Abstract
The authors report the recognition of a G-quadruplex formed by four repeat human telomeric DNA with aminosugar intercalator conjugates. The recognition of G-quadruplex through dual binding mode ligands significantly increased the affinity of ligands for G-quadruplex. One such example is a neomycin-anthraquinone 2 which exhibited nanomolar affinity for the quadruplex, and the affinity of 2 is nearly 1000 fold higher for human telomeric G-quadruplex DNA than its constituent units, neomycin and anthraquinone.
Nucleic acids rich in guanines have been shown to form secondary DNA structures called G-quadruplexes[1] resulting from unimolecular, bimolecular or tetramolecular association of guanine rich nucleic acid strands. In that various telomeric sequences form G-quadruplexes in vitro, they are hypothesized as being essential for regulating the cellular metabolism.[2] Evidence supporting the use of a G-quadruplex structure in eukaryotic biology for developing novel drug therapeutics is now quite significant. Specifically, the formations of G-quadruplex structures has been observed in human cells, and RNA G-quadruplexes have also been identified in translational processes [3] [4].
Targeting G-quadruplex by small molecules is a recognized and effective method for inhibiting telomerase, a ribonucleoprotein.[5] Indeed, telomerase[6] inhibition has provided encouraging results in cancer treatments. The approach relies upon the inactivation of telomerase through the use of small molecules that induce G-quadruplex[5] formation. The G-quadruplex structure effectively blocks telomerase binding which requires the association with single stranded DNA.
Among the host of ligands known to recognize G-quadruplex DNA with varying affinities, most bind with micromolar Kds.[7] The predominance of these mid-to-low affinity binders underscores the need for creating higher affinity G-quadruplex targeting ligands. In this report, we detail our explorations of covalent combination of dual recognition nucleic acid motifs as an effective method for enhancing small molecule G-quadruplex affinity.
Anthraquinones were one of the first ligands shown capable of stabilizing G-quadruplexes and inhibiting telomerase.[8] The major structural characteristic common to G-quadruplex binding ligands is a large planar surface.[9] The diversity within G-quadruplex groove structure, however, makes high affinity groove binders quite rare.[10, 11] Aminosugars have recently identified as non-planar G-quadruplex binding ligand with groove binding as their potential binding mode.[12] Using dual[13, 14, 15, 16, 17] or multiple recognition techniques,[18] we have previously tailored high affinity ligands to recognize specific nucleic acid structures[19] with high affinity.[20, 21, 22, 23] In this paper, we describe our methods for expanding the binding potential of non-planar aminosugars conjugated to planar nucleic acid binding aromatic moieties (anthraquinone, pyrene, napthalenediimide, BQQ), in targeting a human telomeric G-quadruplex.
In that human telomeric DNA consists of a hexamer nucleotide repeat unit d(TTAGGG), a four-repeat 22mer DNA d(AGGGTTAGGGTTAGGGTTAGGG) has been used extensively as a mimic. In the presence of sodium ions, this unimolecular quadruplex adopts an antiparallel conformation (Fig. 1) of four unequal grooves. The individual grooves are important in that the binding of charged molecules such as polyamines takes place within some grooves of human telomeric DNA.[24, 25] Because all the conjugates presented in study contain a common aminosugar (neomycin) conjugated to intercalators with varying surface areas, the study sheds further light on the interplay between the intercalator structure and the planar surface area, with respect to quadruplex binding.
Fig 1.
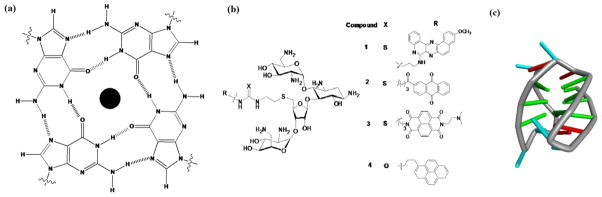
(a) A diagram showing a G-tetrad which is formed by planar arrangement of four guanines in which the guanines associate with each other using Hoogsteen hydrogen bond pairing. The center of G-tetrad cavity is usually occupied by metal cation (b) Chemical structures of neomycin-intercalator conjugates used in the study (c) Structure of human telomeric DNA quadruplex in the presence of sodium ions (adapted from pdbID 143D). Guanine bases are shown in green.
