Summary
The response to endoplasmic reticulum (ER) stress relies on activation of unfolded protein response (UPR) sensors, and the outcome of the UPR depends on the duration and strength of signal. Here we demonstrate a novel mechanism that attenuates the activity of the UPR sensor inositol-requiring enzyme 1α (IRE1α). A resident ER protein disulfide isomerase, PDIA6, limits the duration of IRE1α activity by direct binding to cysteine148 in the luminal domain of the sensor, which is oxidized when IRE1 is activated. PDIA6-deficient cells hyper-respond to ER stress with sustained auto-phosphorylation of IRE1α and splicing of XBP1 mRNA, resulting in exaggerated up-regulation of UPR target genes and increased apoptosis. In vivo, PDIA6-deficient C. elegans exhibits constitutive UPR and fails to complete larval development, a program that normally requires the UPR. Thus, PDIA6 activity provides a mechanism that limits UPR signaling and maintains it within a physiologically appropriate range.
Introduction
Various changes in the cellular environment trigger the Unfolded Protein Response (UPR), from metabolic changes, through limitation of normal protein folding, to the accumulation of misfolded protein in the endoplasmic reticulum (ER). The resulting imbalance between folding requirements and capacity activates the UPR machinery to up-regulate chaperones and folding enzymes, to reduce the flux of newly synthesized secretory proteins and to augment the disposal capacity. One major sensor is the inositol-requiring enzyme 1α (IRE1α), which in response to stress in the ER lumen undergoes trans-auto-phosphorylation and oligomerization (Walter and Ron, 2011), leading to specific splicing of the transcription factor X-box binding protein 1 (XBP1)(Cox and Walter, 1996), more promiscuous cleavage of other transcripts (Hollien and Weissman, 2006) and activation of the c-Jun N-terminal kinase (JNK) pathway (Urano et al., 2000), depending on the nature, duration, and magnitude of the signal (Han et al., 2009; Upton et al., 2012). While some of the activities of IRE1 are adaptive and allow the cells to return to homeostasis, attenuation of IRE1 is similarly critical to cells, since excessive activity of IRE1 can lead to reduced stress tolerance and downstream apoptosis (Chawla et al., 2011; Lin et al., 2007; Rubio et al., 2011).
The ability of UPR to serve as a homeostatic device and not just a stress response depends on the transient nature of its signaling. UPR attenuation is particularly important in normal physiology, as in pancreatic β cells reacting to fluctuations of blood glucose, or during the differentiation of B cells into antibody-secreting cells (Iwakoshi et al., 2003). In both cases, the physiological signal results in large flux of proteins through the secretory pathway and unchecked response to this flux may cause severe pathology.
IRE1 can be deactivated by dephosphorylation (Han et al., 2009; Welihinda et al., 1998), or by interaction with the BCL-2-related proteins and BI-1 (Lisbona et al., 2009), PUMA or BIM (Rodriguez et al., 2012). These attenuators operate on the cytoplasmic domains of IRE1, and it is not well understood how deteriorating folding conditions in the lumen trigger their action. BiP is the only protein currently known to interact with the luminal domain of IRE1, affecting the activation step. Yet, if the initiation of signaling involves communication from the luminal domains of IRE1 to the cytosolic domains, one can postulate that attenuation of signaling may also involve luminal components that restore the sensing mechanism to its basal level. In this work we show that a luminal enzyme, PDIA6, indeed controls the duration of IRE1 activity.
PDIA6, also known as P5 or TXNDC7, is one of more than 20 protein disulfide isomerases (PDIs) in the eukaryotic ER (Hatahet and Ruddock, 2009). It is an active oxidoreductase with similar enzymatic properties to other PDIs (Alanen et al., 2006), yet it does not seem to be involved directly in protein folding. First, a trapping mutant of PDIA6 that can form mixed disulfides but cannot resolve them, failed to identify discrete substrates (Jessop et al., 2009). Second, knockdown of PDIA6 expression in hepatocytes, pancreatic β cell lines and various fibroblastic lines had no effect on the secretion or biosynthesis of a variety of proteins (Rutkevich et al., 2010). In this work we show that PDIA6 is important for limiting UPR signaling within physiologically tolerable range, by binding a specific cysteine in the luminal domain of IRE1, in particular in the activated state. Through this interaction PDIA6 acts to terminate IRE1 signaling.
Results
Depletion of PDIA6 causes higher responsiveness to ER stress and decreased proliferation
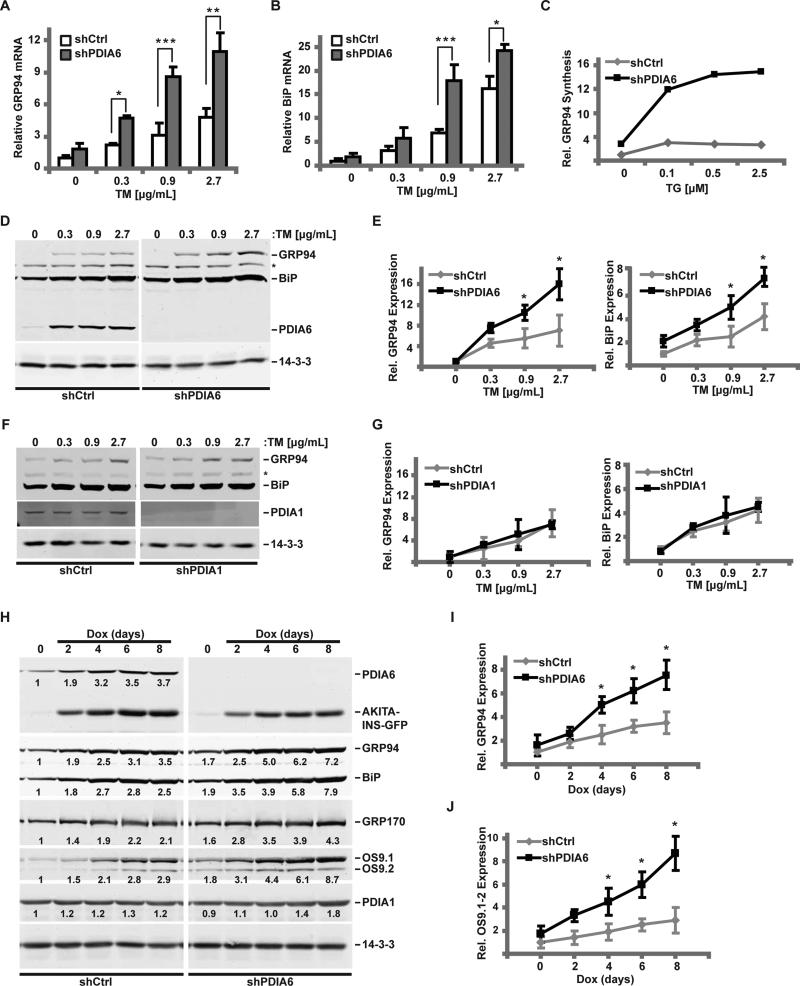
To assess the role of PDIA6 in the UPR we measured induction of several UPR target genes by the glycosylation inhibitor tunicamycin (TM) in 3T3 cells, either sufficient or deficient in PDIA6, by following assays: RT-qPCR of transcripts (Fig. 1A-B), BiP promoter-luciferase reporter activity (Fig. S1A-B), new synthesis of GRP94 by metabolic labeling (Fig. 1C, S1C) and steady state protein levels by immunoblots (Fig. 1D-J). Without overt stress, there was only minimal induction of either BiP or GRP94 expression in PDIA6-depleted cells compared to PDIA6-sufficient cells, by either the transcriptional or the protein readouts. This minimal induction was small compared to that caused by depletion of BiP or GRP94 (Fig. S1G).
Figure 1. Loss of PDIA6 causes an augmented unfolded protein response.
(A-B) 3T3 cells expressing a non-targeting (shCtrl) or a PDIA6-targeting shRNA (clone 1) were treated with the indicated dose of TM for 6hrs. GRP94 (A) and BiP (B) mRNA levels were detected by RT-qPCR, normalized to β-actin, and their expression in shCtrl cells without stress was set at 1. Plots are means±SD, n=3. Significant differences between shPDIA6 and shCtrl conditions are indicated by asterisks. (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 (student t test)).
(C) shCtrl or shPDIA6 3T3 cells were exposed to the indicated dose of TG for 6hrs and labeled with [35S]Met/Cys for 30 min. Relative GRP94 synthesis was determined by immunoprecipitation and normalized to untreated shCtrl cells.
