Abstract
Calorimetric and fluorescence techniques were used to characterize the binding of aminoglycosides-neomycin, paromomycin, and ribostamycin, with 5′-dA12-x-dT12-x-dT12-3′ intramolecular DNA triplex (x = hexaethylene glycol) and poly(dA).2poly(dT) triplex. Our results demonstrate the following features: (1) UV thermal analysis reveals that the Tm for triplex decreases with increasing pH value in the presence of neomycin, while the Tm for the duplex remains unchanged. (2) The binding affinity of neomycin decreases with increased pH, although there is an increase in observed binding enthalpy. (3) ITC studies conducted in two buffers (sodium cacodylate and MOPS) yield the number of protonated drug amino groups (Δn) as 0.29 and 0.40 for neomycin and paromomycin interaction with 5′-dA12-x-dT12-x-dT12-3′, respectively. (4) The specific heat capacity change (ΔCp) determined by ITC studies is negative, with more negative values at lower salt concentrations. From 100 mM to 250 mM KCl, the ΔCp ranges from −402 to −60 cal/(mol K) for neomycin. At pH 5.5, a more positive ΔCp is observed, with a value of −98 cal/(mol K) at 100 mM KCl. ΔCp is not significantly affected by ionic strength. (5) Salt dependence studies reveal that there are at least three amino groups of neomycin participating in the electrostatic interactions with the triplex. (6) FID studies using thiazole orange were used to derive the AC50 (aminoglycoside concentration needed to displace 50% of the dye from the triplex) values. Neomycin shows a seven fold higher affinity than paromomycin and eleven fold higher affinity than ribostamycin at pH 6.8. (7) Modeling studies, consistent with UV and ITC results, show the importance of an additional positive charge in triplex recognition by neomycin. The modeling and thermodynamic studies indicate that neomycin binding to the DNA triplex depends upon significant contributions from charge as well as shape complementarity of the drug to the DNA triplex Watson–Hoogsteen groove.
Keywords: DNA triplex, Major groove, Thermodynamics, Aminoglycoside, Fluorescence, ITC
Considerable attention has been given to oligonucleotide-directed triple helix formation of nucleic acids [1–3] due to its possible regulatory role in vivo [4] and its potential applications in medicine and biotechnology [5,6]. In the intermolecular triplex system, the double helix associates with a single-strand triple helix-forming oligonucleotide (TFO) via Hoogsteen hydrogen bonds in the major groove. Because TFOs bind in the major groove of duplex DNA, they can be used as transcription inhibition agents [7]. TFOs have been used to study the molecular mechanism of triplex-mediated TCR (transcription-coupled repair) in HeLa nuclear extracts, and the formation of a triplex induced much stronger DNA repair activity in promoter-containing plasmids [8]. A 17-mer homopyrimidine oligonucleotide has been shown to bind to the major groove of SV40 DNA, inhibiting enzymatic cleavage [9]. Catapano has shown the suitability of a triplex based approach using phosphorothioate-linked oligos in targeting the C-myc and Ets-2 transcription factors [10–12]. A number of reviews on the potential of DNA triplex in DNA targeted diagnostics and drugs have appeared in the recent literature [13–18].
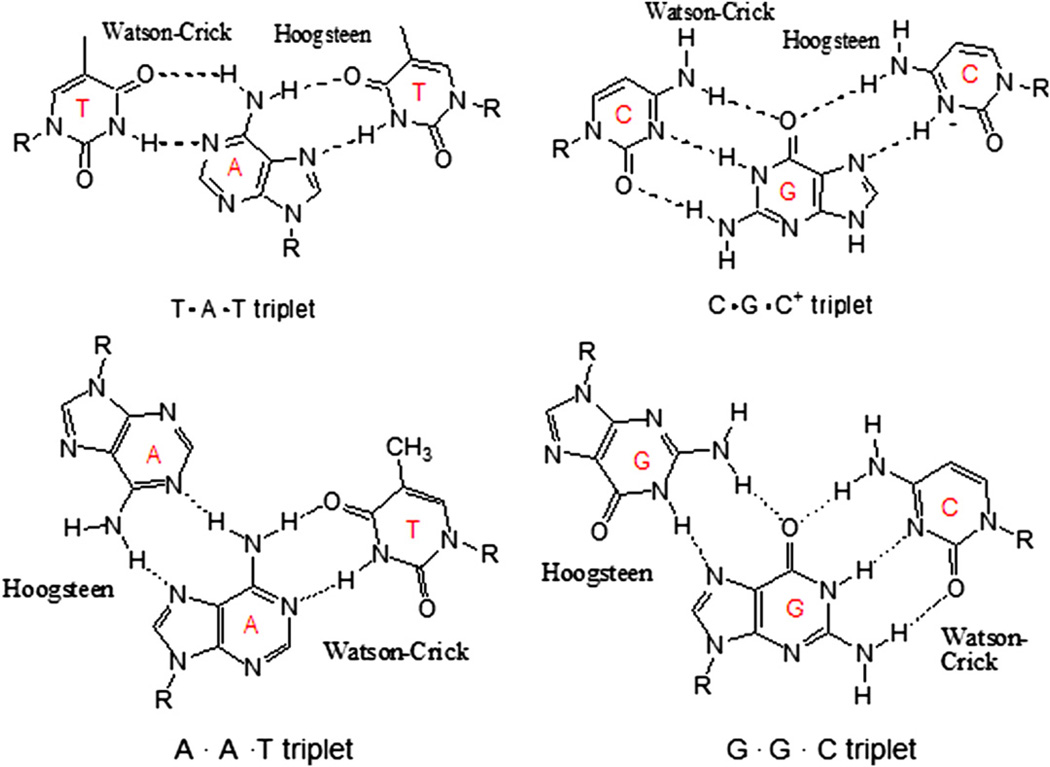
The triplex structure contains two motifs: purine and pyrimidine (Scheme 1). In the purine motif, the homopurine third strand binds antiparallel to the purine strand of the duplex by reverse-Hoogsteen hydrogen bonds [19–21]. In the pyrimidine motif, the third strand composed of pyrimidine bases binds parallel to the purine strand of the Watson–Crick duplex by forming Hoogsteen hydrogen bonds. However, in the intramolecular triplex, first found to exist in biological systems in 1986 [22], triplex formation is different from that of intermolecular triplex. In this case, the homopurine–homopyrimidine mirror repeats within the DNA duplex denature to allow one strand to fold back, forming Hoogsteen hydrogen bonds with the adjacent DNA duplex sequence. The intramolecular triplex, known also as H-DNA [21], is favored under negative DNA supercoiling conditions [23], acidic solutions, or in the presence of divalent cations [24]. The intramolecular triplex has also been reported to have an effect on the transcriptional regulation and replication in vivo [25–27]. H-DNA, found in Escherichia coli and eukaryotic cells [28–30], is found in promoter regions and around recombination hot spots [31]. Therefore, it likely plays a regulatory role in gene expression [32], recombination [33], and in transcription of human viruses [34]. It has also been implicated in the suppression of the human γ-globin gene [35].