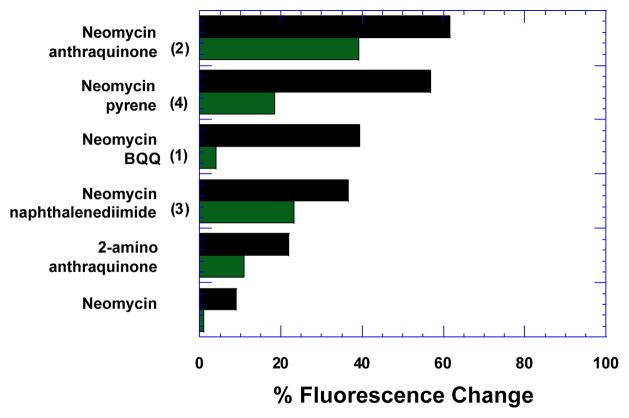
Fluorescent intercalator displacement (FID) assays[26] were initially used to measure the relative binding behaviour of these ligands towards the human telomeric quadruplex (Fig. S1, S2). As shown in Fig. 2, all conjugates exhibited a higher displacement of the fluorescent probe thiazole orange (TO) than neomycin, demonstrating that the conjugation of neomycin to other planar moieties appreciably improves the G-quadruplex binding. Among the ligands studied, neomycin-anthraquinone conjugate (2) exhibited the highest TO displacement (61.6%) at a 1:1 ligand-to-quadruplex ratio (Table 1 and Fig.2). Neomycin-pyrene conjugate (4) followed closely with 56.8% displacement (Table 1 and Fig. 2). Displacement for the neomycin-BQQ conjugate (1) and the neomycin naphthalene diimide conjugate (3) were much poorer. Neomycin displayed the least TO displacement (9%) of all ligands studied (Table 1 and Fig. 2). The ligands that displayed high TO displacement values were further evaluated to obtain DC50 values (DC50 is the amount of the ligand required to displace 50% TO from the quadruplex). As expected, the lowest DC50, was obtained for neomycin-anthraquinone (2) among the ligands studied. Of the four intercalators studied, anthraquinone in fact has the smallest Van der Waals surface area of the aromatic unit (269.16 Å2, Table S1). The next binder compared to (2) was (4) which exhibits very similar displacement values with pyrene as its aromatic moiety. Though pyrene posseses a much larger aromatic Van der Waals surface area (279.09 Å2, Table S1), it is much less polar (nil) than anthraquinone (34.14 Å2, Table S1) as it lacks oxygen atoms which can act as hydrogen bond acceptors with the guanine imino protons. Conversely, the aromatic moieties BQQ and napthalenediimide, with a much greater polar surface area and more hydrogen bond donor/acceptor atoms, displayed poorer binding. These results illustrate that subtle differences in shape/potential complementarity rather than large aromatic surface areas can be a determining factor driving the enhanced binding of these conjugates. Moreover, it is likely that the binding interactions are affected by the strength of the stacking interactions between the individual aromatic units and the G- tetrads. The bases in the loop regions around the top and bottom G-tetrads could possibly affect G-tetrad stacking.
Fig 2.
FID plot showing per cent displacement of TO in the presence of sodium (black bars) and potassium ions (green bars).
Table 1.
Results of FID experiments showing percent displace of TO (ΔF) and DC50 values.
| Ligand | DC50 (μM) | % change in fluorescence
|
|
|---|---|---|---|
| (Na+) | Na+ | K+ | |
| 1 | >2.5 | 39.2 | 4.0 |
| 2 | 0.15 | 61.6 | 39.1 |
| 3 | 1.34 | 36.5 | 23.2 |
| 4 | 0.16 | 56.8 | 18.4 |
| 2-amino anthraquinone | -- | 22.0 | 11.0 |
| neomycin | -- | 9.0 | 1.0 |
We also performed FID titrations in the presence of another relevant biological salt, potassium. While physiologically germane, human telomeric DNA exists as complex quadruplex mixture in the presence of potassium ions.[27] As such, we observed differences in the binding affinity of conjugates (1,3,4) when compared to sodium (Table 1, Fig.2, S3–S4). It is likely that the difference in binding behaviour of these conjugates in potassium results from a mixture of quadruplex structures present in potassium. Even so, similar to the results obtained in the presence of sodium, conjugate (2) afforded the highest TO displacement (39.1%, Table 1, Fig. 2). Since a unique structure is evident in our studies in sodium salt, we conducted further biophysical experiments in sodium buffer to analyse the effects of the highest affinity binder (2) on the G-quadruplex.