(D) shCtrl or shPDIA6 (clone 1) 3T3 cells were treated with varying concentrations of TM for 16 hrs and levels of GRP94, BiP or PDIA6 were determined with αKDEL antibody. 14-3-3 was used as a loading control. *, an unknown KDEL-positive protein whose expression is unchanged.
(E) Quantitation of GRP94 and BiP levels from panel D. Values are means±SD relative to DMSO-treated shCtrl cells (*, p ≤ 0.05; n=3).
(F-G) shCtrl or shPDIA1 3T3 cells were treated with TM and analysed as in panels D-E.
(H-J) Akita-INS cells were transduced with shCtrl or shPDIA6 (clone 1) lentiviruses and Akita insulin expression was induced with 2μg/mL doxycycline (Dox). The relative levels of indicated proteins were measured by immunoblotting and normalized to the loading control, 14-3-3 (numbers below bands). The induction of GRP94 (I) or OS9.1/OS9.2 (J) was quantified as in panels D-E.
See also Figs. S1-S2.
In contrast, BiP or GRP94 induction by TM was significantly higher in PDIA6-depleted cells, shown best by comparing the slopes of the relative expression as a function of stress (Fig. 1). The enhanced UPR responsiveness was also seen with a second ER stressor, the calcium pump inhibitor thapsigargin (TG)(Fig. 1C; Fig. S1C-E). Enhanced responsiveness was not limited to murine 3T3 cells and was also observed in human 293T cells (Fig. S1A,D,E) and in a rat pancreatic β cell line (Fig. 1H-J; Fig. S1F). In both 3T3 and 293T cells (requiring different shRNA sequences for efficient knockdown), BiP promoter was activated more in PDIA6-depleted cells (Fig. S1A-B). The enhanced UPR responsiveness is a direct consequence of PDIA6 depletion, because the same phenotype was observed with 3 distinct targeting sequences (Figs. 1 and S1) and it was rescued by re-expressing shRNA-resistant coding sequence of PDIA6 in the silenced cells (Fig. S1H-I).
Silencing of PDIA1 (P4HB), which is 2.5X more abundant than PDIA6 in 3T3 cells (Schwanhausser et al., 2011), or PDIA3, which has a known role in folding of glycoproteins (Solda et al., 2006), had no effect on UPR responsiveness (Fig. 1F-G and Fig. 3 below). Thus, the ability to modulate UPR responsiveness is not a generic property of PDIs, but a specific function of PDIA6.
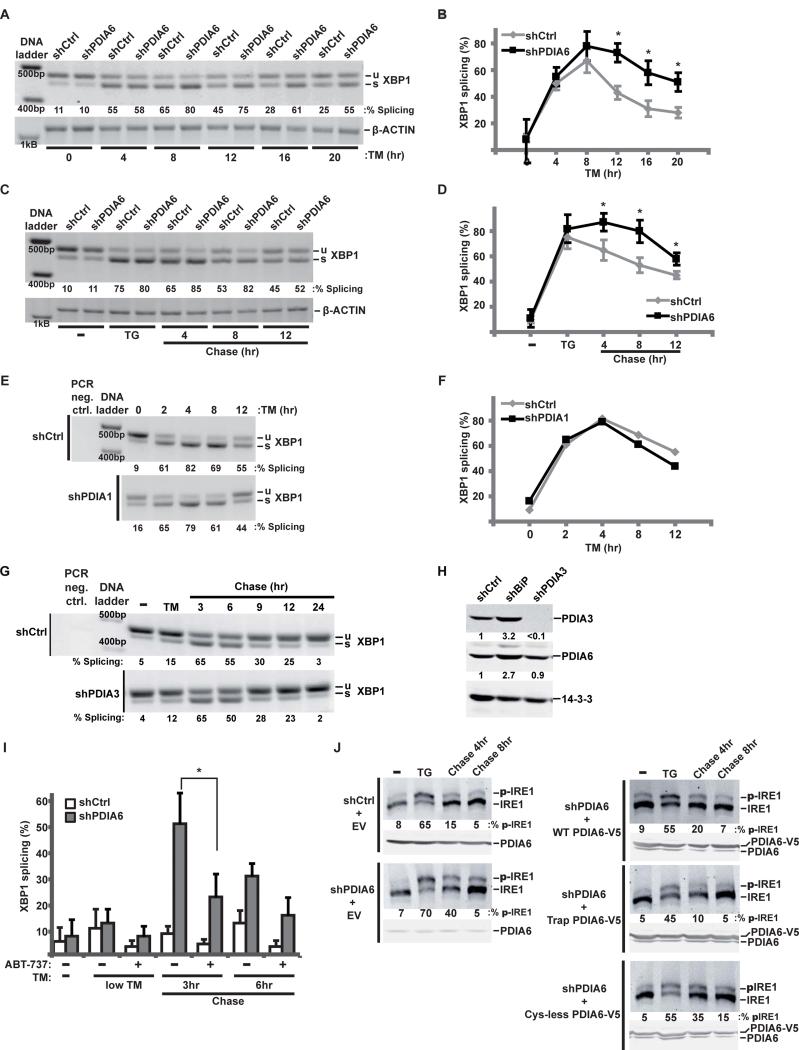
Figure 3. PDIA6 affects the decay of IRE1 signaling.
(A-B) shCtrl or shPDIA6 3T3 cells were incubated with 200 ng/mL TM for the indicated times and unspliced (u) and spliced (s) XBP1 mRNA was amplified by RT-PCR. β-actin served as control for RNA recovery. Means±SD are plotted in B (*, p ≤ 0.05; n=3).
(C-D) shCtrl or shPDIA6 3T3 cells were pulsed with 100nM TG for 4hrs, then “chased” for the indicated times in medium without TG. XBP1 splicing was assayed as in A. Separate experiments were quantified in (D). (*, p ≤ 0.05; n=3).
(E-F) 3T3 cells expressing shCtrl or shPDIA1 were subjected to continuous TM stress and analysed as in A. Percentage of XBP1 splicing is plotted in (F).
(G) ShCtrl or shPDIA3 3T3 cells were first exposed to TM, then chased in fresh growth medium for the indicated times. Total RNA was analysed for XBP1 splicing as in A.
(H) Protein extracts from shCtrl, shBiP or shPDIA3 cells were immunoblotted to determine the level of expression of PDIA3 and PDIA6. 14-3-3 served as loading control. Note that: 1) PDIA3 depletion is not accompanied by induction of PDIA6; 2) both PDIA3 and PDIA6 are over-expressed in BiP-depleted cells, presumably via general UPR.
(I) shCtrl or shPDIA6 3T3 cells were pre-treated (or left untreated) with the Bcl-2 inhibitor ABT-737 (10μM for 30 min). Cells were then pulsed with 50μg/mL TM for 2hrs and chased for the indicated times in fresh medium. XBP1 splicing was assayed as in A. Under such modest ER stress, XBP1 splicing in PDIA6-sufficient cells is minimal and the difference with PDIA6-deficient cells is more dramatic (*, p ≤ 0.05; n=3).
(J) ShCtrl or shPDIA6 (clone 3) 293T cells were transfected with the indicated V5-tagged PDIA6 constructs (EV, empty vector; WT, wild-type PDIA6; Trap, C58A-C193A PDIA6; Cys-less, C55A-C58A-C190A-C193A PDIA6). Cells were incubated with 100 nM TG for 2hrs and then “chased” in fresh medium for 4 or 8 hrs. Phosphorylated IRE1 was resolved by Phos-tag gels. Blots with α-PDIA6 are given below each Phos-tag panel, to show the relative levels of endogenous and exogenous PDIA6. Numbers under the panels, percentages of phosphorylated IRE1 as determined by densitometry. This calculation is conservative, given the variability of the unphosphorylated band across the samples.
See also Figs. S3-S4.
To test whether PDIA6 also regulates the magnitude of UPR induced by misfolded proteins, we used a clone of INS-1 cells that inducibly express misfolded C96Y insulin (Hartley et al., 2010), whose expression causes ER stress and consequent activation of UPR signaling in both the Akita mouse and in human diabetic patients (Steiner et al., 2009). As expected, induction of C96Y insulin caused upregulation of GRP94, BiP and PDIA6, while PDIA1 was unaffected. Silencing PDIA6 further increased the magnitude of this response (Fig. 1H-J). Similarly, there was higher induction (Fig. 1H-J) of the ERAD protein OS-9 (Alcock and Swanton, 2009). These effects were not due to involvement of PDIA6 in the folding or disposal of insulin (Fig. S2D). Thus, PDIA6 controls cellular responsiveness to the stress of expressing an unfolded protein.