Scheme 1.
Hydrogen bonds formed in pyrimidine.purine-pyrimidine triplets (pyrimidine motif) and purine.purine-pyrimidine triplets (purine motif).
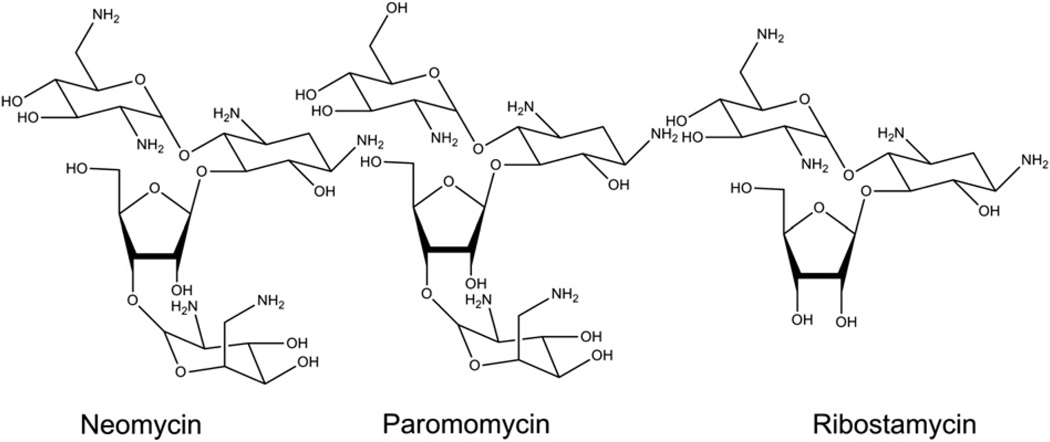
However, the DNA triplex is not as stable as its corresponding duplex. The association of a third strand with a duplex (kon ∼ 10 – 103 M−1 s−1) is a much slower process than the association of two single strands in forming a duplex (kon ∼ 106 M−1 s−1) [36–38]. Many studies have been carried out using small molecules to improve the thermal stability of DNA triple helices [39–49]. However, these ligands do not selectively bind to the DNA triple helices, and some even destabilize triple helices [38]. Our previous work has shown the remarkable ability of neomycin, its conjugates with other intercalators and other aminoglycosides (Scheme 2), to stabilize DNA, RNA and hybrid triple helices [38,39,50–53]and to even aid the delivery of PS-modified TFOs into cancer cells [54]. In particular, neomycin was found to induce the stabilization of a DNA.DNA.DNA [50] triple helix, a DNA.RNA hybrid duplex [39,55], as well as DNA.DNA.RNA. hybrid triple helices [39], significantly adding to the number of nucleic acids (other than RNA) that aminoglycosides have been shown to target [56]. Among the aminoglycosides studied, it was found that neomycin significantly stabilized DNA and RNA triple helices [38]. Herein, we report our observations regarding the role of aminoglycosides in the thermal stabilization of AT-rich triplexes and their interactions with aminoglycosides from a thermodynamic perspective.
Scheme 2.
Structures of the aminoglycosides used in the study.
2. Methods and materials
2.1. Nucleic acids and aminoglycosides
The intramolecular triplex (5′-dA12-x-dT12-x-dT12-3′) was synthesized using an Expedite Nucleic Acid Synthesis System (8909) with standard phosphoramidite chemistry. The oligomer was purified on an anion exchange HPLC column (Water Gen-Pak FAX, 4.6 × 100 mm) with a Tris.HCl buffer system. Buffer A: 25 mM Tris.HCl, 1 mM EDTA, and 10% MeCN (v/v%); Buffer B: Buffer A + 1 M NaCl. Conditions: 2–60% buffer B over buffer A during 0–16 min at a flow rate of 0.75 mL/min. Neomycin sulfate and ribostamycin were obtained from ICN Biomedicals Inc., and paromomycin sulfate was purchased from Sigma. Neomycin sulfate was protected with Boc anhydride, chromatographed on silica gel and deprotected to obtain pure neomycin B. All aminoglycosides were used without further purification. All the polynucleotides were purchased from GE Healthcare Amersham Bioscience. The concentrations of polymer solutions were determined spectrophotometrically using the following extinction coefficients (in units of mol of nucleotide or bp/L−1 cm−1): ε264 = 8520 for poly(dT), ε260 = 6000 for poly(dA)poly(dT); 5′-dA12-x-dT12-x-dT12-3′, ε260 = 341,100.
2.2. UV spectrophotometry
All UV absorbance experiments were conducted on a Cary 1E UV/vis spectrophotometer equipped with temperature programming. Quartz cells with a 1 cm path length were used for all the absorbance studies. For the triplex preparation, all samples were heated at 95 °C for 5 min, then cooled slowly to room temperature and allowed to incubate for 16 h at 4 °C prior to use. Absorbance vs. temperature profiles were recorded at 260 nm and 280 nm. The samples were heated from 5 °C to 95 °C at a rate of 0.2 °C/min, then they were cooled to 10 ° C at a rate of 5.0 ° C/min. Data was recorded at 1° increments. For melting temperature (Tm) determination, two baselines (upper and lower) were drawn, corresponding to folded and unfolded forms, respectively. Thereafter a median line between the two baselines was drawn, and the crossing point between the experimental curve and median line was used for Tm. First derivatives of the denaturation curves were also obtained and were ±2 °C for all reported values in this paper. For all thermal denaturation experiments, DNA concentrations were 1 µM in strand for intramolecular triplex and 15µM for poly(dA).2poly(dT) triplex.
