNP G in Group 1 (P < 0.01) and SNPs A and D in LFS/LFL group (P < 0.01 for both SNPs). When compared with Group 1, the deviation from Hardy–Weinberg equilibrium for SNP D (rs1042522) in LFS/LFL appeared to be due to low numbers of carriers of the C/C (72P/72P) allele combination, whereas numbers of G/G homozygotes and G/C heterozygotes were comparable in both groups. The distribution of polymorphisms was different among the three HapMap series (P < 0.01, t-test). The Brazilian LFS/LFL and the controls of Group 1 (Brazilian population) had a SNP profile similar to HapMap Caucasians (P = 0.47, Mann–Whitney t-test).
Association between polymorphisms in and/or near G4s and age at first diagnosis in LFS/LFL
The summary characteristics of subjects included in the LFS/LFL group are given in Supplementary Table 1, available at Carcinogenesis Online. Among the 145 families, 35 had a germline TP53 mutation (MUT group), 19 of which were carriers of the ‘Brazilian founder’ R337H mutation. The main types of cancer diagnosed in carriers of R337H and in carriers of other mutations, and their mean age at diagnosis, are summarized in Supplementary Table 4, available at Carcinogenesis Online. Overall, R337H carriers developed a range of cancer types that is typical of the LFS spectrum; however, with a non-significant tendency for diagnosis at a later age than carriers of other TP53 mutations. Within families with germline TP53 mutations, subjects who were not found to carry the familial mutation were identified as the WT1 group. Among the 110 families without mutations, 157 cases of cancer were diagnosed among 122 subjects. Subjects from these families without a TP53 mutation, either with or without cancer, were identified as the WT2 group. Types of cancer and ages at diagnosis in this group were compatible with LFS/LFL definitions (Supplementary Table 4, available at Carcinogenesis Online).
We next analyzed the mean age of first cancer diagnosis (± standard deviation) in relation to the carriage of the TP53 SNPs A-G and of rs2279744 in the following subgroups: the MUT group (including subjects with any TP53 germline mutation) and the WT2 group using an adjusted model weighted for family size to take into account a possible familial bias. P values (t-test) were adjusted on family size either for the whole LFS/LFL cohort (‘All’) or for the subgroup of patients considered (‘Group’). Effects on age at diagnosis were observed only with polymorphism C (rs17878362, 16 bp insertion/duplication in intron 3) and polymorphism F (rs17880560, also called rs79948390, rs72526905 or rs66470553, 6 bp insertion/duplication in 3′ flanking region), (Table I; data for other SNPs in Supplementary Table 5, available at Carcinogenesis Online). For each of these two polymorphisms, alleles were identified as A1 (non-duplicated) and A2 (duplicated), defining three genotypes, A1/A1, A1/A2 and A2/A2 at each polymorphism. No significant difference was found for the age at first cancer diagnosis within the WT2 group in relation to the status of these polymorphisms (the age at first diagnosis ranged between 33 ± 19 years in subjects with the rs17880560 A2/A2 allele status and 40 ± 16 in subjects with the rs17880560 A1/A1 allele status). In contrast, in the MUT group, there was a tendency for a difference of over 12.5 years between subjects with either A1/A1 or A1/A2 genotypes, the A1/A2 mutation carriers having their first cancer diagnosis later than the A1/A1 mutation carriers. The difference in age at diagnosis in relation to the genotype was significant in a model adjusted for family size in the whole LFS/LFL cohort (‘All’; P = 0.044 for rs17878362, P = 0.02 for rs17880560) but was at best borderline significance in a model adjusted for the size of families in only the MUT group (‘Group’; P = 0.081 for rs17878362, P = 0.068 for rs17880560). Of note, among subjects carrying a mutation, only one was an A2 homozygote carrier of rs17878362 (with a diagnosis of cancer at age 9), whereas one other subject was an A2 homozygote carrier of rs17880560 (with a diagnosis of cancer at age 14). Thus, it was not possible to evaluate separately the association between the homozygote A2 status of these polymorphisms and the age at first cancer onset. Mean ages at diagnosis in relation to the rs1042522 status, given as comparison in Table I, did not show any significant difference in either WT2 or MUT groups in any of the family weighted models. Results for the MDM2 rs2279744 show a non-significant tendency for later age at diagnosis in carriers of G/G as compared with T/T genotype, but no effect in T/G heterozygotes (Supplementary Table 6, available at Carcinogenesis Online).
Table I.