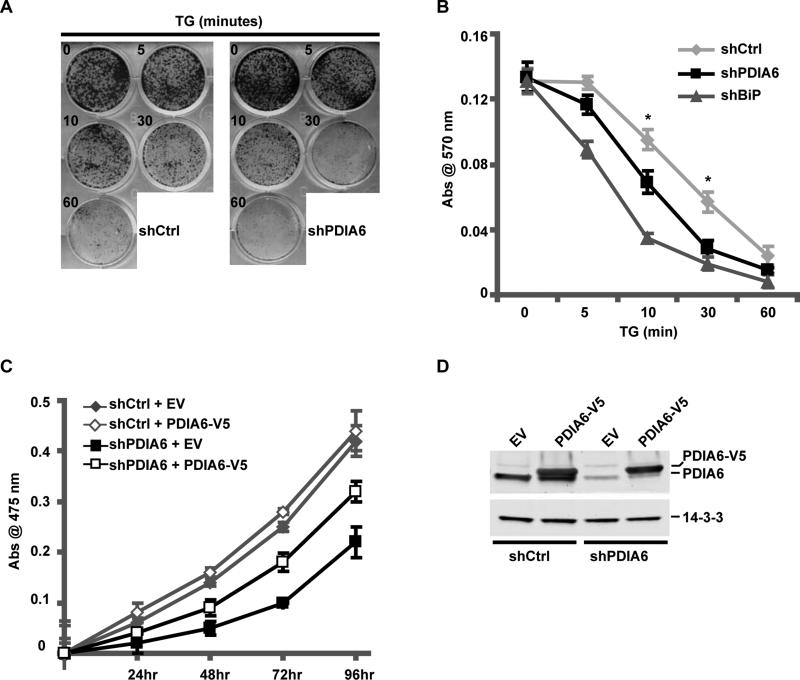
We next asked whether enhanced UPR in PDIA6-silenced cells accentuated the anti-proliferative effect of ER stress, a common UPR outcome (Urano et al., 2000; Yoshida et al., 2000). We found a modest, yet consistent decrease in colony-forming ability of PDIA6-deficient HeLa cells after exposure to brief, mild ER stress (Fig. 2A-B), although the effect was not as detrimental as silencing BiP (Fig. 2B and see (Eletto et al., 2012; Wang et al., 2010). Similarly, PDIA6-depleted 3T3 cells were 1.6x more sensitive to continuous exposure to TG than control cells. Silencing of PDIA6 also caused transient decrease in 293T cell proliferation (Fig. 2C). Complementation of PDIA6-silenced 293T cells with shRNA-resistant PDIA6 improved their proliferation (Fig 2C-D), showing that the growth effect was due to loss of PDIA6 itself. We conclude that PDIA6 expression is important in controlling the magnitude of induction of UPR genes in response to ER stress and that silencing it causes hyper-response to typical ER stress, augmented sensitivity to stress-induced apoptosis, and decreased cell proliferation.
Figure 2. Depletion of PDIA6 leads to slower growth and hyper-sensitivity to ER stress.
(A) HeLa cells, stably expressing shPDIA6 (clone 3) or shCtrl were plated at 5×103 cells/35mm dish, treated with 50nM TG for the indicated time, then returned to growth medium for 6 days. Plates were assessed for colony formation by staining with crystal violet. HeLa cells were used because they form distinct colonies.
(B) Quantitation of crystal violet-stained colonies from HeLa cells expressing shCtrl, shPDIA6 and a shRNA clone targeting BiP (shBiP). Plates from experiments such as in A were lysed in 2% w/v SDS and absorbance at 570 nm was measured. Values are means±SD (*, p ≤ 0.05; n=4).
(C) ShCtrl or shPDIA6 (clone 3) 293T cells were selected for 5 days with 2μg/mL puromycin, and then transfected with either an empty vector (EV) or a plasmid expressing V5-tagged PDIA6 (PDIA6-V5). The proliferation rates were determined by an XTT assay.
(D) Immunoblots of protein extracts from the 24h time point in (C), to measure PDIA6 expression (both endogenous and exogenous).
See also Fig. S2.
PDIA6-dependent UPR hyper-responsiveness is not due to altered protein folding
Because PDIA6 is an oxidoreductase, the heightened UPR responsiveness in its absence could be due to misfolding of proteins that depend on this enzyme. Therefore, we tested how absence of PDIA6 impacted the folding capacity of the ER. Depletion of PDIA6 had minimal effect on the rate of protein synthesis or on the quantity and pattern of secreted proteins (Fig. S2A-C). Past experiments did not find any secretory protein that depended on PDIA6 in either hepatocytes or fibroblasts, using either a physical association (Jessop et al., 2009) or functional (Rutkevich et al., 2010) tests. Nonetheless, we asked whether processing of proinsulin, a disulfide-rich secretory protein, depended on PDIA6 in a β cell line. The absence of PDIA6 did not significantly alter the extent or timing of proinsulin processing (Fig. S2D). Thus, though it remains formally possible that the phenomena observed are due to misfolding of PDIA6 clients, none has been identified using either global or substrate-specific approaches and we surmise that the UPR phenotype is not secondary to protein misfolding.
PDIA6 controls the inactivation of IRE1 and the decay of XBP1 splicing
We next tested the effects of PDIA6 on UPR sensors, in particular IRE1. Interestingly, loss of PDIA6 did not significantly affect the ability to IRE1 to splice XBP1 or the amplitude of the splicing. Instead, it affected the duration of splicing (Fig. 3A-B). XBP1 splicing decayed within a few hours despite continued presence of ER stress, as shown before (Lin et al., 2007); this decay was slower in PDIA6-depleted cells (Fig. 3A-B). To ask if this was due to continued activation of UPR signaling or to decreased attenuation, we followed the kinetics of splicing after the ER stress was removed. The percentage of spliced XBP1 remained high in PDIA6-depleted cells 12 hours after removal of TG, compared to control cells (Fig. 3C-D). The effect on attenuation was a property of PDIA6, and wasn't observed after depletion of either PDIA1 or PDIA3 (Fig. 3E-H).
Modulation of the timing of XBP1 splicing has been reported, but with cytosolic proteins of the BH3-only family (Hetz et al., 2011). To investigate the relationship between the cytosolic and luminal modulators we measured the duration of XBP1 splicing in PDIA6-depleted cells with and without ABT-737, a BH3 domain mimetic that antagonizes the effect of the BH3-only proteins on IRE1 (Rodriguez et al., 2012). We found that while XBP1 splicing was decreased by ABT-737, its duration was still extended by PDIA6 depletion (Fig. 3I), arguing that the luminal and cytosolic modulators of splicing act independently.
XBP1 splicing is preceded by activation of IRE1, through dimerization and self-phosphorylation. Moreover, non-phosphorylating IRE1 mutants can be defective in the attenuation of XBP1 splicing (Rubio et al., 2011). Thus we determined, using Phos-Tag gels (Yang et al., 2010), the time course of IRE1 phosphorylation under stress. In the absence of PDIA6, the phospho-IRE1 species persisted at 4 hours after removal of TG compared to the already attenuated phosphorylation in PDIA6-sufficient cells (Fig. 3J). Conversely, over-expression of the C58A-C193A mutant of PDIA6, which acts as a substrate trap (Jessop et al., 2009) (Fig. S4A), accelerated the dephosphorylation of IRE1. On the other hand, over-expression of a mutant PDIA6 lacking all four Cys in the thioredoxin domains (C55A-C58A-C190A-C193A; Fig. S4A) did not accelerate the inactivation of IRE1 (Fig. 3J). These results indicate that the enzymatic activity of PDIA6 is important for the regulation of IRE1 activation and that PDIA6 affects the most proximal activity of IRE1, auto-trans-phosphorylation.
Since down-regulation of PDIA6 enhanced the induction of UPR target genes by the misfolded Akita insulin (Fig. 1H), we assessed whether this was mediated by prolonged activation of IRE1. Continuous induction of the Akita mutant led to activation of XBP1 splicing after 2 days. In the absence of PDIA6, the magnitude of XBP1 splicing was much higher (Fig. S3A-B). Interestingly, IRE1 signaling did not decay during continuous over-expression of Akita insulin over 8 days, perhaps because accumulation of the Akita mutant is not synchronous. However, like in transient treatment of cells with TG or TM (Fig. 3C, I), transient over-expression of the Akita insulin caused enhanced and prolonged activation of IRE1 activity in PDIA6-depleted Akita INS-1 cells compared to controls (Fig. S3E-F). We next asked whether this exaggerated IRE1 signaling affected the cytotoxic effect of accumulation of misfolded insulin (Steiner et al., 2009). We found that PDIA6 depletion in INS1 cells caused enhanced phosphorylation of JNK (Fig. S3C-D), indicating that PDIA6 affects multiple activities of IRE1 and not just its splicing activity.
