Abstract
This paper reports on the application of an optical fiber biosensor for real-time analysis of cellular behavior. Our findings illustrate that a fiber sensor manufactured from a traditional telecommunication fiber can be integrated into conventional cell culture equipment and used for real-time and label-free monitoring of cellular responses to chemical stimuli. The sensing mechanism used for the measurement of cellular responses is based on the excitation of Surface Plasmon Resonance (SPR) on the surface of the optical fiber. In this proof of concept study, the sensor was utilized to investigate the influence of a number of different stimuli on cells - we tested the effects of trypsin, serum and sodium azide. These stimuli induced detachment of cells from the sensor surface, uptake of serum and inhibition of cellular metabolism, accordingly. The effects of different stimuli were confirmed with alamar blue assay, phase contrast and fluorescence microscopy. The results indicated that the fiber biosensor can be successfully utilized for real-time and label-free monitoring of cellular response in the first 30 minutes following the introduction of a stimulus. Furthermore, we demonstrated that the optical fiber biosensors can be easily regenerated for repeated use, proving this platform as a versatile and cost-effective sensing tool.
Keywords: Cellular analysis, plasmonic fiber sensor, cellular response, real time and label free sensing
1. Introduction
Optical fiber sensors have been extensively employed for the detection of a broad range of biological targets such as, nucleic acids (Delport et al. 2012; Ferguson et al. 2000; Yin et al. 2013), antigens (Jang et al. 2009), antibodies (Lepinay et al. 2013; Ruan et al. 2008) and other low and high molecular weight analytes (Amin et al. 2012; Pollet et al. 2011; Silva et al. 2009; Tierney et al. 2009; Wang and Wolfbeis 2013). Recently there has been an increasing emphasis on the characterization of larger and more complex biological targets such as cells, bacteria (Smietana et al. 2011) and viruses (Bhatta et al. 2010; François et al. 2011). The application of cells as an integral part of the biorecognition scheme is driven by a growing demand for a better understanding of newly discovered cellular signaling pathways, a need for the characterization of drug-cell interactions as well as for a comprehensive evaluation of cellular interactions with their microenvironments (Ghaemmaghami et al. 2012; Khademhosseini et al. 2006).
Optical label-free characterization of cellular behavior can be accomplished by using several approaches. One approach is to apply traditional imaging tools where light propagates in free space and interacts with cells positioned on its path. For instance, imaging using traditional cell phone cameras has been demonstrated as a cost-effective solution that could be integrated into cell culture incubators to monitor cellular responses induced by drugs and other stimuli (Kim et al. 2011; Kim et al. 2012; Zheng et al. 2011).
Another strategy for characterization of cellular responses is to use optical analytical tools where cells would be in a direct contact with the optical elements that confine and guide light. In that case, the detection of cellular reactions is being implemented by utilizing the evanescent field of the light confined by the optical elements. The evanescent field decays away from the surface of an optical element to a distance of a few micrometers, sufficiently deep to penetrate into the cells and probe their optical properties. Changes in the attenuation or phase of the evanescent field penetrating into cells can be used to interpret intra- and extra cellular changes. In particular, changes in the optical field can be used to monitor cellular adhesion (Lin et al. 2006), detachment (Fang et al. 2006), death (Chabot et al. 2009; Maltais et al. 2012), contraction, a response to osmotic stress (Vala et al. 2013) as well as triggering of diverse intracellular reactions (Fang and Ferrie 2008; Fang et al. 2006; Yanase et al. 2013) that are very challenging to monitor by other means. So far, the use of analytical tools for cellular characterization has been limited to a few optical platforms such as, the well-established Surface Plasmon Resonance (SPR) (Chabot et al. 2009; Hide et al. 2002; Robelek 2009; Yanase et al. 2007), resonant waveguide grating EPIC® systems (Fang and Ferrie 2008; Fang et al. 2006; Scibek et al. 2012) and photonics crystal fiber (Lin et al. 2006). The signal captured by these sensors is real-time, dose-dependent, quantitative and label-free. A combination of the plasmonic technique with fluorescent labeling can be used to monitor even finer cellular responses such as cellular reorganization and transient contraction as reported by Chabot et al (2013)(Chabot et al. 2013) if label-free regime is not a requirement.
Although the mentioned optical systems can be used to characterize a wide range of cellular responses, the platforms themselves are large and are not designed to be integrated into traditional tissue culture incubators. Handling cells outside incubators during their analysis severely effects cellular viability due to potential contamination and also affects cellular metabolism. Additionally, the operation of the platforms mentioned above is complicated and requires highly trained personnel. On the other hand, integration of optical elements such as plain optical waveguides, structured optical surfaces or custom-made fiber would require the development of additional optical infrastructure that would ensure delivery of light to an incubator with sensors wired into a Petri dish and optical signal readout. This would increase the complexity and reduce the practical use of such platform.
In this context, optical sensor platforms that are made using optical fiber offer several competitive advantages such as compactness, remote sensing capabilities, simple integration into cell culture equipment, and potential for in situ measurements. When the fiber sensors operate at standard telecommunication wavelengths, as in our case, both the fiber devices and the interrogation equipment become highly cost-effective compared to the more complex and bulkier custom designed optical sensing platforms. Taking into account that the sensitivities reported in the literature for fiber sensors can be similar to those of traditional platforms employing planar optical structures (Sharma and Gupta 2007), fiber sensors position themselves as a versatile solution that can be used in a range of applications including in situ whole cell sensing.
Yanase et al. (2010) showed that a plasmonic fiber biosensor could be used for monitoring cellular response when cells are exposed to a stimulus, i.e. albumin (Yanase et al. 2010). The sensor employed in that study was manufactured using a multimode quartz fiber. However, the use of multimode fiber restricts sensor detection levels as well as hinders the quality of the detected optical signal. In this study we propose to use a completely different sensor manufactured using a standard telecommunication single mode fiber in order to improve the results. The use of a traditional single mode fiber allows the fiber sensor to be robust and achieve a high level of performance. Moreover, this is the first optical fiber sensor that was fully integrated into conventional cell culture equipment making it possible to conduct the experiments in a controlled environment. This sensor was developed not for “field use” but for in situ sensing inside cell culture incubators. Fiber connectorization is very easy to perform with standard low cost instrumentation developed for the telecommunication industry. By conducting all measurements in a conventional incubator, we obtained high cell viability and were able to monitor cellular responses for extended periods of time.