UV thermal denaturation experiments were performed to explore the binding behaviour of conjugate (2) with respect to the human telomeric G-quadruplex. As shown in Table 2, the binding of (2) led to a Tm increase of 23.7 °C while its constituents (neomycin or anthraquinone) afforded < 2 °C of thermal stabilization. The enhanced thermal stability conferred on the target in the presence of conjugate (2) demonstrates a synergistic binding behaviour that produces a greater thermal stabilization of the G-quadruplex when compared to the thermal effect caused by the conjugate’s individual elements.
Table 2.
Thermal denaturation temperature for human telomeric G-quadruplex in the absence and presence of various ligands.
| Ligand | Tm (°C) | ΔTm (°C) |
|---|---|---|
| No Ligand | 57.1 | -- |
| Neomycin | 58.3 | 1.2 |
| 2-amino anthraquinone | 57.3 | 0.2 |
| Neomycin+2-amino anthraquinone | 63.7 | 6.6 |
| 2 | 80.7 | 23.7 |
To further assess the binding of (2) with a human telomeric DNA mimic, we performed circular dichroism (CD) studies. As shown in Fig. 3a, the binding of neomycin did not induce any structural change in DNA conformation. However, binding of (2) produced a change in the CD signal at 295 nm (Fig. 3b). In the absence of ligand, the G-quadruplex DNA signature for antiparallel G-quadruplex DNA. As an increasing amount of ligand was titrated into the quadruplex solution, the CD intensity at 295 nm continuously diminished until 1.6 equivalent of the ligand was added. The subsequent addition of more ligand resulted in sample precipitation. A plot of the change in CD signal at 295 nm was used to obtain the binding stoichiometry between conjugate (2) and the G-quadruplex. As shown in Fig. 3c, the stoichiometry was determined according to the change in CD intensity at 295 nm versus the molar ratio of ligand to G-quadruplex. The resulting plot showed an ~ 1.0 ligand to quadruplex ratio, indicating a 1:1 complex formation. We observe no induced CD in the chromophore absorption region (320–360 nm). The 1:1 binding stoichiometry of the complex, as seen by CD, was also corroborated through FID titration (Fig. S5a). A Scatchard analysis [26] of the binding of neomycin-anthraquinone (2) with G-quadruplex yielded an association constant of Ka = 1.25×107 M−1 (Fig. S5b). Previously, we reported that neomycin binds to a human telomeric quadruplex with a 1:1 stoichiometry and an association constant of (2.93±0.11)×104 M−1.[12] The disparity between association constants represents a nearly thousand-fold increase in the G-quadruplex association constant for the neomycin-anthraquinone conjugate (2) when compared to neomycin alone.
Fig 3.
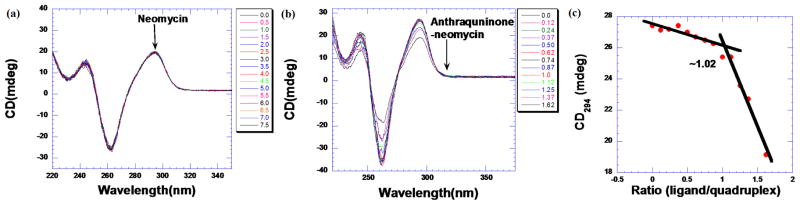
CD titration of human telomeric DNA G-quadruplex with (a) neomycin, (b) an anthraquinone-neomycin conjugate (2), (c) binding stoichiometry plot obtained from the titration of (2) into a human telomeric G-quadruplex.