Our data thus suggest that PDIA6 ablation does not significantly induce IRE1 activation nor does it affect the onset of IRE1 signaling. Rather, it affects the inactivation of IRE1, as measured by attenuation of phosphorylation of the sensor, XBP1 splicing and JNK activation, in response to both chemical stress and misfolded proteins.
PDIA6 interacts directly with IRE1
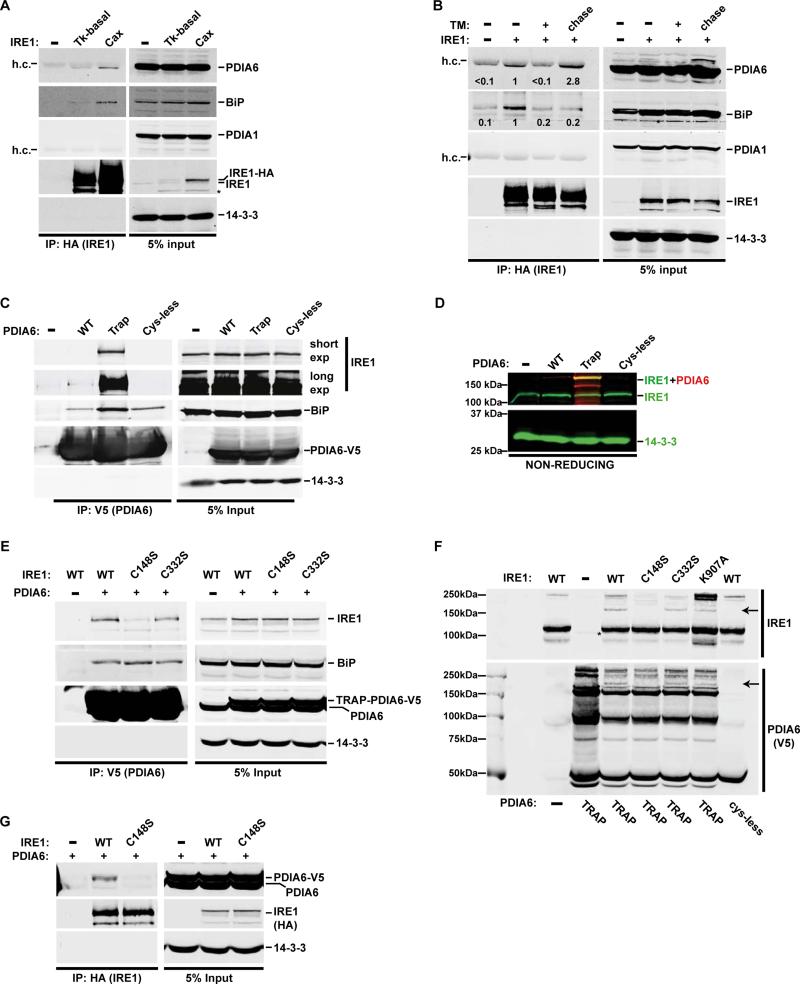
Because PDIA6 controls the decay of IRE1 activity, we tested whether it interacts directly with the sensor. Because normally low abundance of IRE1 is important for its activation properties (see below), exogenous IRE1-HA was expressed in 293T cells under the control of either a weak (pTK) or strong promoter (pCAX). pTK-driven expression of IRE1 was sufficiently low to pre-empt spontaneous activation, while pCAX-driven expression led to strong spontaneous activation even in the absence of overt stress (Fig. S5 and see (Han et al., 2009)). PDIA6 was indeed enriched in IRE1-HA immunoprecipitates in a manner dependent on the level of IRE1 expression (Fig. 4A). Conversely, endogenous IRE1 co-immunoprecipitated with V5-tagged PDIA6 (Fig. 4C). This interaction was specific, as PDIA1 did not co-precipitate with IRE1 (Fig. 4A-B), and it was affected by the state of activity of IRE1: when UPR was transiently activated by a short TM treatment, the PDIA6-IRE1 interaction was initially reduced, and then restored to a higher level after TM was removed (Fig. 4B). Conversely, the interaction of BiP with IRE1 was reduced upon TM treatment (as expected (Urano et al., 2000)), and was not restored within the time frame of this experiment (Fig. 4B).
Figure 4. PDIA6 and IRE1 interact directly through mixed disulfide bonds.
(A) HA-tagged IRE1 was expressed in 293T cells from either weak (tyrosine kinase, Tk-basal) or strong (cytomegalovirus enhancer chicken β-actin, Cax) promoter. PDIA6, BiP, and PDIA1 co-precipitating with IRE1-HA were detected by immunoblotting. 14-3-3 served as a control for non-specific interactions with IRE1. h.c., heavy chain of the αHA antibody.
(B) 293T cells expressing HA-tagged IRE1 (Tk-basal promoter) were either not untreated or acutely exposed to 2 μg/mL TM for 3hrs. The indicated samples were then chased for 16hrs in fresh medium. Lysates were analysed as in A.
(C) 293T cells were either not transfected (−) or transfected with WT, a trapping (Trap) or Cys-less versions of V5-tagged PDIA6 cDNA. IRE1 and BiP co-precipitated with PDIA6-V5 were detected by immunoblotting.
(D) Cell lysates from panel C were analysed by non-reducing gels, followed by dual-color immunoblots with αIRE (green) and αV5 (red) antibodies to detect mixed disulfide between IRE1 and PDIA6. The yellow signal depicts the co-migrated IRE1-PDIA6 bands. 14-3-3 (green) served as a loading control.
(E) The trapping PDIA6-V5 mutant was expressed in 293T cells alongside the indicated IRE1 mutants. Cell lysates were analysed as in (C).
(F) Cell lysates, generated as in (E), were resolved on a non-reducing gel and immunoblotted to detect IRE1-PDIA6-V5 mixed disulfides. The arrows point at the same co-migrating bands. See Fig. S4B for a color version of the same blot.
(G) WT or C148S IRE-HA were expressed in 293T cells with WT PDIA6-V5 and immunoprecipitated with a αHA antibody. IRE1-bound PDIA6 was detected by immunoblotting.
See also Fig. S4.
Because IRE1 attenuation depended on the thioredoxin motifs of PDIA6 (Fig. 3J), we used the trapping and Cys-less mutants as bait for IRE1. Much more endogenous IRE1 was co-immunoprecipitated with the trapping mutant, whereas no IRE1 was detectably co-immunoprecipitated with the Cys-less PDIA6 version (Fig. 4C). Interestingly, BiP interacted with PDIA6 under all conditions, even with the Cys-less mutant (Fig. 4C), as was previously shown (Jessop et al., 2009). Expression of a tagged trap PDIA6, but not the Cys-less mutant of PDIA6, led to appearance of a new 180kD band in lysates on non-reducing gels (Fig. 4D), consistent with a mixed disulfide between the 135kD IRE1 and the 50kD PDIA6. These data indicate that IRE1 and PDIA6 interact directly via a disulfide bond.
PDIA6 controls attenuation of IRE1 signaling by covalent interaction with oxidized Cys148
Mammalian IRE1 has three cysteines in its luminal domain – Cys109, Cys148 and Cys332 (Fig. S4A). None are directly resolved in the crystal structure, though they are clearly in exposed vertices of the domain, accessible for interaction (Zhou et al., 2006). To map the residue which PDIA6 targets we substituted each Cys with Ser. As shown in Fig. 4E, the C148 was the main mediator of the association with PDIA6, while C332 and C109 were less important. Immunoprecipitation with IRE1-HA confirmed that C148S IRE1 mutant did not interact with PDIA6 detectably (Fig. 4G). In non-reducing gels of whole cell lysates, the ~180kD mixed disulfide was seen with WT IRE1, but not with C148S IRE1 nor with the Cys-less PDIA6 (Fig. 4F; Fig. S4B). This adduct was still observed, albeit at lower intensity, with C332S or C109S IRE1 as well as with the ribonuclease mutant K907A (Han et al., 2009). We conclude that PDIA6 primarily targets C148 in the luminal domain of IRE1.