Mean age at first cancer diagnosis depending on TP53 polymorphism genotypes and TP53 mutation family
| WT2 | MUT | |||||||
|---|---|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | |||
| rs17878362 | A1/A1 | 90 | 37.91 | 17.06 | 40 | 30.65 | 19.84 | |
| A1/A2 | 29 | 39.14 | 15.22 | 11 | 43.36 | 19.00 | ||
| A2/A2 | 2 | 35.50 | 12.02 | 1 | 9.00 | — | ||
| P value | Group | 0.959 | 0.081 | |||||
| All | 0.747 | 0.044 | ||||||
| Without | 0.918 | 0.088 | ||||||
| rs17880560 | A1/A1 | 59 | 39.56 | 16.43 | 40 | 32.58 | 21.36 | |
| A1/A2 | 43 | 38.19 | 16.35 | 8 | 45.88 | 11.68 | ||
| A2/A2 | 12 | 32.67 | 18.60 | 1 | 14.00 | — | ||
| P value | Group | 0.410 | 0.068 | |||||
| All | 0.442 | 0.020 | ||||||
| Without | 0.426 | 0.152 | ||||||
| rs1042522 | G/Ga | 68 | 37.96 | 18.09 | 35 | 33.29 | 21.11 | |
| G/Ca | 46 | 37.76 | 14.87 | 16 | 35.63 | 19.89 | ||
| C/Ca | 7 | 42.86 | 9.63 | 2 | 19.50 | 14.85 | ||
| P value | Group | 0.569 | 0.440 | |||||
| All | 0.609 | 0.545 | ||||||
| Without | 0.742 | 0.582 | ||||||
WT2: WT subjects in WT families;
MUT: mutant subjects in mutant families;
Group: P value calculated with weight in the analyzed group. In bold, significative P value;
All: P value calculated with weight in the all cohort. In bold, significative P value;
Without: P value without weight. In bold, significative P value.
aG corresponds to the arginine and C to the proline variants.
We repeated these analyses after separating the MUT group in two subgroups, one with R337H carriers and the second with carriers of other germline TP53 mutations (Supplementary Table 7, available at Carcinogenesis Online). Compatible with Supplementary Table 2, available at Carcinogenesis Online, patients with R337H had their first diagnosis on average 10–12 years later than patients with other TP53 mutations. Nevertheless, an effect on age of cancer onset was seen in both mutation groups. In R337H mutation carriers, the difference in age at diagnosis was, on average, 15 years between rs17878362 A1/A1 and A1/A2 carriers, and 11 years between rs17880560 A1/A1 and A1/A2 carriers. In carriers of other mutations, these differences were of around 10 and 19 years, respectively. However, due to the small numbers in each of the groups and lack of power, these differences were not statistically significant.
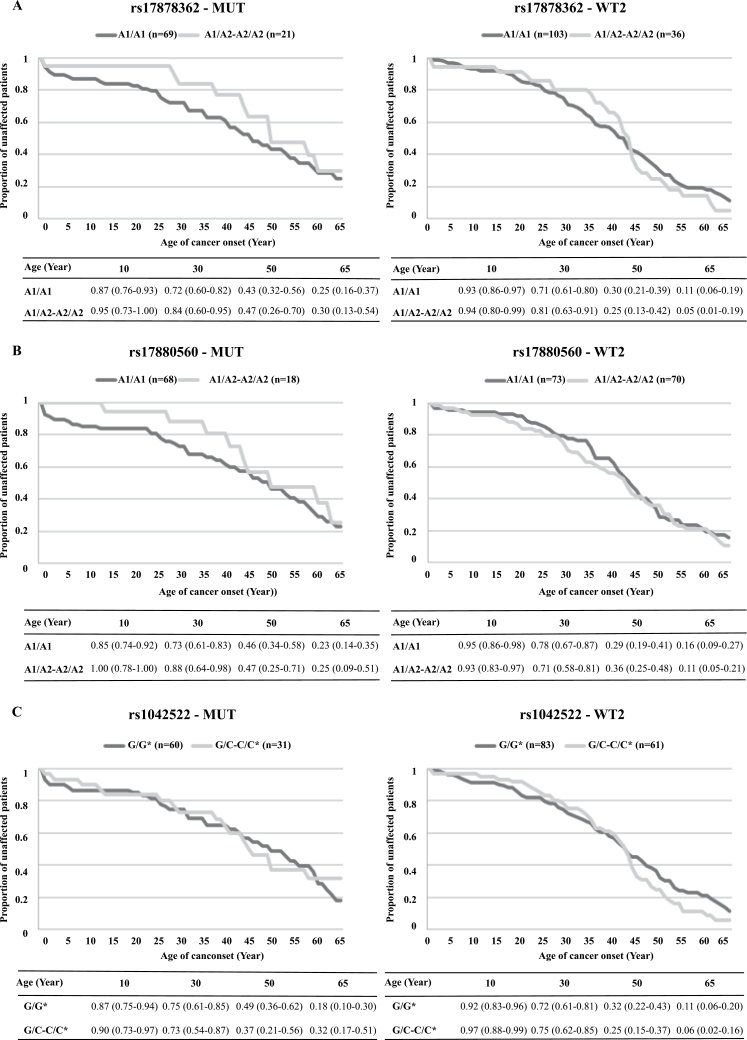
The effects of rs17878362, rs17880560 and rs1042522 genotypes on Kaplan–Meier disease-free probability estimates are shown in Figure 2. No effect of any of the SNPs was detected on accrual of cancer in the WT2 group. In contrast, in the MUT group, the presence of one A2 allele of rs17878362 or of one A2 allele of rs17880560 was associated with a reduced childhood and adolescent cancer risk. Taking as the reference the estimated risk of cancer at age 25 years in A1/A2 carriers, the relative risk for cancer before or at 25 years is 4.0 [95% confidence interval (CI): 2.40–6.40] in rs17878362 A1/A1 carriers and 3.17 (95% CI: 1.83–5.17) in A1/A1 rs17880560 carriers. Again, no effect was seen for the rs1042522 genotypes. These observations suggest that variant alleles of both the rs17878362 and rs17880560 polymorphisms are associated with a substantial protection against early life cancers in carriers of germline TP53 mutations.