Herein we present the application of a fiber biosensor, a miniature and portable sensor platform, for real-time and label-free monitoring of cellular responses. Particularly, we demonstrate that the plasmonic fiber sensor can be used for characterization of a set of fine cellular responses triggered by selected stimuli. The sensor is manufactured using a single mode fiber and works through the excitation of plasmon waves on its gold-coated surface.
2. Experimental
2.1 Materials
Fibronectin was obtained from BioLegend (San Diego, CA, USA). Phosphate buffered saline (PBS), Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), 10× trypsin and penicillin/streptomycin were purchased from Gibco (Carlsbad, CA, USA). Alamar blue, calcein AM, ethidium homodimer, phalloidin (Alexa-Fluor 594), and 4′,6-diamidino-2-phenylindole (DAPI) were obtained from Invitrogen Corp. (Carlsbad, CA, USA). Bovine serum albumin (BSA) and sodium azide was supplied by Sigma-Aldrich (St. Louis, MO, USA). Paraformaldehyde was obtained from Electron Microscopy Sciences (Hatfield, PA, USA). 4-well plastic plates were purchased from Fisher Scientific Co LLC (Pittsburg, PA, USA). All reagents were used as received without further purification.
2.2 Fabrication of the Plasmonic Sensors
The sensors used in this work (Fig. 1A) were manufactured using a traditional telecommunication single mode fiber (CORNING SMF28)(Shevchenko et al. 2010b; Shevchenko and Albert 2007). As reported earlier, the fabrication of sensor elements was completed in a two-step process that involved the fabrication of a resonant grating element followed by a deposition of a gold coating on the surface of the fiber. The gratings were imprinted in the hydrogen-loaded fiber by using UV light passed through a diffractive phase mask, which had a period of 1096 nm. The UV beam was imposed on a fiber with an excimer laser operating at a wavelength of 193 nm. After the inscription process, the gratings were annealed with a heat gun at 400°C for approximately 1 min and thermally stabilized in the oven at 120°C for 12 hours. The final step was the deposition of a 50 nm thick gold coating on the surface of the fiber. The deposition was implemented using an electron beam evaporation setup (Balzer Evaporator system).
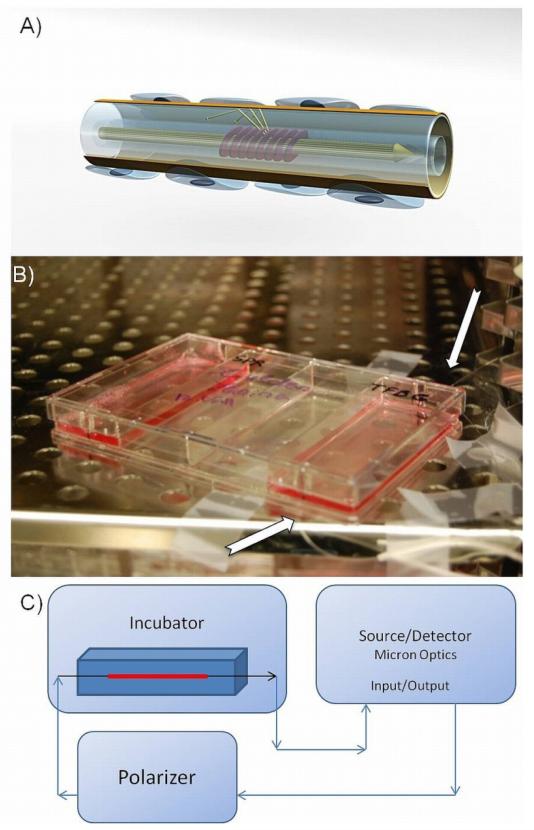
Fig. 1.
Optical fiber configuration. (A) 3D illustration of a fiber sensor with cells attached to the surface of the gold coating. Fringes in the core of the fiber indicate the tilted grating that redirects the light towards the surface of the fiber; (B) Photograph of a 4-well plate with an integrated fiber sensor during a typical experiment. Arrows point at an optical fiber wired outside the plate; (C) Schematic of the optical interrogation setup (all the line arrows represent single mode fiber connections).
2.3 Principle of Operation of the Plasmonic Fiber Biosensor
The fiber sensor operates through the excitation of the SPR effect on the surface of the gold-coated fiber (Albert et al. 2013; Caucheteur et al. 2011; Caucheteur et al. 2013; Shevchenko et al. 2010a; Shevchenko et al. 2011; Shevchenko and Albert 2007). A schematic of the sensor with cells attached to its surface is shown in Fig. 1A. The SPR excitation is achieved by means of a tilted grating imprinted in the core of the fiber. The Tilted Fiber Bragg Grating (TFBG) acts as a resonant element redirecting the light from the guided core to the surface of the fiber. When the wavelength of the light propagating in the fiber is such that a phase matching condition for the excitation of the SPR is satisfied, some of the light escapes from the cladding and is coupled to plasmonic waves on the outer surface of the gold-coated fiber.
The surface plasmon wave is confined at the interface between the gold coating and a dielectric outside a fiber (or sensed medium) with the evanescent tail of the plasmonic field penetrating deeply in the dielectric. It provides the whole excitation system with an enhanced sensitivity towards refractive index changes that occur at the metal surface. When cells are attached to the surface of the gold coating, the plasmonic field penetrates into the cells and enables monitoring of changes inside the cells. The depth of penetration of the plasmonic field is proportional to the wavelength used to excite the SPR (Ligler and Taitt 2002) and is approximately 1400-1500 nm for this sensor. The measured SPR signal results from the average of all chemical changes occurring on the surface of the sensing element which covers 3.92 mm2 (for a 1 cm-long grating) with a depth of penetration of 1500 nm.