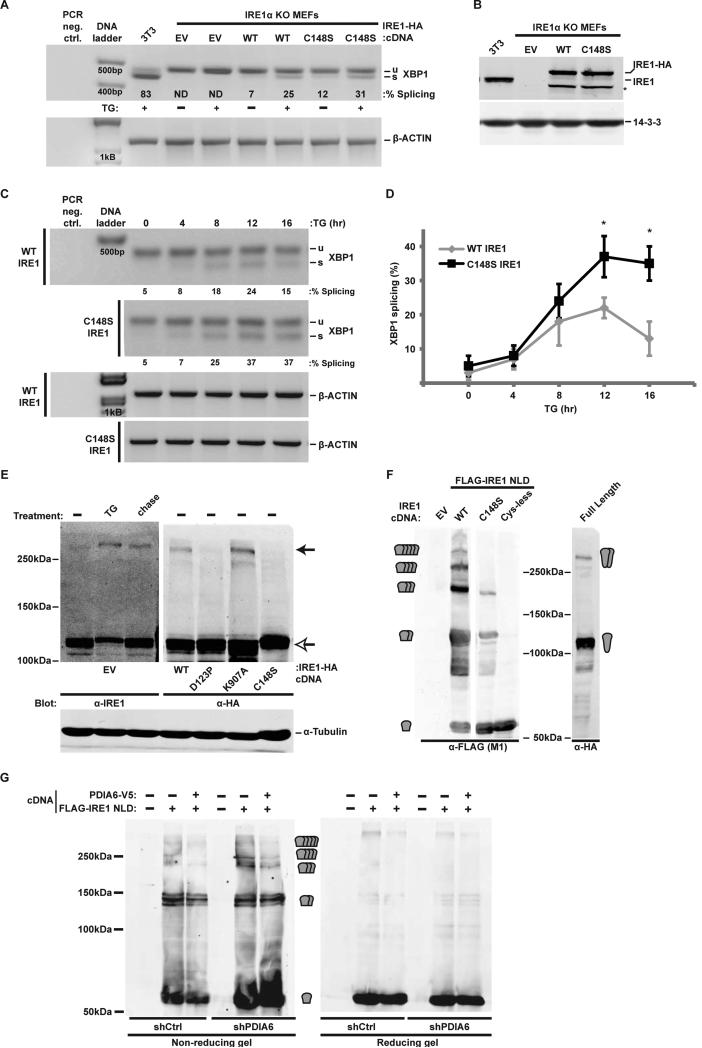
Based on the above result, C148S IRE1 should phenocopy the ablation of PDIA6. To test this prediction we expressed wild type or Cys mutants of IRE1 in IRE1−/− cells (Urano et al., 2000) and measured XBP1 splicing. Both WT- and C148S IRE1-complemented cells displayed basal splicing activity, which increased 3-fold upon challenge with TG (Fig. 5A). The protein levels of WT and C148S IRE1 in these cells were similar (Fig. 5B). In a time course experiment, C148S IRE1 displayed more prolonged splicing activity than WT IRE1 (Fig. 5C-D). Thus, C148S IRE1 was active but attenuated more slowly, as predicted if C148 is the target of PDIA6.
Figure 5. The reactivity of Cys148 is important for the activation kinetics of IRE1.
(A) IRE1 knockout (KO) MEFs stably complemented with WT or C148S IRE1-HA were treated with 300nM TG for 2hrs. RNA samples were then assayed for XBP1 splicing. Percentage of splicing is indicated in each lane. TG-treated 3T3 served as positive controls. EV, empty cDNA vector. ND, not detectable.
(B) Cell lysates from the same samples used in (A) were immunoblotted to determine the level of expression of the rescuing IRE1-HA. 3T3 lysates served as reference for endogenous IRE1 expression. Note that the expression of each rescuing protein as well as the endogenous IRE1 are similar. *, unknown product of the expression plasmid, presumably a cleaved IRE protein.
(C) Extent of XBP1 splicing in IRE1-HA-rescued IRE1 KO MEFs was determined upon persistent exposure to 300nM TG for the indicated times. β-actin served as loading control.
(D) Means±SD of experiments as in (C) are plotted (*, p ≤ 0.05; n=3).
(E) A C148-dependent disulfide forms during activation of IRE1. 293T cells were transfected with either empty vector (EV) or with the indicated IRE1-HA cDNAs (under control of the pCAX promoter). Cells were treated with 100nM TG for 2hrs, followed by 16hr chase in fresh medium where indicated. Reactive cysteines were alkylated with NEM prior to cell lysis and endogenous or HA-tagged exogenous IRE1 were detected by non-reducing gel. α-tubulin (which does not contain reactive cysteines) was used as control for the electrophoretic shift.
(F) 293T cells were transiently transfected with full length IRE1 or with FLAG-tagged IRE1 luminal domain (NLD) that was either WT, C148S or lacked all three cysteines (C109/148/332S). Expression of all NLDs was limited to the ER with a C-terminal KDEL peptide. Protein extracts were resolved by non-reducing PAGE and analysed by immunoblot. EV, empty vector transfection. The predicted sizes of monomeric NLD or multimeric complexes are indicated at the sides of the blots.
(G) shCtrl and shPDIA6 293T cells were transfected with WT IRE1 NLD or mock transfected. Protein extracts were analysed as in F. Co-expressed PDIA6-V5 complemented the silenced enzyme in shPDIA6 cells in reducing multimer formation.
See also Fig. S5.
In addition to complementation of splicing, we tested the ability of C148S IRE1 to affect growth of IRE1−/− MEFs. In a colony formation assay like that shown in Fig. 2, C148S-complemented cells grew significantly less than cells complemented with WT IRE1 (0.81±0.039 vs. 1.15±0.038 A570 units; p<0.01; n=3). Thus, the C148S mutant showed a proliferation defect similar to that of PDIA6-deficient cells.
Finally, we asked what could trigger the association of PDIA6 with C148 so as to only affect the inactivation but not the onset phase of the IRE1 response. Mindful that PDIs engage only thiolates (Hatahet and Ruddock, 2009), we investigated whether C148 is oxidized upon IRE1 activation and therefore engages PDIA6. A ~280kD IRE1-positive non-reduced species, presumably activated IRE1 dimers (Liu et al., 2002), is evident upon treatment with TG and partially disappears upon resolution of the stress (Fig. 5E). This ~280kD band is also seen in cells over-expressing IRE1 even in the absence of overt stress (Fig. 5E), when the ectopic IRE1 is activated spontaneously (Fig. S5). This band is a disulfide species of IRE1 because it is abolished on reducing gels (not shown), depends on Cys148 and is absent in the non-dimerizing mutant D123P (Fig. 5E). Thus, C148 is oxidized during IRE1 activation and becomes a target for PDIA6.
Since the 280kD band is only a modest fraction of total IRE1, we expressed the isolated N-terminal luminal domain (NLD), which was shown to dimerize efficiently (Liu et al., 2003). In response to ER stress, this NLD was robustly oxidized into bands that migrated as dimers and higher orders (Fig. 5F). Mutation of Cys148 inhibited the formation of these oxidized IRE1 forms (Fig. 5F) and overexpression of PDIA6 promoted their collapse to monomeric IRE1 (Fig. 5G). These data suggest that PDIA6 promotes deactivation of IRE1 by reducing its oligomers.