Fig. 2.
Genotype-dependent Kaplan–Meier disease-free probability estimates in LFL/LFS family members with or without TP53 mutations. Kaplan–Meier probability is shown for rs17878362 (A), rs17880560 (B) and rs1042522 (C). In each panel, the left panel corresponds to subjects of the MUT group (TP53 mutation carriers) and the right panel to subjects of the WT2 group (families with no mutation detected). The tables under the graphs show disease-free probability estimates at different ages (10, 30, 50 and 65 years) according to genotype. Only probabilities up to 65 years are shown. *: G corresponds to the arginine and C to the proline variants of rs1042522.
Disease-free probability estimates relation with haplotypes defined by rs17878362 and rs17880560
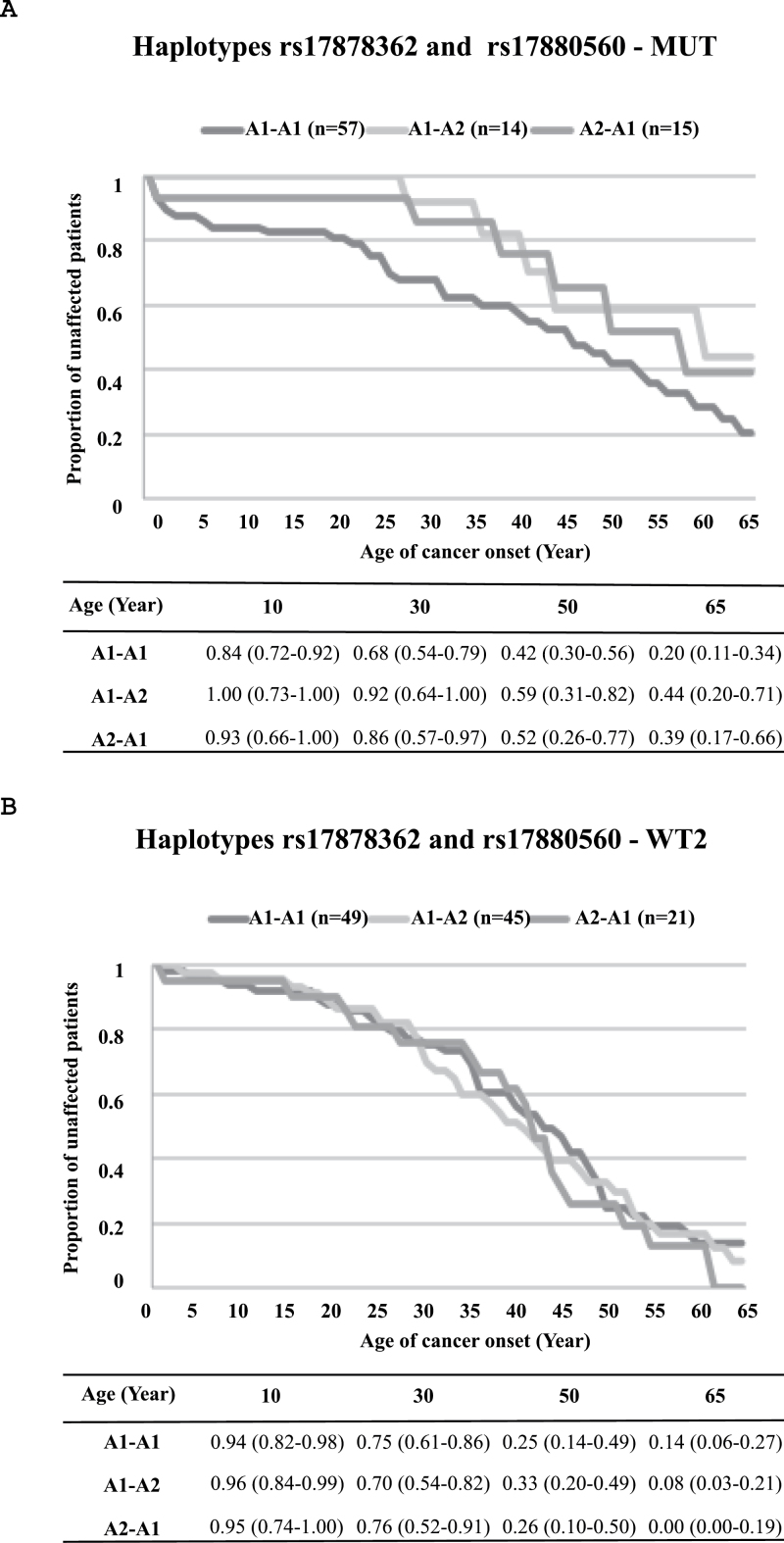
Linkage disequilibrium studies show that rs17878362 and rs17880560 are in strong linkage disequilibrium in all groups analyzed (P = 0.024). To determine whether the two alleles may exert effects either alone or in combination, we reconstructed TP53 haplotypes in subjects of the MUT and WT2 groups using data for the five of the polymorphisms genotyped in this study, plus, in the MUT group, the germline TP53 mutation (Supplementary Table 7, available at Carcinogenesis Online). We next used the genotypes at rs17878362 and rs17880560 to define eight haplotypes (with and without a TP53 mutation). Of these haplotypes, all were represented in the WT2 or MUT series, namely A1-A1 (carrying non-duplicated forms of rs17878362 and rs17880560), A1-A2 (non-duplicated for rs17878362, duplicated for rs17880560), A2-A1 (duplicated for rs17878362 and non-duplicated rs17880560) and A2-A2 (duplicated for both rs17878362 and rs17880560). The ‘Brazilian founder’ R337H mutation was exclusively carried on the A1-A1 haplotype. Of the 16 families with other TP53 germline mutations, 12 carried the mutation on the same, A1-A1 haplotype. Given the predominance of this haplotype as the carrier of the germline mutation, we reasoned that the modifier effect of TP53 polymorphisms might be due to variations in the haplotype of the remaining WT allele, which shows much larger genetic diversity than the mutant haplotype. Figure 3 shows