2.4 Optical Instrumentation and Sensor Integration into an Incubator
All sensors were fixed on the bottom of rectangular 4-well plates using a UV-curable adhesive (Thorlabs, Newton, NJ, USA). The tails of the fiber sensors were guided through two holes made in the sidewalls of the plates (Fig. 1B). Plates with sensing elements were placed in an incubator while the tails of the sensors were wired outside the incubator and connected to the interrogation equipment (Fig. 1C). The interrogation unit consisted of an Optical Sensing Analyzer (model Si720 from Micron Optics) and a Polarization Controller (from JDS Uniphase). Measurements of the optical transmission spectrum of the devices were saved every 0.2 sec with the spectral resolution set at 0.0025 nm. The collected data was internally processed by the Micron Optics unit with a 10 kHz filter.
2.5 Cell Cultures
The NIH-3T3 fibroblast cells were maintained in DMEM medium supplied by 10% FBS and 1% Pen/Strep. Cell cultures were kept in a 5% CO2 regulated incubator at 37 °C. Media was changed every other day during the cell culture period. Upon reaching 70-80% confluence, cells were either passaged into new flasks or harvested by trypsin and counted at the desired cell density (1 million cells/mL) to be used in experiments with the SPR-TFBG sensors.
2.6 Sensor Modification and Monitoring Cellular Response
To ensure cell attachment to the sensor’s surface, the gold coating was modified with fibronectin protein, which is a well known cell-adhesive reagent (Mosher 1984). The fiber sensors were sterilized under UV light and washed with PBS six times before the surface modification. After aspiration of PBS, 500 μL of 100 μg/ml fibronectin was dispensed on the sensors for one hour to facilitate the interaction of the gold coating and charged functionalities of the protein.
Fibronectin was then removed, the sensors were rinsed with PBS and subsequently seeded with 5 mL of NIH-3T3 cells at a 2.1*105 cells/cm2 density. The sensors were then incubated for 14 h at 37 °C to enable the formation of a monolayer of cells on the fiber. The cell number per each sensor was approximately 4×103 cells per fiber. Media was changed 2 h after the incubation started.
The experimental routine included recording of the sensor output for 30 min before the addition of a stimulus, followed by the aspiration of the media and the addition of a stimulus in fresh media. The amount of added stimuli was substantially smaller than the total amount of added medium, therefore no detectable refractive index change was recorded by the sensor. Thus, all SPR signal changes observed right after addition of stimuli were caused by the cellular responses rather than a bulk refractive index mismatch. After aspiration of the old media and the addition of the new media with a stimulus, we recorded the SPR signal for one hour.
After the experiments, sensor surfaces were regenerated by incubation with trypsin and ethanol. Sensors were first rinsed with PBS, then exposed to trypsin (10×) for 30 minutes and later left in ethanol solution (70%) for 10 minutes. The regeneration step was finalized by washing the sensors with PBS.
2.7 Cellular Assays and Sample Imaging
Live/Dead test, alamar blue assay and DAPI/Phalloidin staining were carried out following manufacturer’s protocols. Alamar blue assay was read using a Fluostar fluorescence plate reader supplied by BMG LABTECH GmbH (Offenburg, Germany) at 544 nm/590 nm (Ex/Em). Images were acquired by using an inverted fluorescent microscope (Nikon Model TE 2000-U, Nikon instruments Inc., USA). Spot Advanced Software 4.6.4.6 supplied by Diagnostic Instruments, Inc. (Sterlington Heights, MI, USA) was used for capturing images.
2.8 Statistical Analysis
Statistical analysis of Alamar Blue data was performed using one-way analysis of variance (ANOVA) with Bonferroni post-test processing using the GraphPad software by Prism Inc. (La Jolla, CA, USA). For the analysis, the level of significance was set at p<0.05 (***p<0.001, **p<0.01, *p<0.05).
3. Experimental
3.1 SPR Data Analysis for Cellular Sensing
Fig. 2 shows typical transmission spectra acquired with an SPR-TFBG sensor as well as an example of applied data analysis.
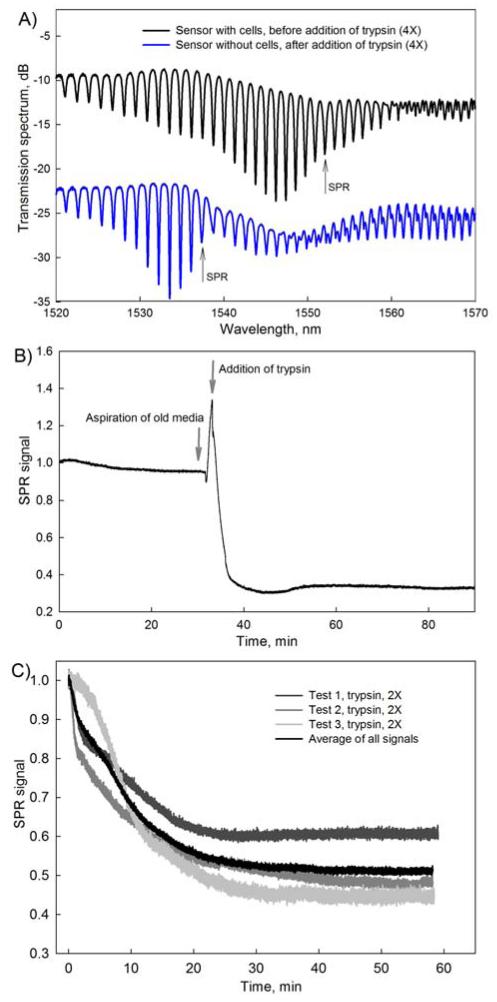
Fig. 2.
Typical SPR curves obtained with the fiber sensor. (A) SPR-TFBG transmission spectra. The black spectrum is obtained for a sensor with NIH-3T3 cells on its surface. The blue spectrum is from the same sensor after the cells were detached using trypsin (4×). Quantitative information is extracted by measuring the amplitude change of the most sensitive resonance indicated by an arrow; (B) Change in the amplitude of the most sensitive SPR resonance following the addition of trypsin (4×); (C) Reproducibility of experiments with trypsin at 2× concentration. The black curve is the average of the all SPR signals with a confidence interval of σ=0.578.