PDIA6 also affects PERK, but not ATF6 signaling
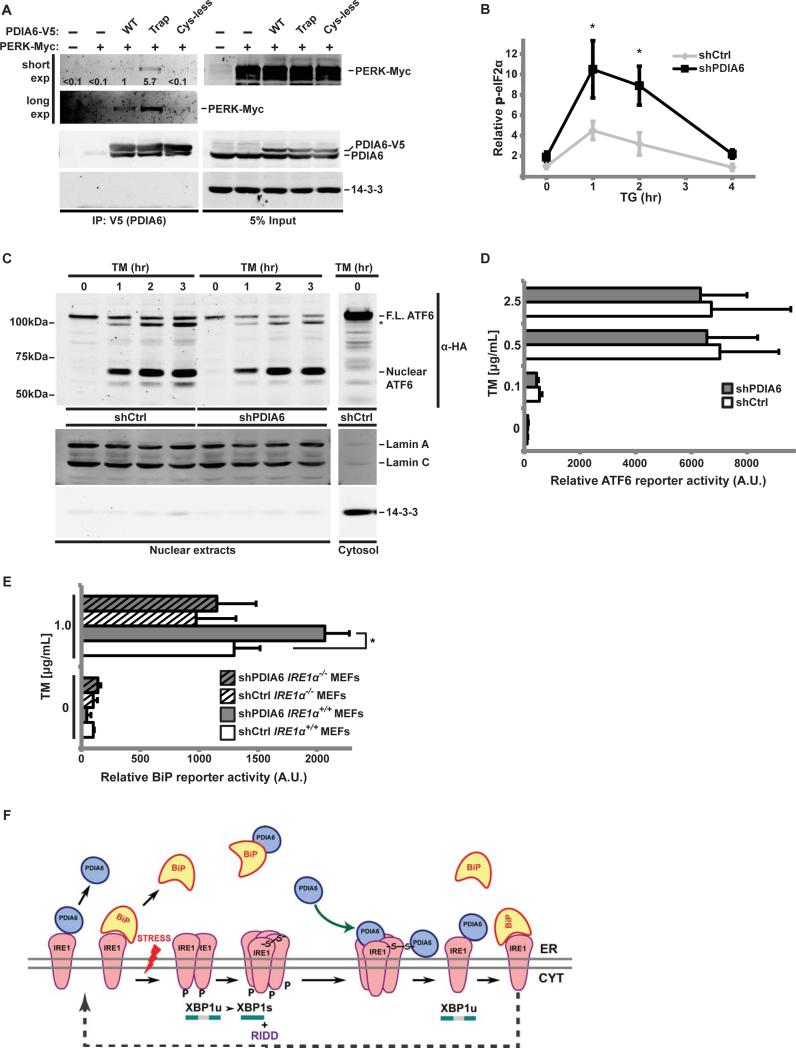
Because the luminal domains of IRE1 and PERK are homologous (Zhou et al., 2006), we next assessed whether PDIA6 affects only IRE1 or also the other UPR branches. When exogenous myc-tagged PERK was expressed in 293T cells together with wild type, the trapping mutant, or Cys-less V5-tagged PDIA6, PERK was detected in V5-PDIA6 immunoprecipitates and its co-association increased with the trap mutant, but not the Cys-less PDIA6 (Fig. 6A). More importantly, there was functional association of PDIA6 with PERK, as depletion of PDIA6 caused increased and prolonged phosphorylation of eIF2α, a proximal mediator of PERK signaling (Walter and Ron, 2011)(Fig. 6B). Thus, PDIA6 associates with the luminal domains of two homologous sensors. Further studies of the PDIA6-PERK interaction are ongoing.
Figure 6. PDIA6 also affects PERK signaling but not ATF6 activity.
(A) Extracts of 293T cells expressing the indicated versions of PDIA6-V5 (WT, client-trapping (Trap) or Cys-less mutants), along with a Myc-tagged PERK, were immunoprecipitated with αV5 antibody and blotted to detect co-precipitated Myc-tagged PERK. 14-3-3 served as specificity control.
(B) shCtrl or shPDIA6 3T3 cells were exposed to 1μM TG for the indicated times. Cell lysates were then immunoblotted to detect total or Ser-51-phosphorylated eIF2α. Means±SD of the phospho-signal/total eIF2α ratios are plotted (*, p≤0.05; n=3). Untreated shCtrl samples served as internal reference.
(C) 293T cells stably expressing HA-tagged ATF6 were infected with lentivirus containing shCtrl or shPDIA6 (clone 3). After exposure to TM for the indicated times, nuclear fractions were isolated as in (Li et al., 2000) and immunoblotted to detect the active, nuclear fragment of ATF6. LaminA/C and 14-3-3 were used as nuclear or cytosolic markers, respectively. Cells were pretreated with 1μM MG-132 to prevent proteasomal degradation of nuclear ATF6. F.L, full length ATF6.
(D) The 5X ATF6 site luciferase reporter was expressed in shCtrl and shPDIA6 3T3 cells, which were then treated with the indicated doses of TM for 18hrs. Means±SD of relative Firefly luciferase activity, normalized to Renilla activity, are shown (n=4).
(E) IRE1α+/+ and IRE1α−/− MEFs stably expressing shCtrl or shPDIA6 were transiently transfected with a luciferase reporter driven by a minimal BiP promoter, exposed to TM and analysed as in (D) (*, p ≤ 0.05).
(F) Proposed model for the attenuation of IRE1 by PDIA6. The luminal domain of inactive IRE1 is occupied by BiP. Some fraction of IRE1 is independently bound reversibly by PDIA6. Upon ER stress BiP dissociates, IRE1 dimerizes, oligomerizes and becomes an active kinase and a ribonuclease, splicing XBP-1. We propose that during the activation sequence, IRE1 becomes disulfide-bonded via its luminal Cys148. This makes activated IRE1 a target for PDIA6, which then reduces the disulfide and converts oligomeric IRE1 back to the monomeric form, which is available for another cycle of activation. PDIA6 is also known to bind BiP, but this interaction is Cys-independent.
See also Fig. S6.
In contrast to PERK and IRE1, several lines of evidence show that PDIA6 does not affect ATF6 signaling. First, the generation of the active nuclear fragment of ATF6 is not augmented in PDIA6-silenced cells (Fig. 6C). Second, an ATF6-sensitive reporter shows no increased activity in shPDIA6 cells, whether or not they are treated with TM (Fig. 6D; Fig. S6A). The BiP promoter is known to depend on ATF6 (Adachi et al., 2008; Shoulders et al., 2013) as well as IRE1 branch (Lee et al., 2003). Indeed, the PDIA6-dependent exaggerated responsiveness is reduced in IRE1−/− MEFs, as shown by BiP promoter assays (Fig. 6E and Fig. S6B). We conclude that IRE1 is a major contributor to the PDIA6–mediated enhanced UPR.
All together, we conclude that PDIA6 affects UPR signaling via the IRE1 and PERK sensors but not via ATF6. Because PERK and IRE1 do not hetero-dimerize, and expression or activation of PERK does not impact the ribonuclease activity of IRE1 (Lin et al., 2009), the data suggest that PDIA6 impacts each sensor independently.
Loss of PDIA6 in C. elegans leads to larval lethality and hyper-activation of UPR
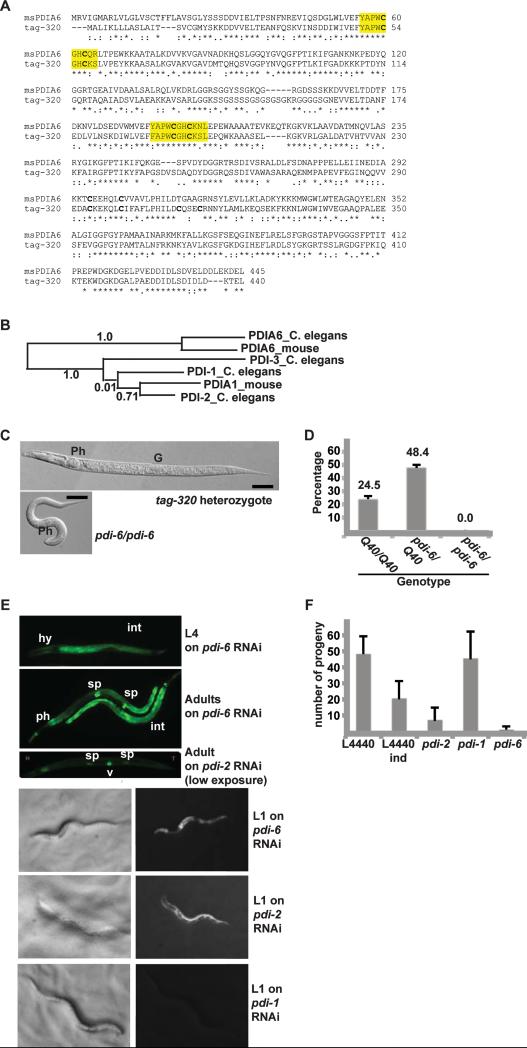
To ask whether regulation of IRE1 by PDIA6 was physiologically important in an intact organism, we searched for its homologue in C. elegans. Three C. elegans PDI genes have been characterized (Winter et al., 2007). A fourth, temporarily assigned gene 320 (tag-320) shows high sequence homology to the other three PDI proteins, in particular through its two thioredoxin domains. Sequence alignment and phylogenetic analysis (Fig. 7A-B) show that tag-320 (PDIA6_C. elegans) forms a distinct branch with the mammalian PDIA6 and is much closer to murine PDIA6 than to murine PDIA1, or any of the other three C. elegans PDIs (Fig. 7B). Henceforth, this gene is designated pdi-6.
Figure 7. The C. elegans PDIA6 homologue is an essential gene whose silencing induces UPR.
(A) Sequence alignment of murine PDIA6 and C. elegans tag-320, using Lalign v.2.1.30. The CXXC motifs and their flanking residues are highlighted. Symbols under the alignment indicate identity/conservation of residues.
(B) Phylogenetic analysis (Phylogeny.fr) of C. elegans PDI-1, -2, -3 and the PDIA6 homologue, and mouse PDIA1 and PDIA6. Line lengths and numbers indicate relative sequence relatedness.