Kaplan–Meier disease-free probability estimates in MUT and WT2 groups, given one ‘fixed’ A1-A1 TP53 haplotype (carrying a germline mutation in subjects of the MUT group and a WT TP53 in subjects of the WT2 group) and considering the effect of the other TP53 allele. This analysis showed that, in patients where the haplotype carrying the germline mutation was A1-A1, the presence of either A1-A2 or A2-A1 on the remaining WT allele was associated with a delay in the age at first cancer diagnosis (Table II and Figure 3). A significant difference was observed in mean age at first diagnosis in the MUT group in relation to the haplotype status of the WT allele after weighting for family size (‘Group’: P = 0.019; ‘All’: P = 0.035) (Table II). Importantly, none of the individuals with a WT A1-A2 haplotype, and only one with a WT A2-A1 haplotype, had developed cancer by age 25 years among carriers of a germline mutation, whereas 25% (95% CI: 15–38) of subjects with a WT A1-A1 haplotype had developed a cancer by that age. Thus, the presence of a WT allele carrying a duplication of either rs17878362 or rs17880560 appears to exert a strong protective effect toward early cancer occurrence in this cohort of patients with germline TP53 mutations.
Fig. 3.
Kaplan–Meier disease-free probability estimates based on the haplotype of the WT allele. Kaplan–Meier probabilities in MUT (TP53 mutation carriers in LFL/LFS family members) (A) and WT2 groups (LFL/LFS families with no mutation detected) (B) are shown. In the MUT group, the TP53 mutation is present on a haplotype defined as A1A1 (non-duplicated variant for both polymorphisms; rs17878362 andrs17880560), and the remaining WT haplotype is shown. In the WT2 group, in which subjects carry two WT alleles, one of the alleles has been ‘fixed’ as the A1A1 haplotype and the effect of the other haplotype is shown. Tables under the graphs show disease-free probability estimates at different ages (10, 30, 50 and 65 years) according to haplotypes (A1A1: non-duplicated for both polymorphisms; A1A2: non-duplicated for rs17878362 and duplicated for rs17880560; A2A1: duplicated for rs17878362 and non-duplicated for rs17880560). Only probabilities up to 65 years are shown.