Both spectra shown in Fig. 2A were obtained with the same SPR-TFBG sensor. The upper response was measured in the presence of cells attached to the sensor surface while the bottom response was measured after cells were detached by trypsin. The difference in the shape of the output signal can be attributed to removal of cells from the fiber resulting in a change in the refractive index at the surface of the sensor. SPR-coupled portions of the spectra responsive to cellular behavior are indicated with arrows pointing at the most sensitive SPR-coupled resonances. The experimental data was analyzed using the method previously reported by Shevchenko et al. (2010)(Shevchenko et al. 2010a), which is based on tracking the relative amplitude of the most sensitive SPR-coupled cladding mode resonance. The most sensitive SPR-coupled resonance for each of the sensors was determined by analyzing the amplitude change of all SPR-coupled cladding modes and selecting the resonance with the largest relative amplitude change. The relative change in the amplitude of a resonance is proportional to the SPR effective index change and therefore could be used to evaluate the alterations in refractive index on the sensor surface. Fig. 2B shows a typical SPR response during the process of trypsinization. Arrows indicate the points in time when the original medium surrounding the sensor was aspirated and a fresh solution with trypsin was added. The strong SPR signal changes observed in between these two points is a transient result that reflects the emptying of the Petri dish and can be simply omitted from the data analysis.
To illustrate the reproducibility of SPR measurements, three sets of SPR responses were obtained from three different sensors exposed to the same conditions (Fig. 2C). All three sensors were seeded with five million NIH-3T3 cells, incubated for 12 h, and exposed to trypsin (2×) solution for 60 minutes. Standard deviation σ between the average signal and the three sets of data is 0.0578 indicating the appropriate level of reproducibility of the results.
In addition to seeding sensors with cells and exposing them to a stimulus, it was also possible to regenerate the surface of used sensors using ethanol and trypsin solutions. The sensor surface was regenerated as described in the Experimental section. Exposure to ethanol, trypsin and PBS cleaned the surface from cellular residues and enabled the reuse of same sensors in subsequent experiments.
3.2 Detection of Cell Detachment
Following 12 h of incubation period with NIH-3T3 cells, the SPR-TFBG sensors were exposed to different concentrations of trypsin solution. Trypsin, a serine protease, is regularly used to cleave cells from the surfaces to which they adhere through destruction of peptide bonds. Presented in Fig. 3A are SPR responses of fiber sensors coated with NIH-3T3 cells after exposure to three different concentrations of trypsin (0.5×, 2× and 4×). It can be observed from Fig. 2C and Fig. 3A that SPR signals induced by the addition of trypsin are always negative and saturate within first 20 minutes after the initial exposure. The amplitude of the recorded negative SPR signals decrease accordingly to the concentration of the applied solution, which is consistent with results reported by Fang et al 2006 (Fang et al. 2006).
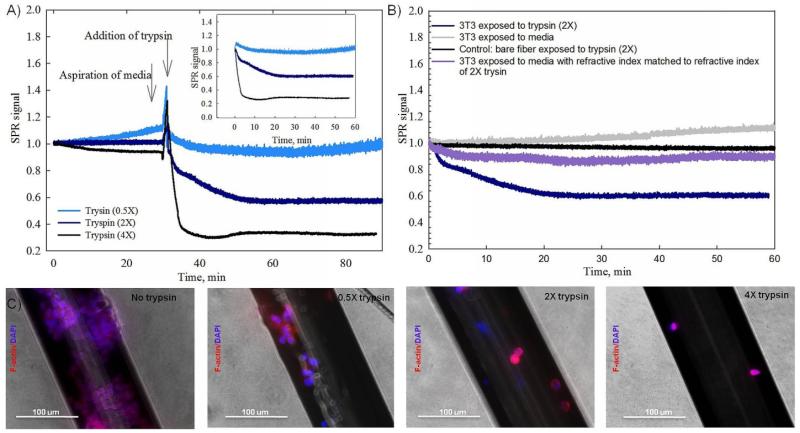
Fig. 3.
Real-time SPR signals obtained during cellular exposure to trypsin solutions, control SPR measurements, and cytoskeleton/nuclei imaging. (A) The SPR response was measured while NIH-3T3 cells were exposed to trypsin solutions (0.5×, 2× and 4× concentrations). Arrows indicate points at which the old medium was aspirated and the new solution was added. Inset shows SPR responses starting from the moment of addition of stimuli. (B) Various control experiments were performed to monitor SPR responses in the presence and absence of trypsin (2×). (C) Cytoskeleton/nucleus staining (F-actin/DAPI) demonstrates the remaining cells on the sensor surface after exposure to different concentrations of trypsin.
Fig. 3B presents results for a set of SPR controls, which illustrate that the measured SPR responses presented in Fig. 3A were only due to a reduction in the number of attached cells and not due to other background. The SPR response caused by cellular detachment induced by trypsin (navy curve) is compared to the response of a sensor with cells exposed to a regular medium (grey curve). As expected, no change in the SPR signal was recorded when standard medium was added to the sensor. The violet SPR curve represents the sensor response to medium with an adjusted refractive index. In this case, the refractive index of the solution was lowered by the addition of PBS in order to match it to the refractive index of the 2× trypsin solution. Difference between two responses (dark navy and violet curves) confirms that the measured SPR signal is due to a specific cellular interaction with trypsin solution and is not due to the refractive index mismatch. Additionally Fig. 3B provides the SPR response of a sensor without cells while being exposed to 2× trypsin solution (black curve). The difference between a sensor with 3T3 (navy curve) and a sensor without 3T3 cells attached on its surface (black curve), of which both were exposed to trypsin, proves that the measured SPR signal should be attributed to the change in the number of attached cells and was not affected by the change of the bulk background refractive index induced by the addition of trypsin. The flat response during the control test without cells (black curve) also confirms that there is no non-specific adsorption to the surface of the sensor upon exposure to trypsin. The absence of non-specific interactions proves that the decay in the SPR signal for the sensor with cells exposed to trypsin is not distorted by extracellular non-specific processes. It should be noted that all SPR curves including controls were measured by using different sensors.
The effect of different concentrations of trypsin on the degree of cellular detachment was also confirmed with fluorescence staining of cytoskeleton and nuclei. As can be observed in Fig. 3C, the density of cellular coverage was affected by the addition of trypsin at each concentration. A higher concentration of trypsin solution resulted in lower number of attached cells, which correlates well with the real-time SPR data shown in Fig. 3A.