2.3. Isothermal titration calorimetry (ITC)
All isothermal calorimetric measurements were performed on a MicroCal VP-ITC (MicroCal, Inc., Northampton, MA) at 10 °C, except for temperature dependence experiments. In ITC studies, DNA concentrations were varied from 10 µM/base triplet to 50 µM/ base triplet to obtain reliable signal intensity. In every titration, 5 or 10 µL aliquots of aminoglycoside solution were injected into a sample cell containing 1.42 mL of DNA triplex solution. After each experiment, a corresponding control experiment was performed by titrating the same drug solution into experimental buffer. The injection spacing was either 240s or 300s, syringe rotation rate was 260 rmp, and duration of each injection was 20s. The resulting data were processed by Origin version 5.0. Every heat burst curve was due to a drug injection. Integrating the area under each heat curve yielded the heat given off upon each injection, from which the corresponding control data was subsequently subtracted to obtain the ITC binding profile associated with the drug–DNA binding.
2.4. Differential scanning calorimetry (DSC)
The DNA melting temperature and melting enthalpy changes in the absence of drug were obtained using a MicroCal VP-DSC (MicroCal, Inc., Northampton, MA). The scan rate was 1 ° C/min, and the operating temperature range was 5 °C−95 °C. After each DSC experiment, a corresponding control experiment was conducted with only sample buffer in the sample cell. The corrected DSC profile was obtained by subtracting the control data from the sample data. The enthalpy changes for the melting of triplex in the absence of drug (ΔHHS) were calculated by integrating the area under the heat capacity curves using Origin version 5.0.
2.5. Circular Dichroism Spectropolarimetry (CD)
All CD experiments were conducted at 10 °C on a JASCO J-810 spectropolarimeter equipped with a thermoelectrically-controlled cell holder. A quartz cell with a 1 cm path length was used in all CD studies. CD spectra were recorded as an average of 3 scans from 300 nm to 200 nm in 0.1 nm increments. In isothermal CD titration experiments, small aliquots of concentrated drug solutions were added to a solution of DNA, inverted twice, and allowed to equilibrate for at least 10 min prior to scanning. In each CD titration, small aliquots (0.6–40 µL) of a concentrated aminoglycoside solution (500 µM) were added to a 2 mL solution of DNA triplex. The initial DNA solution was allowed to equilibrate for at least 30 min prior to the first addition of drug.
2.6. Fluorescence intercalator displacement assay (FID)
FID assay was done using a Photon Technology International instrument (Lawrenceville, NJ) at 10 °C. A 2 mL quartz cuvette was filled with sodium cacodylate buffer (10 mM SC, 150 mM KCl,0.5 mM EDTA, pH 6.8, 5.5) and thiazole orange (700 nM final concentration) was added. Thiazole orange was excited at 504 nm and the emission was recorded from 520 to 600 nm. The triplex deoxynucleotide hairpin 5′-dA12-x-dT12-x-dT12-3′ was added (100 nM in strand final concentration) and the fluorescence was measured again and normalized to 100% relative fluorescence. A concentrated solution of compound, aminoglycoside, (100 µM–10 mM) was added, and the fluorescence was measured after 5 min of incubation at 10 °C. The addition of compound was continued until the fluorescence reached saturation. For all titrations, final concentrations were corrected for dilution (less than 5% of the total volume).
The solution of poly(dA).2poly(dT) was incubated with thiazole orange for 30 min prior to use for FID titration. Each well of 96-well plate was loaded with poly(dA).2poly(dT) solution (200 µL each). A concentrated solution of compound, aminoglycoside, (9.35 µM–935 µM) was added, and the fluorescence was measured after 5 min of incubation. The 96-well plate was read in triplicate on Carry eclipse plate reader fluorometer with advanced reads software (ex. 504, em. 532 nm, cutoff filter at 430–1100 nm). (No aminoglycoside = 100% fluorescence, no DNA = 0% fluorescence). Fluorescence readings are reported as % fluorescence relative to control wells.
2.7. Molecular modeling
The structures of the dA110.2dT10 triplex, neomycin, and paromomycin structure have been built and optimized with Macro-Model program[57]using the AMBER* force field [58,59]. The all atom AMBER* force field was used since it reproduces X-ray and NMR-derived DNA structures. The continuum GB/SA model of water [60,61], as implemented in MacroModel, has been used in all calculations. The force field atomic charges were used for triplex. The ligands were built and optimized in MacroModel and the RESP charges were derived using ab initio calculations with 6–31G* basis set in Jaguar program [57]. The structure of the dA10.2dT10 triplex was built starting from the experimental (NMR) solution structure of related DNA triplex [62,63]. Twenty seven sodium ions were added to neutralize the initial structure. The sodium ions were removed and the structure was reoptimized to within a gradient of 0.05 kJ/(mol Å). Only the movement of atoms of external bases was restricted during minimization. The structure of neomycin was built by MacroModel and submitted to conformational searching in order to find the global minimum structure and the other low energy conformations. For conformational searching, the Monte Carlo (MC) routine[64,65]from MacroModel was used. Three MC runs (for all single bond torsional angles and 1000 steps each) starting from different initial conformations were performed, yielding the same global minimum conformation. All the flexible bonds were selected.
Several of the lowest conformations of neomycin were manually docked to the DNA triplex groove in different orientations. The conformations of the molecules were adjusted to allow maximum base-ligand, and phosphate-ligand H-bond formation with no significant unfavorable van der Waals interactions. Neomycin was manually docked into the triplex grooves avoiding any unfavorable steric clashes, and positioned approximately central in the entrance to the grooves. For a more detailed analysis of ligand groove interactions, appropriate contacts between protonated amines and phosphate oxygens on the backbone were selected. The minimized complexes were then re-minimized with the distance constraints removed to an RMS gradient of 0.08 kcal/(mol Å) to eliminate unfavorable contacts. In the next step, all the restrictions were removed except the movement of ring atoms of the terminal bases, and the structure was optimized to a gradient of 0.05 kJ/(mol Å) or lower. To test the stability of the complex, the low energy complexes were submitted to 500 ps MD simulation using reported procedures (stochastic dynamics, constant temperature of 300 K, with coupling constant of 0.2 ps, and a time step of 1.0 fs) [66]. The SHAKE procedure was used for all bonds. Electrostatic potential was generated from viewerLite 5.0 for windows (Accelrys Inc., CA, USA) using Gasteiger charges.