In conclusion, we show that combination of dual recognition motifs leads to stronger G-quadruplex binding ligands. Conjugation of neomycin to four well-known intercalators produced a higher affinity binding than neomycin alone. An analysis of highest affinity ligand, neomycin-anthraquinone (2), showed a nearly thousand-fold increase in the G-quadruplex association constant. An increase in the intercalator’s planar surface area does not necessarily lead to enhanced G-quadruplex binding. Rather, an appropriate combination of surface area, hydrogen donor/acceptor interactions, and steric interactions within the loop regions led to an optimum binding to the G-quadruplex. Further enquiries are currently underway with neomycin-anthraquinone conjugates in which varied linker length constraints are examined to determine the role that spacers play in dual recognition of quadruplexes.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support provided by the National Institute of Health (Grant 15CA125724 and R41GM097917).
Footnotes
Electronic supplementary information (ESI) available. Details of experimental procedure and additional figures of biophysical studies. See DOI: XXXXXX
Notes and references
- 1.Keniry MA. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, Komiyama M. Methods. 2012;57:100–105. doi: 10.1016/j.ymeth.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Mergny JL, Helene C. Nature Medicine (New York) 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 6.Zahler AM, Williamson JR, Cech TR, Prescott DM. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 7.Pagano B, Mattia CA, Giancola C. Int J Mol Sci. 2009;10:2935–2957. doi: 10.3390/ijms10072935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 9.Monchaud D, Teulade Fichou MP. Org Biomol Chem. 2008;6:627–36. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 10.Cosconati S, Marinelli L, Trotta R, Virno A, De Tito S, Romagnoli R, Pagano B, Limongelli V, Giancola C, Baraldi PG, Mayol L, Novellino E, Razzo A. J Am Chem Soc. 2010;132:6425–6433. doi: 10.1021/ja1003872. [DOI] [PubMed] [Google Scholar]
- 11.Cosconati S, Marinelli L, Trotta R, Virno A, Mayol L, Novellino E, Olson AJ, Razzo A. J Am Chem Soc. 2009;131:16336–16337. doi: 10.1021/ja9063662. [DOI] [PubMed] [Google Scholar]
- 12.Ranjan N, Andreasen KF, Kumar S, Hyde-Volpe D, Arya DP. Biochemistry (NY) 2010 doi: 10.1021/bi101517e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue L, Charles I, Arya DP. Chem Commun (Camb) 2002;(1):70–71. doi: 10.1039/b108171c. [DOI] [PubMed] [Google Scholar]
- 14.Arya DP, Xue L, Tennant P. J Am Chem Soc. 2003;125:8070–8071. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 15.Arya DP, Willis B. J Am Chem Soc. 2003;125:12398–12399. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 16.Willis B, Arya DP. Biochemistry-Us. 2006;45:10217–10232. doi: 10.1021/bi0609265. [DOI] [PubMed] [Google Scholar]
- 17.Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP. Biochemistry. 2010;49:5540–5552. doi: 10.1021/bi100071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis B, Arya DP. Biochemistry. 2010;49:452–469. doi: 10.1021/bi9016796. [DOI] [PubMed] [Google Scholar]
- 19.Xue L, Ranjan N, Arya DP. Biochemisry. 2011;50:2838–2849. doi: 10.1021/bi1017304. [DOI] [PubMed] [Google Scholar]
- 20.Shaw NN, Xi H, Arya DP. Bioorg Med Chem Lett. 2008;18:4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 21.Willis B, Arya DP. Adv Carbohydr Chem Biochem. 2006;60:251–302. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 22.Willis B, Arya DP. Curr Org Chem. 2006;10:663–673. [Google Scholar]
- 23.Arya DP. Top Curr Chem. 2005;253:149–178. [Google Scholar]
- 24.Rossetti L, Franceschin M, Bianco A, Ortaggi G, Savino M. Bioorg Med Chem Lett. 2002;12:2527–2533. doi: 10.1016/s0960-894x(02)00504-8. [DOI] [PubMed] [Google Scholar]
- 25.Rossetti L, Franceschin M, Schirripa S, Bianco A, Ortaggi G, Savino M. Bioorg Med Chem Lett. 2005;15:413–420. doi: 10.1016/j.bmcl.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 26.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J Am Chem Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 27.Dai J, Carver M, Yang D. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.