3.3 Detection of Cellular Uptake
In addition to the detection of cell detachment, the optical fiber sensor was used to monitor the uptake of serum by the cells (Fig. 4). Fetal Bovine Serum (FBS) is widely used as a supplement for in vitro cell cultures. Often used at 10% (v/v) concentration, it provides cells with nutrients required for sustained cell growth and proliferation. In this experiment, FBS was used at concentrations of 10%, 30% and 50% (v/v), to monitor its effects on SPR response due to possible cellular uptake of nutrients.
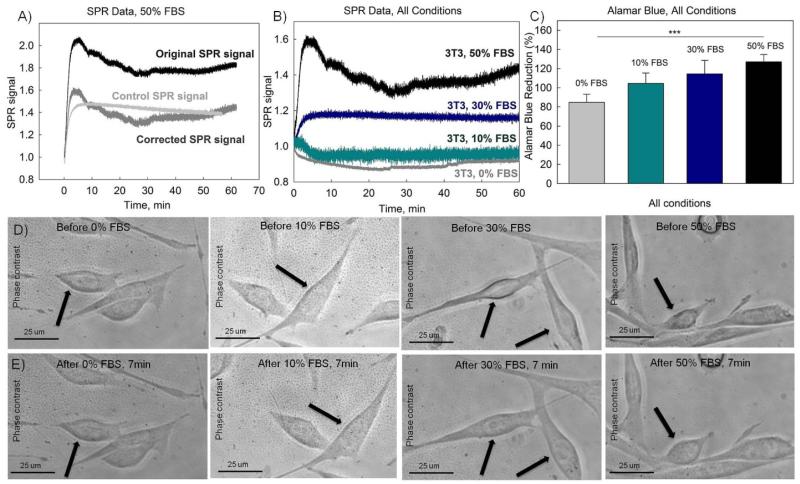
Fig. 4.
The addition of serum and its uptake by cells monitored in real-time with SPR fiber sensors confirmed with alamar blue assay and phase contrast imaging. (A) SPR responses during exposure to 50% (v/v) FBS solution. The black curve is the original SPR signal obtained in the presence of cells, the grey curve is a control SPR signal obtained in the absence of cells. The dark grey curve is the corrected SPR signal found as a difference between the original SPR and the control SPR signals. (B) Corrected SPR responses during exposure to different serum solutions (10%, 30% and 50% (v/v)) versus response to medium without FBS. (C) Alamar blue assay results for cells exposed to different concentrations of FBS; (D-E) Phase contrast images of the NIH-3T3 cells exposed to FBS solutions (50% FBS, 30% FBS, 10%FBS and 0%FBS). (D) Images were taken before the addition of FBS. (E) Images were taken 7 minutes after the addition of FBS.
It was found that serum, which contains a substantial number of proteins, resulted in a considerable non-specific adsorption on the surface of the sensors. Fig. 4A shows a comparison between original SPR signal measured in the presence of cells exposed to serum and a control SPR signal measured by a sensor without cells, also exposed to the same concentration of serum. The SPR signal increase observed in the case of the sensor without cells is due to non-specific adsorption of proteins from serum on the surface of the sensor. In order to eliminate the contribution from non-specific adsorption, each SPR signal presented on Fig. 4B was found as the difference between the SPR signal measured in the presence of cells and the SPR signal measured in the absence of cells, as illustrated in Fig. 4A.
Fig. 4B demonstrates cellular responses to 50%, 30% and 10% (v/v) FBS solutions. It can be seen from Fig. 4A-B that the SPR signal increased within the first five minutes following the addition of 50% FBS. The addition of 30% of FBS also resulted in a slight positive change. On the contrary, the addition of medium with 10% and 0% of FBS resulted in a slightly negative SPR change (Fig. 4B).
Alamar blue assay was used to evaluate effect of the addition of FBS on metabolic activity of cells. Results presented in Fig. 4C indicate a consistent increase in the alamar blue fluorescence signal, which is consistent with the concentration of applied FBS solution. Additionally, phase contrast images were acquired in order to investigate the effect of the addition of FBS solution on cellular morphology (Fig. 4D-E). It can be seen that seven minutes after the addition of 50% (v/v) FBS solution, an increase in the cellular size took place. There was also a slight increase in the cellular size for 30% (v/v) FBS solution. On the other hand, the addition of medium with 10% (v/v) FBS and medium without FBS did not result in any observable change of cellular shapes. Therefore both alamar blue assay and phase contrast imaging data are consistent with obtained SPR results indicating that there are considerable cellular morphological changes at higher serum concentrations while the changes are barely detectable at lower concentrations.
3.4 Detection of Inhibition of Cellular Metabolism
This work was concluded by testing an agent that affected cellular metabolism and acted as a toxin. Sodium azide (NaN3) is a crystalline salt that is known for inhibition of metabolic activity in mitochondria. By interacting with Fe3+ ions, sodium azide interrupts oxidative phosphorylation and thus inhibits respiration and metabolism (Ishikawa et al. 2006).