3. Results and discussion
3.1. Study of KCl on the melting temperature of AT-rich triplexes
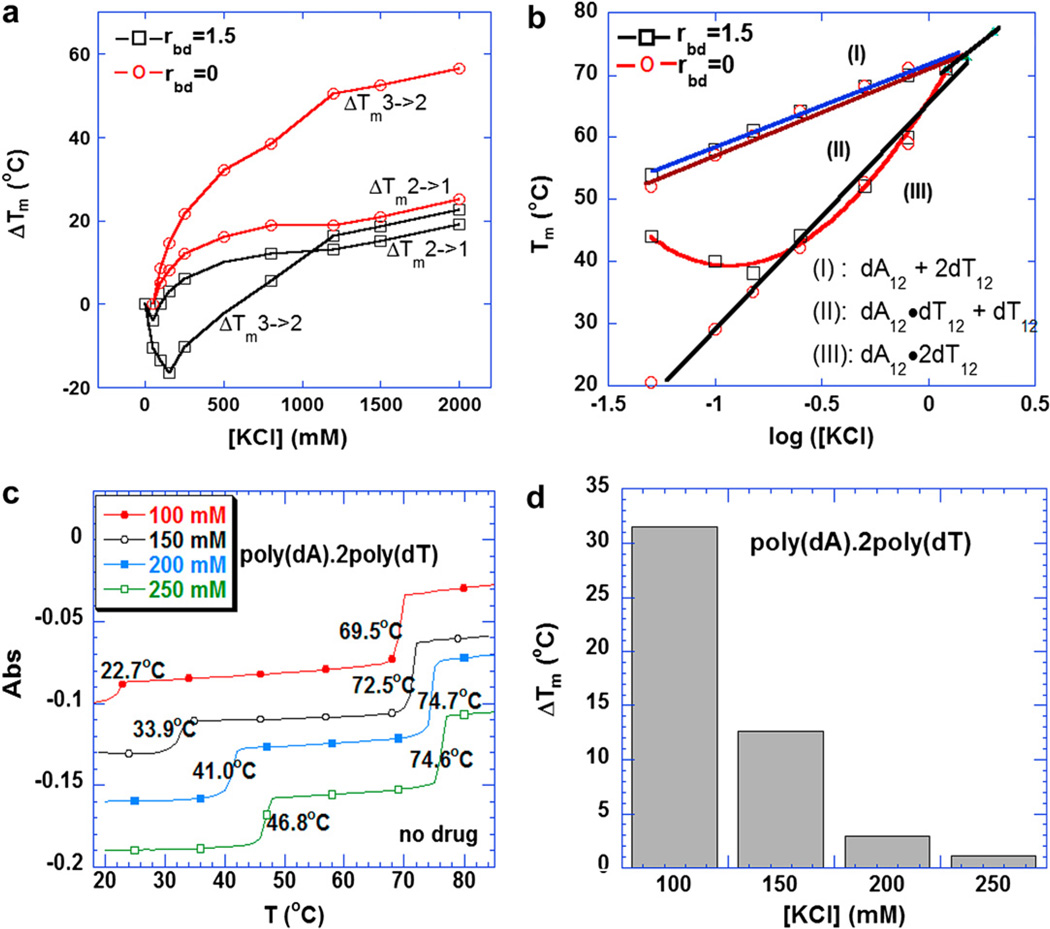
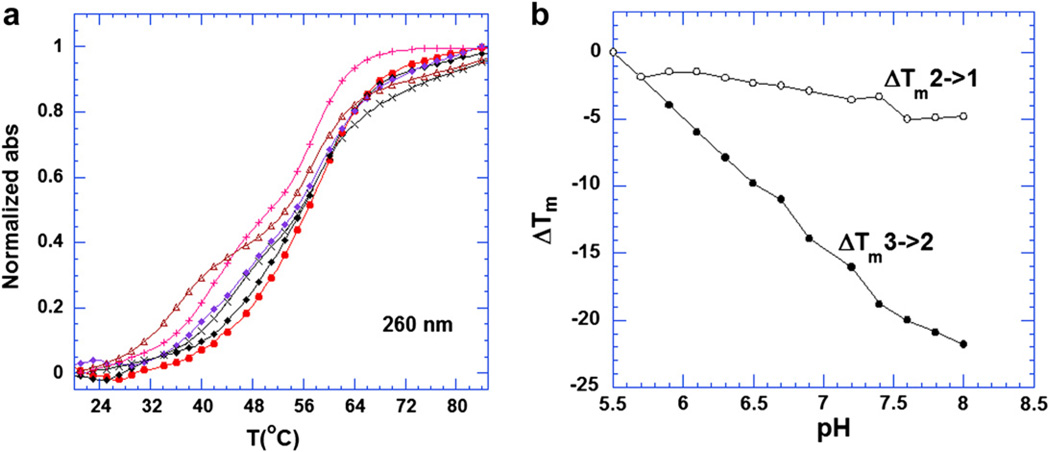
It has been previously demonstrated that at 260 nm both Hoogsteen (HS) and Watson–Crick (WC) transitions can be observed [66]. It has also been reported that at 280 nm there is a clear hyperchromic shift, corresponding to the triplex–duplex transition [66]. In the absence of KCl, a monophasic transition is observed, due to duplex denaturation. When the ionic strength was increased from 50 mM to 250 mM KCl, two transitions could clearly be observed at 260 nm (see supporting information fig. 1). The transition at the lower temperature corresponds to the HS transition, which is consistent with the melting temperature at 280 nm. The transition at the higher temperature is due to the WC melting. When the ionic strength was increased above 800 mM, the melting profiles became monophasic with only one transition observed at 260 nm (see supporting information fig. 1). This transition presumably corresponds to the denaturation of the triplex directly to the coil state due to the high salt conditions. The increase of the ionic strength raised the ΔTm of the triplex more than that of the corresponding duplex. This phenomenon is seen for both 5′-dA12-x-dT12-x-dT12-3′ (Fig. 1a) and poly(dA).2poly(dT) (Fig. 1c).
Fig. 1.