Table II.
Mean age at first cancer diagnosis depending on TP53 haplotypes and TP53 mutation family status
| rs17878362 | rs17880560 | WT2 | MUT | ||||
|---|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | ||
| A1 | A1 | 42 | 39.36 | 17.36 | 35 | 29.31 | 20.15 |
| A1 | A2 | 35 | 37.11 | 15.89 | 6 | 46.47 | 15.05 |
| A2 | A1 | 18 | 37.17 | 15.05 | 7 | 41.71 | 22.69 |
| A2 | A2 | 3 | 45.33 | 10.69 | 1 | 50.00 | — |
| P value | Group | 0.864 | 0.019 | ||||
| All | 0.869 | 0.035 | |||||
| Without | 0.800 | 0.138 | |||||
One allele, corresponding to the frequent allele (rs17878362-A1 and rs17880560-A1), is fixed and the haplotype frequencies correspond to the second allele. WT2: WT subjects in WT families;
MUT: mutant subjects in mutant families;
Group: P value calculated with weight in the analyzed group. In bold, significative P value;
All: P value calculated with weight in the all cohort. In bold, significative P value;
Without: P value without weight. In bold, significative P value.
Discussion
LFS/LFL is a heterogeneous familial predisposition syndrome with risk of multiple cancers and ~90% penetrance over lifetime in carriers of a germline TP53 mutation. However, there are considerable interindividual and familial variations in cancer patterns and age at disease onset, raising the hypothesis that a number of other genetic, epigenetic or lifestyle factors may affect the course of the disease. Guanine-rich tracts involved in G4 motifs may represent important targets for genetic polymorphisms with significant functional effects. The enrichment of G4s in key chromosomal regions has suggested a functional role for these motifs in genomic regulation. Data collected from human SNP databases indicated that guanine triplets involved in G4s are more conserved and less polymorphic than their neutral counterparts (40). However, a recent analysis of SNPs occurring within predicted G4 motifs in the promoter of 48 genes has demonstrated a strong correlation between G4 SNPs and expression of the corresponding gene at the individual level (41). These observations support the hypothesis that G4 SNPs may play significant roles in the mechanisms of variations in gene expression among individuals. In this study, we have systematically searched the sequence of the TP53 locus for areas that may form G4s and we have analyzed polymorphisms in these areas in search of genetic variations that may modulate disease onset in TP53 mutation carriers and therefore account for at least part of individual and familial variations in LFS/LFL.
Among five regions predicted to form G4s in TP53, our results identified two polymorphisms, rs17878362 and rs17880560, with a frequency of above 4% that appeared to modulate the age at first cancer onset in TP53 mutation carriers, but not in subjects with LFS/LFL traits that do not carry a mutation in TP53. rs17878362 occurs within a previously demonstrated G4 located in intron 3 (28,33) and rs17880560 overlaps with a predicted G4 in the 3′ flanking region of the pre-mRNA of p53. Interestingly, the 3′ flanking region contains a succession of several G4s. Of these, one has been experimentally demonstrated (33) but is distinct from the predicted G4 structure overlapping with rs17880560. However, the presence of a G4 overlapping with the sequence of rs17880560 remains to be demonstrated using biophysical or molecular biological methods.
Both the rs17878362 and rs17880560 polymorphisms consist of duplications introducing additional runs of guanines, which may change the composition, position, structure or stability of the G4s. For both G4 regions, effects were seen in association with the duplicated A2 alleles. Compared with the non-duplicated A1 alleles, the presence of one A2 allele of either rs17878362 or rs17880560 retarded by ~15 years (range: 12.5–18.0 years) the age at first cancer diagnosis in TP53 mutation carriers. As a comparison, we used a large group of subjects from families with cancer patterns corresponding to LFS/LFL traits but in whom no familial TP53 mutation had been identified. Neither of the two polymorphisms had a detectable impact on age at first cancer diagnosis or on cancer-free survival in subjects without mutations. To assess the statistical significance of the effects observed in mutation carriers, we performed a t-test on average age at diagnosis, using a model weighted for family structure and number of members in each family, in order to rule out bias due to family-specific traits that may influence the clinical presentation of LFS/LFL. Although this approach decreases statistical power, it allows the clarification of effects that are solely due to the presence of particular allele combinations. Using this method, we detected that each of the polymorphisms had a borderline significant effect on age at first cancer diagnosis (P = 0.082 for rs17878362; P = 0.067 for rs17880560). However, the magnitude of the effect was large (12.5–18.0 years) and separated subjects with cancer diagnosis in childhood or adolescence from subjects with diagnosis in adulthood. Taking as reference the risk of developing cancer before the age 25 years in carriers of one A2 allele of either polymorphism, the relative risk of early cancer in mutation carriers was 4.0 (95% CI: 2.40–6.40) for rs17878362 A1/A1 and 3.17 (95% CI: 1.83–5.17) for rs17880560 A1/A1. By comparison, the common non-synonymous SNP at codon 72 (rs1042522, TP53 PEX4, R72P) had no effect on age at first cancer diagnosis in this cohort.