(C) Comparison of heterozygous pdi-6/Q40::YFP and pdi-6/pdi-6 homozygous larva at an L2 stage. Ph, pharynx. G, gonadal precursor. The pdi-6/pdi-6 homozygous larva is at the same age post-hatching as the heterozygous larva, without apparent morphological defects. Bars, 50μ.
(D) Developmental arrest of pdi-6 homozygous larvae. Eggs laid by pdi-6/Q40::YFP adults were allowed to develop for 3 days and the number of L4 or older animals of each genotype was scored. The values are percentages of the starting numbers of embryos, means±SD (n=4). No pdi-6/pdi-6 animals developed to L4 stage.
(E) Induction of a UPR reporter in larvae and adult animals by RNAi. YAR72 worms harboring the phsp-4::GFP reporter (zcIs4) on an rrf-3 (ok1426) RNAi-sensitized background were exposed from eggs to pdi-1, -2 or -6 RNAi and representative animals were imaged. Note reporter induction in worms depleted of pdi-6 and the distinct cellular pattern (intestine and pharynx for pdi-6 vs hypodermis for pdi-2 in L1 animals). The exposure of the adult pdi-2 panel was adjusted to show the individual vulval cells. Ph, pharynx. Sp, spermatheca. Hy, hypodermis. Int, intestine.
(F) Fecundity of animals subjected to RNAi of the indicated genes. YAR72 animals (as in E) were grown on the indicated RNAi bacteria for 1-2 generations, singled and their progeny enumerated.
L4440, empty vector control; L4440 ind, control animals with spontaneously induced reporter. Note the correspondence between low brood size on pdi-2, pdi-6 RNAi plates and in spontaneously induced animals and the induction of the UPR reporter. RNAi of pdi-1 does not induce this reporter significantly. Data are means±SD (n >7 individual animals per genotype).
See also Fig. S7.
The C. elegans pdi-6 is an essential gene, since a strain homozygous for a deletion (ok1373) is larval lethal (WormBase version WS238, Fig. 7C-D and Fig. S7A-B). The deletion removes much of exons 3-4, retaining only the N-terminal 200 amino acids of PDIA6 (out of 440, WormBase). We maintain this deletion as a balanced strain across from a wild type X chromosome marked by a Q40::YFP transgene (Gidalevitz et al., 2006). Heterozygous animals segregate non-fluorescent pdi-6/pdi-6 L1 homozygotes at a frequency of 15.1+1.0%, a lower transmission ratio of the deleted allele than the 25% expected from simple Mendelian segregation. This does not appear to be due to the failure of embryos to hatch, but we cannot yet exclude maternal contribution, germ cell or fertilization effects.
To assess the organismal effects of pdi-6 deletion, we scored development of embryos produced by heterozygous animals. Although at the first larval stage, L1, pdi-6/pdi-6 homozygotes appeared grossly normal compared to pdi-6/Q40::YFP heterozygotes (Fig. S7C), none reached L4 or young adult stages (Fig. 7D). Instead, they arrested at early larval stages (Fig. 7C and Fig. S7A-B), indicating that PDIA6 is necessary during larval development. Interestingly, many arrested larvae appeared small and had molting defects, varying from an arrest at the L1/L2 molt to the inability to fully shed the old cuticle after the molt (Fig. S7A-B).
Since C. elegans development requires functional UPR (Shen et al., 2001), and lack of PDIA6 in mammalian cells leads to augmented rather then deficient IRE1 signaling, we asked if depletion of PDIA6 leads to hyper-induction of the physiological UPR in C. elegans larvae. UPR-dependent induction of the hsp-4 (BiP) gene depends on active IRE1 and XBP-1 as described in (Calfon et al., 2002; Shen et al., 2001). Induction of an hsp-4 transcriptional reporter, zcIs4 (Calfon et al., 2002) was assessed upon down-regulation of various PDI genes by RNAi. pdi-6 RNAi induced this UPR reporter in intestinal, hypodermal and pharyngeal cells, and also increased its constitutive expression in spermathecae (Fig. 7E). Down-regulation of two other distinct PDI genes, pdi-1 or pdi-3, did not induce the UPR reporter (pdi-1 RNAi shown in Fig. 7E), while RNAi to pdi-2, the closest homologue to the murine PDIA1, gave rise to strong reporter induction but in different tissues compared to pdi-6 (Fig. 7E). Thus, loss of PDIA6 leads to UPR induction in C. elegans. Since no overt stress was needed for this phenotype, it is likely that the observed reporter induction corresponds to enhancement of the physiological UPR, similar to that observed in mammalian INS-1 cells (Fig. 1H).
Finally, we observed a marked reduction in fecundity of pdi-6 RNAi animals (Fig. 7F), consistent with the lethality of the pdi-6 deletion. Loss of fecundity appears to be a consequence of chronic UPR induction, as also it was also observed in pdi-2 RNAi animals and in those with a spontaneous induction of the reporter. We conclude that pdi-6 is an essential gene in C. elegans that has been conserved in evolution, perhaps because of its role in controlling the UPR. The sensitivity of C. elegans larval development to both exaggerated IRE1 activity (due to deletion of pdi-6) and to absence of UPR signaling (Shen et al., 2001) suggests that development of the organism requires precise control over UPR signaling.
Discussion
In this study we show that a luminal protein disulfide isomerase, PDIA6, attenuates UPR signaling by interacting with the luminal domain of activated IRE1 and the related PERK, but not with ATF6. The primary target residue of PDIA6 on IRE1 is Cys148 (Fig. 6F), which forms a disulfide bond upon activation-induced oligomerization. PDIA6 apparently reduces this disulfide, thus promoting the inactivation of IRE1. In the absence of either PDIA6 or its target Cys148, the inactivation of IRE1 is significantly delayed, as reflected by sustained self-phosphorylation and XBP1-splicing activity and by increased JNK phosphorylation. Loss of PDIA6 does not in itself cause significant ER stress and does not impair secretory protein folding (this work and (Rutkevich et al., 2010)). Thus, PDIA6 affects IRE1 (and PERK) directly, limits its activation and helps return it to the inactive form. PDIA6 is the first selective attenuator of UPR that acts from the lumen of the ER, where the stress signal is generated.
Like PDIA6-deficient cells, ATF6α−/− cells also display prolonged IRE1 activity (Wu et al., 2007). Yet, we find that PDIA6 does not directly affect ATF6 processing. We think that the phenotypic similarity is due to functional overlaps between the UPR branches in transcriptional activation of targets (Wu et al., 2007). Although many UPR genes are mostly ATF6 targets (Adachi et al., 2008; Shoulders et al., 2013), they are also activated through the IRE1 branch (Lee et al., 2003). For example, we show that the mammalian BiP promoter is regulated by IRE1.
The absence of PDIA6 exaggerates several of the activities of IRE1. This effect appears to be maladaptive, as PDIA6-deficient cells are more sensitive to chemical ER stressors or expression of mutant insulin, and PDIA6-deficient worms fail to complete their development. This maybe counter-intuitive, particularly since spliced XBP1 is often regarded as protective (Han et al., 2009). However in C. elegans, spliced XBP1 can be either protective or detrimental, based on the tissue where it is expressed (Taylor and Dillin, 2013). The negative outcome can be explained in part by activation of apoptotic mediators like JNK as the strength of IRE1 signal increases. Because PDIA6 senses a disulfide that characterizes IRE1 oligomers, and these mediate toxic rather than adaptive outcomes (Han et al., 2009; Upton et al., 2012), the effect of PDIA6 on RIDD activity of IRE1 (Hollien and Weissman, 2006) should be investigated.
In C. elegans, ablation of PDIA6 causes exaggerated physiological UPR, as judged by the hsp-4 reporter, much like it does in mammalian cells, and results in developmental arrest. Thus, the action of PDIA6 on the UPR is evolutionarily conserved. The UPR signaling was shown previously to be required for development, with IRE1 deficiency causing mild inhibition of larval development, and combined IRE1 and PERK deficiency causing arrest (Shen et al., 2001; Shen et al., 2005). Because the augmented UPR signaling seen here upon PDIA6 depletion is also associated with larval arrest, it appears that both activating the UPR through IRE1 and PERK, and attenuating it through PDIA6 must be balanced within a narrow range to execute proper development.