(a) Plots of ΔTm3→2 and ΔTm2→1 of the 5′-dA12-x-dT12-x-dT12-3′ triplex as a function of increasing KCl with or without 8 mM neomycin. (b) Phase diagram of Tm as a function of log [K+] for 5′-dA12-x-dT12-x-dT12-3′ triplex with or without 8 µM neomycin. The data points represent the melting temperature. Each phase labeled as I, II, and III refers to the single strand, duplex and triplex structural states, respectively, that DNA adopts. (c) UV melting profile of poly(dA).2poly(dT) at various KCl. (d) A bar graph of triplex melting temperature increase (ΔTm) of poly(dA).2poly(dT) at saturated amount of neomycin. Buffer conditions: 10 mM sodium cacodylate, 0.5 mM EDTA, and pH 6.8. DNA triplex concentration was 1 µM/strand for oligomer and 15 µM/base triplet for polymer.
3.2. UV denaturation of 5′-dA12-x-dT12-x-dT12-3′ triplex
Fig. 1a shows the melting temperature increase (ΔTm) with respect to the KCl concentration. The increase of KCl concentration from 50 mM to 2 M resulted in a 55 °C increase in the HS transition, with a corresponding increase in the WC transition of 25 °C in the absence of neomycin. However, in the presence of neomycin (rbd:1.5, rbd = the ratio of base triplet to drug), KCl exhibited a smaller effect on the HS transition. As shown in Fig. 1a, both the duplex (when the KCl concentration is below 50 mM) and the triplex (when the ionic strength is below 150 mM) were destabilized by the addition of KCl in the presence of neomycin, with a 20 °C decrease in the HS transition and only a 4 °C decrease in the WC transition. When the ionic strength was increased above 50 mM for the duplex or 150 mM for the triplex, the stability of both structures increased. The observation of destabilization of both triplex and duplex by added KCl is likely due to the competitive binding of neomycin ammonium groups and K+ ions to DNA. With increasing K+ concentration, the apparent affinity of neomycin for duplex and triplex decreases, resulting in a decrease in duplex and triplex stabilization by neomycin. Since the interaction of neomycin with the triplex groove will be more sensitive to competition with added electrolytes than the interaction of neomycin with the duplex (due to a higher negative potential-three negatively charged backbones for triplex vs. two for duplex), the effects are larger for the triplex. The experiment in question is a thermal denaturation of DNA and the effect of the drug is being monitored indirectly by looking at the stability of the DNA. Since the triplex melts at a lower temperature than the duplex, temperature dependence of drug binding likely effects the changes in duplex and triplex Tm as well.
The phase diagram (Fig. 1b) also reveals three different phases in the triplex melting process. In Region I, the solution contained only single strands of dA12 and dT12; in Region III, the triplex was formed completely; while Region II represents the coexistence of duplex dA12.dT12 and single strand dT12. The transition lines meet when the concentration of KCl was above 1200 mM (triple point), and together, they formed the boundary between Region I and Region III. Similar results can be seen for the triplex poly(dA).2poly(dT) (see supporting information Fig. 7). Increased ionic strength also increased the thermal stability of triplex, but with less effect on the duplex as shown in Fig. 1c.
Triplex thermal stability at a saturated amount of neomycin was investigated; the results are shown in Fig. 1d. In this study, the rbd ratio used were 6.2, 6.8, 5.9 and 5.8 under 100, 150, 200, and 250 mM KCl, respectively. The comparison of the stabilization effect of neomycin on intramolecular and polynucleotide triplex lead to the following conclusions: (1) At low ionic strengths (up to 150 mM KCl), the intramolecular triplex is more stable than the intermolecular polynucleotide triplex in the absence of neomycin, most probably because of the vicinity of three strands held by a linker (Fig. 1). When the KCl concentration was increased above 150 mM, the stabilization effect of the salt on the polynucleotide is much more pronounced. (2) Under the same ionic strength, neomycin stabilized the poly(dA).2poly(dT) triplex much more significantly than the 5′-dA12-x-dT12-x-dT12-3′ triplex, with a 31.5 °C increase in Tm for poly(dA).2poly(dT) (rbd of 6.2) (Fig. 1d) but only a 11.0 °C increase for 5′-dA12-x-dT12-x-dT12-3′ triplex at a much higher concentration of neomycin (rbd of 1.5) (Fig. 1b). (3) For both poly (dA).2poly(dT) and 5′-dA12-x-dT12-x-dT12-3′ triplex, the stability of duplex was relatively unchanged in the absence or presence of neomycin or any aminoglycoside (Figs. 1 and 2). (4) There was no significant effect of neomycin on the stability of triplex at ionic strength above 150 mM (Fig. 1).
Fig. 2.
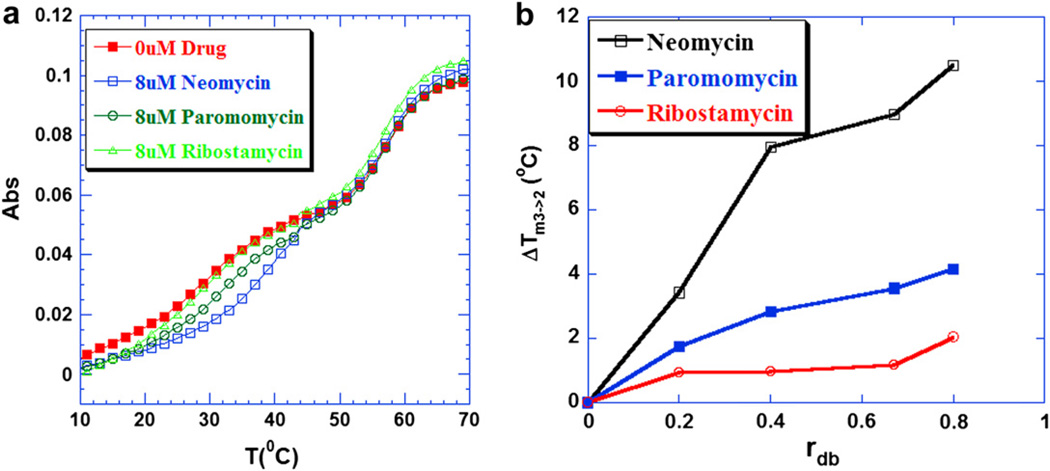
(a) UV melting profiles of 5′-dA12-x-dT12-x-dT12-3′ triplex at 260 nm in the presence of aminoglycosides. From left to right, the UV melting profiles correspond to 0 µM drug, 8 µM ribostamycin, paromomycin and neomycin respectively. (b) A plot of ΔTm3→2 of 5′-dA12-x-dT12-x-dT12-3′ triplex as a function of increasing rdb values [rdb = drug/base triplet ratio]. Buffer conditions: 10 mM sodium cacodylate, 0.5 mM EDTA, 100 mM KCl, and pH 7.2. DNA triplex concentration was 1 µM/strand.