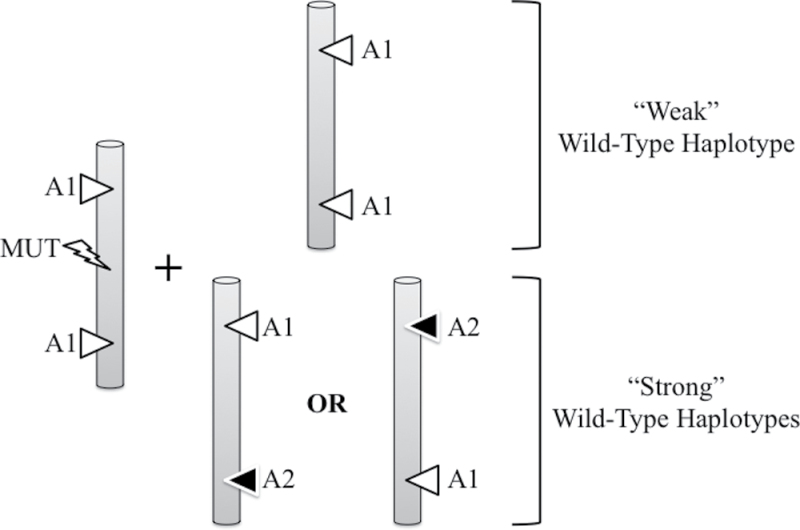
Given that rs17878362 and rs17880560 are in linkage disequilibrium, we reconstructed TP53 haplotypes to estimate the joint effects of these polymorphisms. We made the assumption that the effects of these polymorphisms were associated with the haplotype of the WT allele, rather than the mutant allele. This assumption is based on the observation that, in our series of Brazilian families with TP53 mutation, the mutation is almost systematically located on a haplotype carrying the A1 alleles of both rs17878362 and rs17880560. Thus, the effect of alleles carrying A2 haplotypes, if any, is associated with the properties of the residual WT allele and with its capacity to compensate the functional defect caused by the mutant allele. Using this framework, we found that presence of haplotypes defined by either A1 rs17878362–A2 rs17880560 or A2 rs17878362–A1 rs17880560 had a significant effect on delaying the age at first cancer diagnosis. Among 91 documented subjects carrying a germline mutation, only 1 of 29 having one of these two haplotypes developed cancer before 25 years of age (3.4%), whereas 15 of 57 having the WT A1-A1 haplotype developed cancer by this age (26.3%). We therefore conclude that, when the mutation is present on the A1-A1 haplotype (the most common haplotype in all populations, in particular Caucasians and Asians), the presence of one A2 variant of either rs17878362 or rs17880560 on the residual WT allele is protective against the risk of cancer in childhood and adolescence. This effect is compatible with a simple model in which, in the presence of a mutant allele with partial or total loss of function, TP53 suppressor activity is contributed by the residual WT allele (Figure 4). Thus, WT haplotypes carrying one A2 allele of either rs17878362 or rs17880560 would define ‘strong’ TP53 haplotypes, contributing WT TP53 activity sufficient to suppress cancer until at least early adulthood, whereas A1-A1 WT haplotypes would be ‘weak’ TP53 haplotypes, unable to compensate the loss of function of the mutant haplotype and thus being permissive for early cancer onset.
Fig. 4.
Effect of different WT haplotypes in TP53 mutation carriers: a model. TP53 alleles are represented as rods. Left, mutant allele occurring on a haplotype carrying A1 variants of both rs17878362 and rs17880560 (A1A1). Right, different types of WT haplotypes. The WT haplotype defined by A1A1 is considered as a ‘weak’ haplotype (associated with early cancer, indicative low capacity to compensate the loss of p53 function of the mutant allele). The WT haplotypes defined by A1A2 or A2A1 are considered as ‘strong haplotype’ (associated with later cancer onset, thus providing at least partial compensation for the loss of function of the mutant allele). Of note, our data do not predict the effect of WT A2A2 haplotypes, or the effects of these haplotypes when the mutation occurs on another haplotype than A1A1.