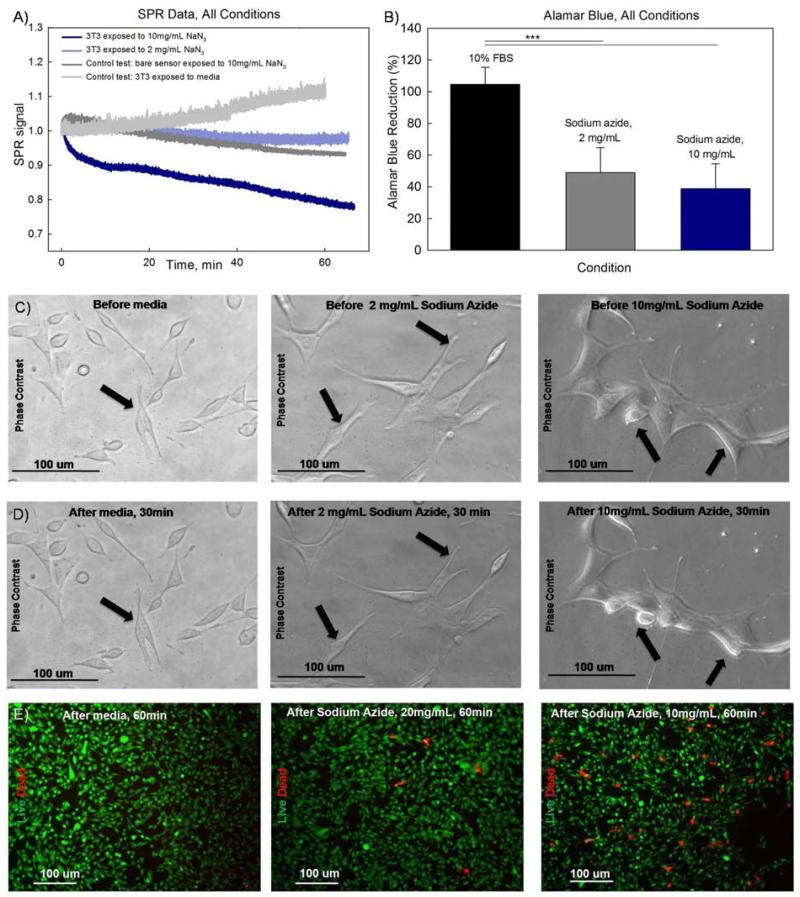
Fig. 5A shows the measured SPR responses while NIH-3T3 cells were exposed to two different concentrations of sodium azide (10 mg/ml and 2 mg/ml) and medium. They are compared with a control SPR response measured in the absence of cells. The measured SPR signature of cellular response to sodium azide exhibits a slow SPR decay. In both cases, the decay in the SPR signal correlates with the concentration of the applied toxin. As expected, the response to the toxin in the absence of cells and the response of cells to medium alone are both flat.
Fig. 5.
Real-time SPR response during cellular exposure to sodium azide complemented with alamar blue assay results and imaging confirming the effect of the toxin on cellular mortphology and viability. (A) SPR responses during cellular exposure to sodium azide (10 mg/ml, 2 mg/ml) and medium versus SPR control response to sodium azide (10mg/ml) in the absence of cells. (B) Alamar blue assay results for the applied concentrations of sodium azide. (C-D) Phase contrast images of NIH-3T3 cells exposed to sodium azide (10mg/ml and 2mg/ml) and standard medium. (C) Images of cells right before addition of sodium azide. (D) Images of the same cells 30 minutes after addition of sodium azide. (E) Live/dead staining of NIH-3T3 cells exposed to sodium azide (10mg/ml, 2mg/ml) and standard medium for one hour.
Phase contrast images were taken to observe the changes in cellular morphology upon activation by sodium azide (Fig. 5C-D). The phase contrast images compare cells before and 30 minutes after addition of the toxin. A higher concentration of toxin resulted in a significant reduction in the cellular size. A smaller concentration of sodium azide only slightly affected the cellular shape while also causing some cellular shrinkage. No change in cellular morphology was observed in the case of the addition of standard medium. Additionally, live/dead staining showed that the addition of sodium azide resulted in an increase in the number of dead cells after one hour of exposure (Fig. 5E).
3.5 Discussion
SPR signals are generally proportional to refractive index (RI) changes on the surface of the sensor and therefore can be influenced by different processes leading to RI change. The experiments reported in this study were conducted in a controlled environment where all sensors were exposed to identical sets of conditions. Moreover, the number of factors that could change RI was either reduced to one (the cell perturbation under study) or if not possible, additional control experiments were performed. The SPR results shown in this study present three distinct optical signatures that correspond to very specific cellular responses induced by applied stimuli. When coated with NIH-3T3 fibroblast cells, SPR-TFBG sensors measure a local refractive index change that is strongly influenced by the density of cells attached to the surface and also to reactions within the attached cells, which lead to changes in the mean refractive index of the cell interior. The 1 cm-long fiber biosensor has a sensitive surface area of 3.92 mm2 and a significant depth of penetration of the plasmonic field, which is approximately 1500 nm for this sensor configuration. As a result, the measured response is the average of the responses from all the cells attached to its sensing area. The density of the cells attached to the fiber surface, changes of the cellular morphology or changes to the intracellular content have considerable effects on the sensor response as the combination of these factors determine the average refractive index seen by the plasmon wave over its propagation length along the fiber.
The optical signature measured during cellular detachment induced by trypsin had a negative SPR signal change with semi-exponential character (Fig. 3A). The signal change was consistently dose-dependent and was not caused by refractive index mismatch or by non-specific adsorption, as confirmed by various controls (Fig. 3B). It was previously reported (Strasser et al. 2011) that the refractive index of intracellular content is on average higher than that of medium, therefore a decrease of the number of attached cells should result in a decrease in the effective SPR index. The observed negative SPR signal change confirms the anticipated refractive index decrease, it is also correlates well with fluorescence images showing a reduction of the number of attached cells.
The optical signature measured during exposure of cells to the FBS solutions had the opposite response, it was positive with a very rapid saturation time (5-10min). The rapid SPR increase in the case of 50% and 30% (v/v) FBS could be attributed to the cellular uptake of nutrients from the added solution and the subsequent cellular spreading. Phase contrast images confirm that there is a slight increase of the cellular shapes right after the addition of FBS solutions with 30% and 50% (v/v) concentrations. The absence of positive SPR signal change after addition of 10% and 0% (v/v) FBS could be explained by the fact that right before the addition of a stimulus the sensors were immersed in a medium solution with the 10% (v/v) concentration of FBS, thus cells did not react to the same or lower concentrations of serum. The decrease in the SPR signal after the addition of medium without FBS could be caused by the cellular uptake of freshly added medium and the subsequent intracellular refractive index decrease.
The addition of sodium azide also produced a distinct optical signature (Fig. 5A) that can be characterized by a SPR signal decay within the first 20 minutes. The recorded negative decay of the SPR signal is consistent with the SPR signature of sodium azide reported by Chabot et al. (Chabot et al. 2009). Phase contrast imaging indicates that there was a significant change in the cellular shape due to exposure to sodium azide at both concentrations. Live/dead fluorescence staining shows that there is also a consistent increase in the number of dead cells after exposure to sodium azide. Alamar blue assay confirms that there is a reduction in the metabolic activity after exposure to sodium azide that correlates well with the SPR data. These findings are making apparent the immediate effect of sodium azide that is expressed through a negative SPR signal change, a decrease of metabolic activity and an increase in the number of dead cells. Thus it can be concluded that the observed SPR signature is representative of immediate cellular response to a toxin, which results in an inhibition of the metabolic activity and cellular death.