3.3. Study of aminoglycosides on the thermal stability ofAT-rich triplexes
To investigate the stabilization effect of different aminoglycosides on the intramolecular triplex, the UV melting studies were conducted in the presence of three aminoglycosides (neomycin, paromomycin and ribostamycin). As shown in Fig. 2a, the representative UV melting profiles (rbd: 1.5) reveal the different abilities of these aminoglycosides to stabilize the intramolecular triplex. The melting temperatures for the 5′-dA12-x-dT12-x-dT12-3′ triplex increased in the presence of drugs at 100 mM KCl (Fig. 2a). However, there was no effect observed on the thermal stability of the corresponding duplex (the melting temperature remained around 57 °C), suggesting that these aminoglycosides selectively stabilized the intramolecular triplex.
As shown in Fig. 2a, among the three aminoglycosides studied here, neomycin was found to be the most effective in enhancing the thermal stability of the intramolecular triplex, with an 11 °C increase in the melting temperature at the saturated rdb studied; paromomycin exhibited a 6 °C lower triplex melting temperature than neomycin; no significant change in the melting temperature was observed for the triplex in the presence of ribostamycin, as shown in Fig. 2b. This observation is consistent with previous studies showing that neomycin stabilizes the poly(dA).2poly(dT) triplex most effectively and selectively among the aminoglycosides [38,50].
3.4. Study of pH on thermal stability of AT-rich triplexes
UV melting analysis showed the effect of pH on the stabilization of 5′-dA12-x-dT12-x-dT12-3′ triplex. These UV experiments were conducted at increments of 0.2 pH units, ranging from pH 5.5 to pH 8.0 for 5′-dA12-x-dT12-x-dT12-3′ (Fig. 3a). In the absence of neomycin, the melting temperature of the triplex (29 °C) and duplex (58 °C) remained unchanged over the pH range studied. However, in the presence of neomycin, the melting temperature of the triplex was found to be dependent on pH. With increasing pH values, the melting temperature decreased gradually (Fig. 3b). At a low pH (5.5–5.9), the triplex exhibited nearly monophasic transitions at 260 nm, suggesting a 3 → 1 transition while biphasic transitions are observed at pH above 5.9 (Fig. 3a). These results indicate that neomycin is more effective in stabilizing the intramolecular triplex at lower pH values as more amino groups of neomycin are protonated. However, an opposite trend was observed when studying poly(dA.2poly(dT) triplex stability at pH 5.5 and 6.8 under the same salt conditions. The potency of neomycin in stabilizing the polynucleotide triplex at pH 5.5 was not as strong as at pH 6.8 (see supporting information Fig. 2). The thermal stability of poly(dA).2poly(dT) (as monitored by ΔTm3→2) was raised by neomycin (rbd 6.2) by approximately 31.5 °C at pH 6.8, but only 8.5 °C at pH 5.5. The reason for this difference observed between the intramolecular triplex 5′-dA12-x-dT12-x-dT12-3′ and intermolecular triplex poly(dA).2poly(dT) may be due to the fact that the rigid intramolecular triplex 5′-dA12-x-dT12-x-dT12-3′ structure may be quite different from the intermolecular triplex. It has been reported that the duplex conformation in the intramolecular triplex structure is somewhat different from the free duplex conformation [67], suggesting that the covalent linkage between the two strands could affect the conformation of triplex.
Fig. 3.
(a) UV melting profiles at 260 nm of the 5′-dA12-x-dT12-x-dT12-3′ triplex in the presence of 8 µM neomycin (rdb = 0.75). From left to right, the pH values of solutions were 7.4, 6.9, 6.7, 6.3, 5.9 and 5.5, respectively. (b) Plots of the ΔTm3→2 and ΔTm2→1 of the 5′-dA12-x-dT12-x-dT12-3′ triplex as a function of increasing pH value in the presence of 8 µM neomycin (rdb = 0.75). The Tm for the triplex was determined from the profiles at 280 nm ΔTm = Tm(any pH)-Tm(5.5). All the buffer solutions contained 10 mM sodium cacodylate, 0.5 mM EDTA and 100 mM KCl. DNA triplex concentration was 1 µM in strand.
3.5. ITC studies of neomycin, paromomycin and ribostamycin binding to 5′-dA12-x-dT12-x-dT12-3′ and poly(dA).2poly(dT) triplex
3.5.1. 5′-dA12-x-dT12-x-dT12-3′
Isothermal titration calorimetry (ITC) is a technique that allows one to extract a complete thermodynamic profile of a bimolecular interaction, was then applied. In a sample ITC titration, a macro-molecule (DNA triplex) solution was filled in a sample cell, and the interacting ligand (neomycin) is titrated into the solution through a titration syringe gradually. The heat evolved or absorbed resulting from the reaction is measured by the instrument.
3.5.2. At pH 5.5
As discussed previously, the stabilization effect of aminoglycosides toward intramolecular triplex is more potent at a low pH than at a higher pH. It is important to study the interaction at a pH value in which the aminoglycosides are highly protonated, to minimize drug protonation contributions upon binding. Inspection of the pKa values of six amino groups on neomycin indicates that neomycin ammonium groups are substantially protonated at pH 5.5 [68]. The merit of studying the complex at this pH is that the observed binding enthalpy reflects a more intrinsic interaction between the ligand and the DNA triplex. However, study at such a low pH leads to practical problems since the ITC signal intensity and derived binding enthalpy is much lower than the enthalpy at a higher pH (Fig. 4d, e). In order to obtain enough signal, a much larger amount of drug and DNA triplex sample is required. Both 5′-dA12-x-dT12-x-dT12-3′ and poly(dA).2poly(dT) triplex were found to be stable at a pH of 5.5.
Fig. 4.