The present study differs from previous ones on modifier polymorphisms in TP53 by several characteristics. First, we have used a weighted model to take into account family size and control for possible bias cause by specific familial traits. Second, we have used as reference a group of LFS/LFL families with no mutation (WT2) rather than a heterogeneous group of ‘wild type’ subjects pooling the members of TP53 families who have not inherited the mutation (and therefore should be considered as low risk for cancer) and members of families with no mutations (some of them likely to be at high risk due to germline mutation in yet unidentified gene(s) other than TP53). Third, our series is constituted of LFS/LFL families recruited in Brazil, whereas previous studies were based on Western European or North American cohorts (9,13,14). A common mutant haplotype, R337H, is present in 0.3% of the population of South-Eastern Brazil and represents over half of the germline TP53 mutations in Brazilian families with LFS/LFL traits (19,35). This mutation is carried by the same haplotype in all families and is considered to result from a widespread founder effect. Since its initial identification in the germline of children with adrenal cortical carcinoma (42), there has been debate as whether R337H should be considered as predisposing to LFS/LFL or only to specific tumor types. Our results on LFS/LFL families have shown that carriers of R337H are prone to develop a spectrum of cancers that covers the whole LFS/LFL spectrum, although the average age at first diagnosis for adolescent and adult cancers tend to be up to 10 years later than in carriers of classic, DNA-binding domain TP53 mutations commonly associated with LFS (19). In the present study, we can rule out that the modifier effects of rs17878362 and rs17880560 polymorphisms are restricted to this ‘Brazilian founder’ allele because we observed effects of similar type and amplitude in Brazilian carriers of “classic” TP53 mutations, who have cancer patterns and age at onset identical to Caucasian TP53 mutation carriers from North America or Western Europe (data not shown).
The molecular mechanisms responsible for the association of the A2 alleles of rs17878362 or rs17880560 with late age at first cancer onset are a matter of conjecture. The G4s containing rs17878362 (intron 3) has been shown to exert a regulatory effect on the alternative splicing of intron 2 and on the generation of p53I2, a p53 mRNA that encodes the p53 protein isoform Δ40p53. In animal models, homologs of Δ40p53 downregulate stem cells and promote aging (in mice) and induce abnormal development (in zebrafish) exclusively when coexpressed together with full-length, WT p53 (43,44). The precise structure and biological effect of the predicted polymorphic G4 in the 3′ flanking region remain to be demonstrated.
To conclude, this study has identified polymorphisms associated with G4 structures, which modify the penetrance of germline TP53 mutation in LFS/LFL. They have a general effect on the mean age at first cancer diagnosis but, more importantly, they distinguish patients who are at high risk of developing childhood/adolescent cancers from those who develop mainly, if not exclusively, cancers during adulthood. These findings require evaluation and replication in independent LFS/LFL cohorts but may provide an important marker for developing personalized strategies for surveillance of cancer risk in carriers of germline TP53 mutations.
Supplementary material
Supplementary Figure 1 and Tables 1–7 can be found at http://carcin.oxfordjournals.org/
Funding
French National Cancer Institute (INCa 2009–192 ‘TP53 intron3’ to J.H., P.H. and J.-L.M.); research in Inserm U612 is also supported by funding from Institut Curie and Inserm; C.S. has a PhD fellowship from the French Ministry of Research and V.M. and L.P. were supported by funding from EU FP7 (249689) for the network of excellence DoReMi (low dose research toward multidisciplinary integration); INSERM (D.G.C.); Conselho Nacional de Pesquisa (CNPq 40.0949/2005–9 to P.P.) and Susan G Komen for the Cure (POP 0403033 to P.P.). Research on G-quadruplex is supported by Fondation ARC, Région Aquitaine and ANR program "Quarpediem" (J.-L.M.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- CI
confidence interval
- G4
G-quadruplex
- LFS/LFL
Li-Fraumeni/Li-Fraumeni-like syndrome
- mRNA
messenger RNA
- RFLP
restriction fragment length polymorphism
- SNP
single-nucleotide polymorphism.