All three stimuli, trypsin, FBS, and sodium azide, have had different effects on NIH-3T3 fibroblast cells and resulted in distinguishable optical signatures that are reflective of cellular changes. While exposure to FBS produced the smallest SPR signal change, trypsin caused the strongest negative SPR signal change. SPR-TFBG sensors having several tens of cladding resonances and as a result of it, a wide operating range, are an appropriate choice for tracking such big chemical changes. At the same time, a wide operating range does not compromise the sensitivity of the sensor. The high sensitivity (500 nm/refractive index units (Shevchenko and Albert 2007)) allows for the measurement of intracellular changes of smaller magnitudes, such as those obtained in the case of serum uptake that could be barely visualized using standard imaging tools. In the future, if necessary, a slightly different interrogation setup reported by Caucheteur et al 2011(Caucheteur et al. 2011) may enable the detection of even finer intracellular changes that could be triggered inside cells ensuring that the sensor platform can be employed for a wide spectrum of triggered intracellular responses.
4. Conclusion
This paper presents the findings from one of the first fiber biosensors for real-time analysis of cellular behavior. The SPR-TFBG sensor is a compact and robust fiber biosensor that is manufactured out of a standard telecommunication single-mode fiber coated with a thin layer of gold and interrogated with traditional fiber optic device measurement instrumentation. The presented sensor completely integrated into a conventional cell incubator. To enable cellular adhesion to the surface of the fiber, the sensors were coated with fibronectin. This step was followed by incubation of sensors with NIH-3T3 fibroblast cells and they were exposed to different stimuli, namely, trypsin, FBS and sodium azide, producing three distinct optical signatures. Trypsin cleaved attached cells from the fiber sensors resulting in a sharp decrease of the SPR signal. On the other hand, the addition of serum resulted in a sharp increase of the SPR signal, which could be caused by the uptake of nutrients from the serum solution. The addition of sodium azide resulted in a slow SPR signal decay that could be explained by the inhibition of cellular metabolism upon penetration of sodium azide inside the cells. In all cases, the measured cellular response was immediate and the amplitude of SPR change was correlated with the concentration of applied stimuli. Responses to all stimuli were compared to the set of controls where sensors were exposed to solutions without stimuli, or where only stimuli were applied to the sensors in the absence of cells. Rigorous control experiments enabled the confirmation of measured SPR signals resulting from a specific cellular response to the added stimuli. It was also proven that surface of the sensors can be regenerated for repeated use.
Overall, the results illustrate that it is possible to use the presented fiber biosensor technology to characterize a wide range of cellular responses right from the moment of stimulation. The information is obtained in real-time and the whole measurement method is label-free, providing a unique platform for non-invasive monitoring of cellular behavior. Furthermore, this sensor technology has the potential to be used in the characterization of less pronounced and more complex cellular reactions, such as for triggering very specific signaling pathways that would be challenging to detect using other analytical tools.
Highlights.
Plasmonic fiber sensor can be easily integrated into traditional cell culture equipment.
Plasmonic fiber senor allows for real-time and label-free monitoring of cellular behavior.
Fiber sensor can be used to track cellular response to sodium azide, tripsin and serum.
Surface Plasmon Resonance fiber sensor is a promising approach towards monitoring of intracellular changes.
Acknowledgements
This project was supported by the National Institutes of Health (HL092836; HL099073; AR057837; DE019024; EB008392; GM095906), the National Science Foundation (DMR0847287), the Institute for Soldier Nanotechnologies, the US Army Corps of Engineers, the Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs program, and the Canadian Institute for Photonic Innovations.
Abbreviations
- SPR
Surface Plasmon Resonance
- RI
Refractive Index
- TFBG
Tilted Fiber Bragg Grating
- FBS
Fetal Bovine Serum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
The work presented in the manuscript was a result of collaboration between all authors. AK, GCU, JA and YS outlined the project’s objective. GCU, DFC and YS were involved in the laboratory experiments, troubleshooting and interpretation of the results. YS was responsible for the data analysis. AK, MD, GCU, YS and JA were involved in the interpretation of results, revising and finalizing of the manuscript.
References
- Albert J, Lepinay S, Caucheteur C, DeRosa MC. Methods. 2013 doi: 10.1016/j.ymeth.2013.07.007. accepted. [DOI] [PubMed] [Google Scholar]
- Amin R, Kulkarni A, Kim T, Park SH. Current Applied Physics. 2012;12(3):841–845. [Google Scholar]
- Bhatta D, Stadden E, Hashem E, Sparrow IJG, Emmerson GD. Sensors and Actuators B-Chemical. 2010;149(1):233–238. [Google Scholar]
- Caucheteur C, Shevchenko Y, Shao LY, Wuilpart M, Albert J. Optics Express. 2011;19(2):1656–1664. doi: 10.1364/OE.19.001656. [DOI] [PubMed] [Google Scholar]
- Caucheteur C, Voisin V, Albert J. Optics Express. 2013;21(3):3055–3066. doi: 10.1364/OE.21.003055. [DOI] [PubMed] [Google Scholar]