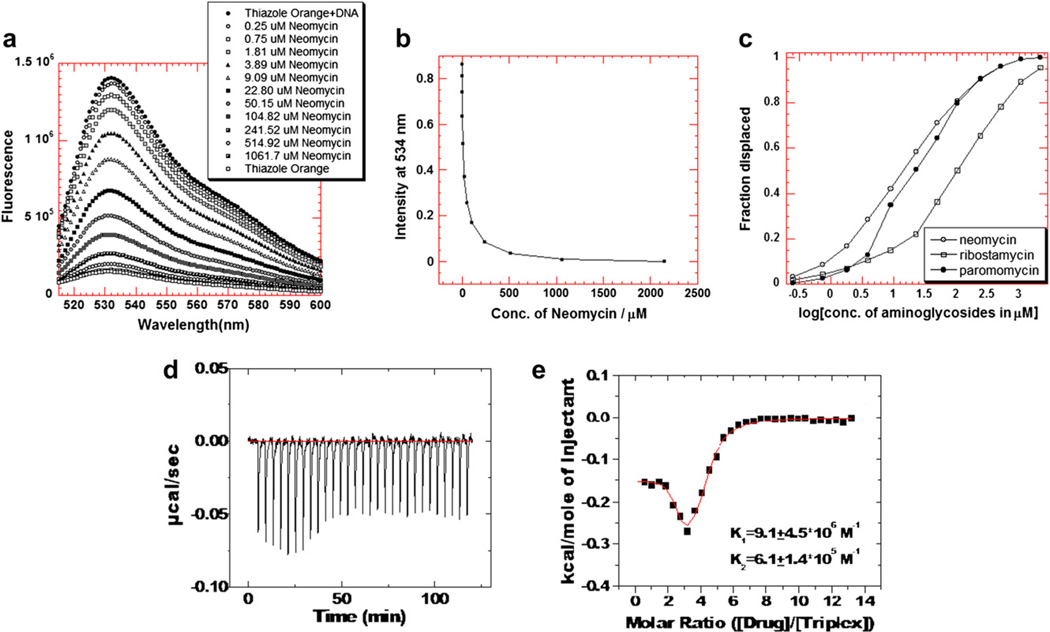
(a–c). A graphical representation of thiazole orange displacement assay (a) A raw emission data of 1.25 µM thiazole orange upon excitation at 534 nm with buffer only (open circles) and after addition of 100 nM/strand triplex deoxynucleotide hairpin 5′-dA12-x-dT12-x-dT12-3′ (black circles). Neomycin was then titrated from 0.25 µM to 1.06 mM. (b) The decrease in the fluorescence intensity of the complex (DNA-thiazole orange) upon addition of neomycin aliquots. (c) Assuming a linear relationship between the changes in fluorescence intensity with the fraction of thiazole orange displaced results in S-shaped binding isotherm. This graph allows the determination of concentration of ligands needed to displace half of the thiazole orange from triplex deoxynucleotide hairpin 5′-dA12-x-dT12-x-dT12-3′. (d–e) ITC titration of neomycin with intrramolecular triplex at pH 5.5. (d) ITC profiles of neomycin binding with 5′-dA12-x-dT12-x-dT12-3′ triplex. (e) Corrected injection heat as a function of [drug]/[triplex] ratio. The data points represent the experimental injection heat and the solid lines were corresponding to the calculated fits of the data using a model with two sets of binding sites (Origin 5.0). Buffer condition: 10 mM sodium cacodylate, 0.5 mM EDTA, 150 mM KCl, pH 5.5. T= 10 °C.
Model fitting with two binding sites yields a binding affinity of (9.1 ±4.5) × 106 M−1, a binding enthalpy of−0.15 ± 0.01 kcal/mol, and a binding entropy of 9.7 kcal/mol K for the first binding event. Approximately 98% of the binding results from entropy, with only 2% contribution from enthalpy [69]. The second binding site contains a binding affinity of (6.1 ± 1.4) × 105 M−1, a binding enthalpy of −0.34 ± 0.03 kcal/mol, and a binding entropy of 7.2 kcal/mol K. Our thermodynamic studies can not offer a more detailed explanation of the nature of the second binding site, as it differs from the first site.
It has previously been reported that in the absence of the effect of drug protonation, paromomycin binding to the A-site of 16S rRNA is entropy-driven, providing 72% of the driving force for binding, with only 28% coming from the enthalpic contribution at pH 5.5 [68]. The factors causing this favorable entropic contribution in our study could include a conformational change upon drug-DNA triplex complexation, release of counterions caused by the electrostatic interactions between drug and DNA triplex, and the desolvation of both the drug and DNA triplex upon binding [68].
Since the ITC signal-to-noise ratio becomes larger when the binding signal intensity decreases to less than 0.1, it is desirable to apply more than one technique to confirm and validate the ITC derived binding constant for a system. Fluorescence intercalator displacement (FID) method, a technique complementing the limitations of ITC in calculating the intrinsic affinities, has been introduced by Boger [70,71]. In this method, an intercalator such as ethidium bromide or thiazole orange intercalates into the DNA triplex base triplets. Titration of the ligand into the solution will displace the intercalator. The amount of displaced intercalator, measured with decrease in intercalator fluorescence, is then directly proportional to the amount of bound ligand. Ethidium bromide, which intercalates into nucleic acid with relatively small affinities [72], can be easily displaced by a subsequently added ligand. Thiazole orange was used in our assay as its high fluorescence enhancement upon intercalation has been reported to yield a 3000-fold increase (only a 20-fold increase for ethidium bromide) [72].
The displacement assays were first performed as complete titrations on a fluorometer (Fig. 4a) and were then run (as triplicates) on a 96-well plate reader at pH 5.5 and at pH 6.8 at a salt concentration of 150 mM KCl (Fig. 4a – c). The AC50 values reported in Table 1c–d are the aminoglycoside concentrations required to displace 50% of thiazole orange from the triplex, as measured by a decrease in 50% of the fluorescence. At pH 5.5, the neomycin AC50 (AC50 = 0.42 µM) is 22-fold lower than paromomycin (AC50 = 11.5 µM) and approximately 50 fold lower than ribostamycin (AC50 = 26.7 M). A similar trend, with neomycin (AC50 = 13µM) <paromomycin (AC50 = 157 M) <ribostamycin (AC50 =459 µM) is observed for the 5′-dA12-X-dT12X-dT12-3′ triplex. The AC50 values for the smaller intramolecular triplex are about an order of magnitude higher than the AC50 values observed for the polynucleotide triplex.