If the binding avidity of PDIA6 to IRE1 is regulated by ER stress, what feature changes during IRE1 activation to allow PDIA6 to engage Cys148? Since PDIs engage either disulfides or thiolates (Hatahet and Ruddock, 2009), the redox state of Cys148 must be different in activated IRE1. Indeed, we show that C148 becomes engaged in disulfide bonds during activation-induced dimerization of IRE1, converting it into a potential substrate for PDIA6. This finding is consistent with the crystal structure of IRE1: C148 is located at the edge of the molecular surface that mediates IRE1 dimerization and remains exposed even in the oligomeric forms (Credle et al., 2005; Zhou et al., 2006). Furthermore, the luminal domain of overexpressed IRE1 forms a dimer by hydrophobic interaction that is stabilized by intermolecular disulfide bridges between Cys148 and Cys332 (Liu et al., 2003). Remarkably, these cysteines are not required for the stress-sensing function of IRE1 (Liu et al., 2003). We show that chemical reactivity of at least one of these Cys determines the duration of IRE1 activation, and propose that these cysteines are conserved in evolution because they help regulate IRE1 in vivo.
PDIA6 had been shown to binds to the chaperone BiP when BiP is engaged in folding clients (Jessop et al., 2009). We verified this association and its non-covalent nature, which suggests that the mode of association of PDIA6 with BiP is different from that of PDIA6 and IRE1. Moreover, because the PDIA6-IRE1 complex is maintained even upon complete RNAi depletion of BiP (Fig. S4C), and because BiP still binds C148S IRE1, the current data predicts the existence of two separate populations of PDIA6, one that modulates IRE1 and one that is a cofactor of BiP (Jessop et al., 2009). These populations may exist in equilibrium that may affect the availability of PDIA6 towards IRE1 (and PERK).
Aside from BiP and PDIA6, a number of proteins bind to IRE1 and regulate the duration of its activity (reviewed in (Hetz et al., 2011). The pro-apoptotic proteins BAK and BAX are necessary for full activation of IRE1 (Hetz et al., 2006) and the chaperone Hsp72 enhances XBP1 splicing and inhibits the apoptotic pathway (Gupta et al., 2010). On the other hand, recruitment of phosphatases (Qiu et al., 2010) and the Bcl-2 family protein BI-1 limit IRE1 activity (Lisbona et al., 2009) and the BH3 domain proteins BIM and PUMA are needed to prolong its splicing activity (Rodriguez et al., 2012). All these modulators affect IRE1 by binding to its cytosolic domains, while BiP and PDIA6 regulate IRE1 signaling from the luminal domain and therefore can be sensitive to the folding conditions in the lumen. The evidence in this work is consistent with independent action of the effectors that inactivate IRE1 on each side of the ER membrane, separately ensuring termination of signaling to guard against excessive UPR.
This work highlights the multilayer regulation required to match both the magnitude and the type of UPR to the nature of ER stress (Walter and Ron, 2011). Imbalance in this regulation, whether in failing to respond to the changing folding conditions or in failing to attenuate the response, has dramatic consequences to cellular health and to organismal development. As we show, the consequences of sensor activation are not monolithic and we propose that regulating the duration of signaling is one aspect of determining whether the response is adaptive or deleterious. Given the coordination of growth and differentiation signals in metazoan, the activities of the UPR sensors have an even more complex, cell non-autonomous regulation (Taylor and Dillin, 2013), akin to that shown for the heat shock response (Prahlad et al., 2008), whose mechanisms and implications are yet to be uncovered. Answers to these questions are particularly important when UPR is aimed at promoting physiological functions, such as growth and differentiation, or at coping with chronic ER stress, like in protein misfolding diseases. In these situations, the function of PDIA6 as an IRE1 attenuator that limits excessive UPR is essential.
Experimental Procedures
Lentiviral shRNA clones (Sigma) were used for gene knockdowns as previously reported (Eletto et al., 2012). See Supplemental Information for the target sequences.
Expression and association of proteins were determined by co-immunoprecipitation and immunoblotting as in (Eletto et al., 2012). IR-labeled secondary antibodies and Odyssey imaging system (Licor) were used for quantitation. XBP1 splicing was assayed by the PCR assay of (Calfon et al., 2002). The procedures for tissue culture, molecular biology, IRE1 activity measurements and immunoblots have been described before; all procedures are detailed in the Supplemental Information.
Supplementary Material
Highlights.
The ER resident enzyme PDIA6 is an attenuator of the Unfolded Protein Response
PDIA6 interacts covalently with cysteine 148 in the luminal domain of activated IRE1
The interaction facilitates the decay of active IRE1 without affecting its onset
The worm orthologue of PDIA6 is an essential gene and its loss causes excessive UPR
Acknowledgments
We thank Drs. D. Ron (Univ. of Cambridge), L. Hendershot (St. Jude Children's Hospital), A. Volchuk (Univ. of Toronto), N. Bulleid (Univ. of Glasgow), E. Snapp (A. Einstein College of Medicine of Yeshiva Univ.), T. Iwawaki (Gumna Univ.) and R. Morimoto (Northwestern Univ.) for generous gifts of cell lines, bacteria, plasmids and antibodies. Some strains were provided by the CGC, funded by the NIH (P40 OD010440). We also thank S. Boyle and R. Nobel for technical help, Drexel Univ. Cell Imaging Center and Dr. J. Burkhardt for the use of microscopes and Drs. Ron, Burkhardt, C Thorpe, A. Gentilella, M. Marzec, and R. Kalb for comments and suggestions. The authors declare no financial conflicts of interest. This work was funded by NIH grants AI-18001 and GM-77480.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- Alanen HI, Salo KE, Pirneskoski A, Ruddock LW. pH dependence of the peptide thiol-disulfide oxidase activity of six members of the human protein disulfide isomerase family. Antioxid Redox Signal. 2006;8:283–291. doi: 10.1089/ars.2006.8.283. [DOI] [PubMed] [Google Scholar]
- Alcock F, Swanton E. Mammalian OS-9 is upregulated in response to endoplasmic reticulum stress and facilitates ubiquitination of misfolded glycoproteins. J Mol Biol. 2009;385:1032–1042. doi: 10.1016/j.jmb.2008.11.045. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Nat Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Maganty A, Eletto D, Dersh D, Makarewich C, Biswas C, Doroudgar S, Glembotski CC, Argon Y. Limitation of Individual Folding Resources in the ER Leads to Outcomes Distinct from the Unfolded Protein Response. Journal of Cell Science. 2012;125:4865–4875. doi: 10.1242/jcs.108928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010;8:e1000410. doi: 10.1371/journal.pbio.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley T, Siva M, Lai E, Teodoro T, Zhang L, Volchuk A. Endoplasmic reticulum stress response in an INS-1 pancreatic beta-cell line with inducible expression of a folding-deficient proinsulin. BMC Cell Biol. 2010;11:59. doi: 10.1186/1471-2121-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J Cell Sci. 2009;122:4287–4295. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee AS. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PloS one. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Wong HN, Schauerte JA, Kaufman RJ. The protein kinase/endoribonuclease IRE1alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J Biol Chem. 2002;277:18346–18356. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- Liu CY, Xu Z, Kaufman RJ. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1alpha. J Biol Chem. 2003;278:17680–17687. doi: 10.1074/jbc.M300418200. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding Q, Chen Y, Wu D, Xie D, Lin X, et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal. 2010;3:ra7. doi: 10.1126/scisignal.2000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez DA, Zamorano S, Lisbona F, Rojas-Rivera D, Urra H, Cubillos-Ruiz JR, Armisen R, Henriquez DR, Cheng EH, Letek M, et al. BH3-only proteins are part of a regulatory network that control the sustained signalling of the unfolded protein response sensor IRE1alpha. EMBO J. 2012;31:2322–2335. doi: 10.1038/emboj.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol. 2011;193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkevich LA, Cohen-Doyle MF, Brockmeier U, Williams DB. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Mol Biol Cell. 2010;21:3093–3105. doi: 10.1091/mbc.E10-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR, 3rd, Su AI, Kelly JW, Wiseman RL. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda T, Garbi N, Hammerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Park SY, Stoy J, Philipson LH, Bell GI. A brief perspective on insulin production. Diabetes Obes Metab 11 Suppl. 2009;4:189–196. doi: 10.1111/j.1463-1326.2009.01106.x. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang M, Ye R, Barron E, Baumeister P, Mao C, Luo S, Fu Y, Luo B, Dubeau L, Hinton DR, et al. Essential role of the unfolded protein response regulator GRP78/BiP in protection from neuronal apoptosis. Cell Death Differ. 2010;17:488–498. doi: 10.1038/cdd.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18:1967–1977. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AD, McCormack G, Page AP. Protein disulfide isomerase activity is essential for viability and extracellular matrix formation in the nematode Caenorhabditis elegans. Dev Biol. 2007;308:449–461. doi: 10.1016/j.ydbio.2007.05.041. [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yang L, Xue Z, He Y, Sun S, Chen H, Qi L. A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PloS one. 2010;5:e11621. doi: 10.1371/journal.pone.0011621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Nat Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.