References
- 1. Li F.P., et al. (1988). A cancer family syndrome in twenty-four kindreds. Cancer Res., 48, 5358–5362 [PubMed] [Google Scholar]
- 2. Mai W.J., et al. (2012). Characterization of the tilapia p53 gene and its role in chemical-induced apoptosis. Biotechnol. Lett., 34, 1797–1805 [DOI] [PubMed] [Google Scholar]
- 3. Birch J.M., et al. (1994). Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res., 54, 1298–1304 [PubMed] [Google Scholar]
- 4. Eeles R.A. (1995). Germline mutations in the TP53 gene. Cancer Surv., 25, 101–124 [PubMed] [Google Scholar]
- 5. Chompret A. (2002). The Li-Fraumeni syndrome. Biochimie, 84, 75–82 [DOI] [PubMed] [Google Scholar]
- 6. Malkin D., et al. (1990). Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science, 250, 1233–1238 [DOI] [PubMed] [Google Scholar]
- 7. Srivastava S., et al. (1990). Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature, 348, 747–749 [DOI] [PubMed] [Google Scholar]
- 8. Varley J.M., et al. (1997). Germ-line mutations of TP53 in Li-Fraumeni families: an extended study of 39 families. Cancer Res., 57, 3245–3252 [PubMed] [Google Scholar]
- 9. Ruijs M.W., et al. (2010). TP53 germline mutation testing in 180 families suspected of Li-Fraumeni syndrome: mutation detection rate and relative frequency of cancers in different familial phenotypes. J. Med. Genet., 47, 421–428 [DOI] [PubMed] [Google Scholar]
- 10. Tinat J., et al. (2009). 2009 version of the Chompret criteria for Li Fraumeni syndrome. J. Clin. Oncol., 27, e108–9; author reply e110. [DOI] [PubMed] [Google Scholar]
- 11. Bougeard G., et al. ; French LFS working group (2008) Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J. Med. Genet., 45, 535–538 [DOI] [PubMed] [Google Scholar]
- 12. Villani A., et al. (2011). Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol., 12, 559–567 [DOI] [PubMed] [Google Scholar]
- 13. Bond G.L., et al. (2004). A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell, 119, 591–602 [DOI] [PubMed] [Google Scholar]
- 14. Bougeard G., et al. (2006). Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J. Med. Genet., 43, 531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marcel V., et al. (2009). TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: impact on age at first diagnosis. J. Med. Genet., 46, 766–772 [DOI] [PubMed] [Google Scholar]
- 16. Ruijs M.W., et al. (2007). The single-nucleotide polymorphism 309 in the MDM2 gene contributes to the Li-Fraumeni syndrome and related phenotypes. Eur. J. Hum. Genet., 15, 110–114 [DOI] [PubMed] [Google Scholar]
- 17. Wu C.C., et al. (2011). Joint effects of germ-line TP53 mutation, MDM2 SNP309, and gender on cancer risk in family studies of Li-Fraumeni syndrome. Hum. Genet., 129, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knappskog S., et al. (2011). Effects of the MDM2 promoter SNP285 and SNP309 on Sp1 transcription factor binding and cancer risk. Transcription, 2, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Achatz M.I., et al. (2007). The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett., 245, 96–102 [DOI] [PubMed] [Google Scholar]
- 20. Bochman M.L., et al. (2012). DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet., 13, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ribeyre C., et al. (2009). The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo . PLoS Genet., 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paeschke K., et al. (2011). DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell, 145, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paeschke K., et al. (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature, 497, 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cayrou C., et al. (2012) New insights into replication origin characteristics in metazoans. Cell Cycle, 11, 658–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Millevoi S., et al. (2012). G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA, 3, 495–507 [DOI] [PubMed] [Google Scholar]
- 26. Halder R., et al. (2012). Bisquinolinium compounds induce quadruplex-specific transcriptome changes in HeLa S3 cell lines. BMC Res. Notes, 5, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomez D., et al. (2004). Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res., 32, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marcel V., et al. (2011). G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis, 32, 271–278 [DOI] [PubMed] [Google Scholar]
- 29. Ghosh A., et al. (2004). Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell. Biol., 24, 7987–7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Courtois S., et al. (2002). DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene, 21, 6722–6728 [DOI] [PubMed] [Google Scholar]
- 31. Yin Y., et al. (2002). p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol., 4, 462–467 [DOI] [PubMed] [Google Scholar]
- 32. Marcel V., et al. (2011). Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ., 18, 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Decorsière A., et al. (2011). Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3’-end processing and function during DNA damage. Genes Dev., 25, 220–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petitjean A., et al. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat., 28, 622–629 [DOI] [PubMed] [Google Scholar]
- 35. Garritano S., et al. (2010). Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum. Mutat., 31, 143–150 [DOI] [PubMed] [Google Scholar]
- 36. Kikin O., et al. (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res., 34(Web Server issue), W676–W682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scaria V., et al. (2006) Quadfinder: server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res., 34(Web Server issue), W683–W685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huppert J.L., et al. (2005). Prevalence of quadruplexes in the human genome. Nucleic Acids Res., 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stacey S.N., et al. ; Swedish Low-risk Colorectal Cancer Study Group (2011) A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet., 43, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakken S., et al. (2009). The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res., 37, 5749–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baral A., et al. (2012). Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res., 40, 3800–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ribeiro R.C., et al. (2001). An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc. Natl Acad. Sci. U S A, 98, 9330–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maier B., et al. (2004). Modulation of mammalian life span by the short isoform of p53. Genes Dev., 18, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davidson W.R., et al. (2010) Differential regulation of p53 function by the N-terminal ΔNp53 and Δ113p53 isoforms in zebrafish embryos. BMC Dev. Biol., 10, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.