- Chabot V, Cuerrier CM, Escher E, Aimez V, Grandbois M, Charette PG. Biosensors and Bioelectronics. 2009;24(6):1667–1673. doi: 10.1016/j.bios.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Chabot V, Miron Y, Charette PG, Grandbois M. Biosensors & Bioelectronics. 2013;50:125–131. doi: 10.1016/j.bios.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Delport F, Pollet J, Janssen K, Verbruggen B, Knez K, Spasic D, Lammertyn J. Nanotechnology. 2012;23(6) doi: 10.1088/0957-4484/23/6/065503. [DOI] [PubMed] [Google Scholar]
- Fang Y, Ferrie A. FEBS Letters. 2008;582(5):558–564. doi: 10.1016/j.febslet.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Fang Y, Ferrie AM, Fontaine NH, Mauro J, Balakrishnan J. Biophysical Journal. 2006;91(5):1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JA, Steemers FJ, Walt DR. Analytical Chemistry. 2000;72:5618–5624. doi: 10.1021/ac0008284. [DOI] [PubMed] [Google Scholar]
- François A, Boehm J, Oh SY, Kok T, Monro TM. Biosensors and Bioelectronics. 2011;26(7):3154–3159. doi: 10.1016/j.bios.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Drug discovery today. 2012;17(3-4):173–181. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hide M, Tsutsui T, Sato H, Nishimura T, Morimoto K, Yamamoto S, Yoshizato K. Analytical Biochemistry. 2002;302(1):28–37. doi: 10.1006/abio.2001.5535. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Zhu BL, Maeda H. Toxicology and Industrial Health. 2006;22:337–341. doi: 10.1177/0748233706071737. [DOI] [PubMed] [Google Scholar]
- Jang HS, Park KN, Kang CD, Kim JP, Sim SJ, Lee KS. Optics Communications. 2009;282(14):2827–2830. [Google Scholar]
- Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, Khademhosseini A. Lab on a Chip. 2011;11(10):1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SB, Bae H, Koo KI, Dokmeci MR, Ozcan A, Khademhosseini A. Journal of laboratory automation. 2012;17(1):43–49. doi: 10.1177/2211068211426695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepinay S, Staff A, Ianoul A, Albert J. Biosensors and Bioelectronics. 2013 doi: 10.1016/j.bios.2013.08.058. in press. [DOI] [PubMed] [Google Scholar]
- Ligler SF, Taitt CR. Gulf Professional Publishing. 2002. [Google Scholar]
- Lin B, Li P, Cunningham BT. Sensors and Actuators B: Chemical. 2006;114(2):559–564. [Google Scholar]
- Maltais JS, Denault JB, Gendron L, Grandbois M. Apoptosis. 2012;17(8):916–925. doi: 10.1007/s10495-012-0737-y. [DOI] [PubMed] [Google Scholar]
- Mosher DF. Annual Review Medicine. 1984;35:561–575. doi: 10.1146/annurev.me.35.020184.003021. [DOI] [PubMed] [Google Scholar]
- Pollet J, Delport F, Janssen KPF, Tran DT, Wouters J, Verbiest T, Lammertyn J. Talanta. 2011;83(5):1436–1441. doi: 10.1016/j.talanta.2010.11.032. [DOI] [PubMed] [Google Scholar]
- Robelek R. Bioanalytical Reviews. 2009;1(1):57–72. [Google Scholar]
- Ruan Y, Foo TC, Warren-Smith S, Hoffmann P, Moore RC, Ebendorff-Heidepriem H, Monro TM. Optics Express. 2008;16(22):18514–18523. doi: 10.1364/oe.16.018514. [DOI] [PubMed] [Google Scholar]
- Scibek JJ, Randle DH, Bunch TA, Fang Y, Frutos AG. Corning Inc.; 2012. p. 17. [Google Scholar]
- Sharma RAK, Gupta BD. Journal of Optics A: Pure and Applied Optics. 2007;9(7):682–687. [Google Scholar]
- Shevchenko Y, Ahamad UN, Ianoul A, Albert J. Optics Express. 2010a;18(19):20409–20421. doi: 10.1364/OE.18.020409. [DOI] [PubMed] [Google Scholar]
- Shevchenko Y, Chen C, Dakka M, Albert J. Optics Letters. 2010b;25(5):637–639. doi: 10.1364/OL.35.000637. [DOI] [PubMed] [Google Scholar]
- Shevchenko Y, Francis TJ, Blair DAD, Walsh R, DeRosa MC, Albert J. Analytical Chemistry. 2011;83(18):7027–7034. doi: 10.1021/ac201641n. [DOI] [PubMed] [Google Scholar]
- Shevchenko YY, Albert J. Plasmon resonances in gold-coated tilted fiber Bragg gratings. Optics Letters. 2007;32(3):211–213. doi: 10.1364/ol.32.000211. [DOI] [PubMed] [Google Scholar]
- Silva LIB, Ferreira FDP, Freitas AC, Rocha-Santos TAP, Duarte AC. Talanta. 2009;80(2):853–857. doi: 10.1016/j.talanta.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Smietana M, Bock WJ, Mikulic P, Ng A, Chinnappan R, Zourob M. Optics Express. 2011;19(9):7971–7978. doi: 10.1364/OE.19.007971. [DOI] [PubMed] [Google Scholar]
- Strasser SD, Shekhawat G, Rogers GD, Dravid VP, Taflove A, Backman V. Optics Letters. 2011;37(4):506–508. doi: 10.1364/OL.37.000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney S, Falch BMH, Hjelme DR, Stokke BT. Analytical Chemistry. 2009;81(9):3630–3636. doi: 10.1021/ac900019k. [DOI] [PubMed] [Google Scholar]
- Vala M, Robelek R, Bockova M, Wegener J, Homola J. Biosensors & Bioelectronics. 2013;40(1):417–421. doi: 10.1016/j.bios.2012.07.020. [DOI] [PubMed] [Google Scholar]
- Wang XD, Wolfbeis OS. Analytical Chemistry. 2013;85(2):487–508. doi: 10.1021/ac303159b. [DOI] [PubMed] [Google Scholar]
- Yanase Y, Araki A, Suzuki H, Tsutsui T, Kimura T, Okamoto K, Nakatani T, Hiragun T, Hide M. Biosensors and Bioelectronics. 2010;25(5):1244–1247. doi: 10.1016/j.bios.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Yanase Y, Hiragun T, Yanase T, Kawaguchi T, Ishii K, Hide M. Allergology International. 2013;62(2):163–169. doi: 10.2332/allergolint.12-RA-0505. [DOI] [PubMed] [Google Scholar]
- Yanase Y, Suzuki H, Tsutsui T, Hiragun T, Kameyoshi Y, Hide M. Biosensors and Bioelectronics. 2007;22(6):1081–1086. doi: 10.1016/j.bios.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Yin MJ, Wu C, Shao LY, Chan WKE, Zhang AP, Lu C, Tam HY. Analyst. 2013;138(7):1988–1994. doi: 10.1039/c3an36791f. [DOI] [PubMed] [Google Scholar]
- Zheng GA, Lee SA, Antebi Y, Elowitz MB, Yang CH. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):16889–16894. doi: 10.1073/pnas.1110681108. [DOI] [PMC free article] [PubMed] [Google Scholar]