Table 1.
| a | ||||
|---|---|---|---|---|
| Thermodynamic profiles of aminoglycosides binding to the 5′-dA12-x-dT12-x-dT12-3′ triplex in 10 mM sodium cacodylate or MOPS, 0.5 mM EDTA and 150 mM KCl with pH 6.8. T= 10 °C. | ||||
| Parameter | Cacodylate buffer |
MOPS buffer |
||
| neomycin | paromomycin | neomycin | paromomycin | |
| Kobs(M −1) | 0(3.8 ±0.4×105 | (8.7 ±0.2)×104 | (1.6 ±0.3)×105 | (6.7 ±3.7)×104 |
| ΔHobs (kcal/mol) | −5.9±0.2 | −4.8±0.2 | −4.4±0.3 | −2.7±0.8 |
| ΔG (kcal/mol) | −7.0±0.2 | −6.4±0.2 | −6.7±0.3 | −6.2±0.8 |
| (drug/triplex) | 1.1±0.0 | 1.2±0.2 | 1.1±0.1 | 1.2±0.3 |
| C | 2.1 | 0.8 | 1.3 | 0.7 |
| b | |||
|---|---|---|---|
| ITC derived binding constants and binding enthalpy for neomycin binding to 5′-dA12-x-dT12-x-dT12-3′ triple helix at pH 5.5,6.8 and 7.4 in10 mM sodium cacodylate, 0.5 mM EDTA and 150 mM KCl. T= 10 °C. | |||
| pH | 5.5 | 6.8 | 7.4 |
| Kobs1 (M −1) | (9.1 ±4.5) ×106 | (3.8 ±0.4) ×105 | (2.7 ±0.81)×105 |
| ΔG1 (kcal/mol) | −9.8±0.01 | −7.1±0.2 | −7.0±0.2 |
| c | ||
|---|---|---|
| Fluorescence derived AC50 values for aminoglycoside binding to 5′-dA12-x-dT12-x-dT12-3′ triple helix at pH 5.5, 6.8 in 10 mM sodium cacodylate, 0.5 mM EDTA and 150 mM KCl. T= 10 °C, [DNA triplex] = 100 nM/strand, [Thiazole Orange] = 700 nM. | ||
| Aminoglycoside | AC50 (pH 6.8) (µM) | AC50 (pH 5.5) (µM) |
| Neomycin | 35.5 | 13.3 |
| Paromomycin | 179.0 | 157.9 |
| Ribostamycin | 486.0 | 459.0 |
| d | ||
|---|---|---|
| Fluorescence derived AC50 values for aminoglycoside binding to Poly(dA.2poly(dT) triplex at pH 5.5, 6.8 in 10 mM sodium cacodylate, 0.5 mM EDTA and 150 mM KCl. T= 25 °C, [polydA.2polydT] = 0.88 mM/bp, [Thiazole Orange] = 1.25 µM. | ||
| Aminoglycoside | AC50 (µM) at pH 5.5 | AC50 (µM) at pH 6.8 |
| Neomycin | 0.42 ± 0.05 | 3.0 ± 0.6 |
| Paromomycin | 11.5 ± 2.0 | 21.0 ± 1.2 |
| Ribostamycin | 26.7 ± 2.4 | 34.6 ± 3.5 |
All data are derived from ITC experiments. Kobs determined from fits of ITC profile using Origin 5.0 using a model with two sets of binding sites. The results shown here correspond to the high affinity first binding event.
3.5.3. At pH 6.8
FID assay was first used to determine the AC50 values for the three aminoglycosides with poly(dA)·2poly(dT) and with 5′-dA12-x-dT12-x-dT12-3′ triplex. As seen in Table 1d (see supporting information Figs. 11–13), neomycin shows a seven fold higher affinity (AC50 = 3 µM) than paromomycin (AC50 = 21 µM) and an eleven fold higher affinity than ribostamycin (AC50 = 34.6 µM) at pH 6.8. A similar trend, with neomycin (AC50 = 35.5 µM) < paromomycin (AC50 = 179 µM) < ribostamycin (AC50 = 486 µM) is observed for the 5′-dA12-x-dT12-x-dT12-3′ triplex (Table 1c). The AC50 values for the smaller intramolecular triplex are about an order of magnitude higher than the AC50 values observed for the polynucleotide triplex. When the AC50 values for the aminoglycosides are compared are different pHs, neomycin shows a larger decrease at lower pH (3–6 fold), when compared to paromomycin (2 fold). A very little change in ribostamycin AC50 is observed at different pH values signifying that ring IV amino protonation at lower pH contributes to increased affinity of paromomycin and neomycin. When the FID experiments were carried out with poly(dA).poly(dT) duplex, thiazole orange could not be displaced up to 10 mM neomycin, suggesting that the affinity of the aminoglycoside is 2–3 orders of magnitude lower for the AT-rich DNA duplexes, when compared to its affinity for the AT-rich DNA triplex.
As indicated previously, the complexation of aminoglycosides with DNA triplex involves the protonation of aminoglycosides. To investigate drug protonation upon binding, ITC was used to study the binding of neomycin and paromomycin to the 5′-dA12-x-dT12-x-dT12-3′ triplex in two different buffers, sodium cacodylate and MOPS. Ribostamycin binding was too weak to study using ITC. Both buffers contained either 10 mM sodium cacodylate or MOPS, 0.5 mM EDTA and 150 mM KCl at pH 6.8. The ITC profile and corresponding data fits are shown in Fig. 5 for the two aminoglycosides in the sodium cacodylate buffer (see supporting information, Fig. 6 for the ITC profile in the MOPS buffer). Panels (a) and (b) in Fig. 5 correspond to injections of neomycin and paromomycin into the 5′-dA12-x-dT12-x-dT12-3′ triplex solution. The corrected injection heats as a function of the [drug]/[triplex] ratio are listed in the panels (c) and (d) in Fig. 5, the data points reflecting the experimental injection heats and the solid line reflecting the model fitting. The model yielding a reasonable fit of experimental data is the one having two binding sites. The size of the binding site were obtained through CD titration of oligonucleotides and polynucleotides (see supporting information Figs. 3 and 8). Polynucleotide titrations yield an apparent single binding site of ∼6 base triplet/neomycin (see supporting information Fig. 8). CD titrations of the oligomer triplex show an apparent binding site of 2.2 neomycin/triplex or ∼6 base triplets/neomycin (see supporting information Fig. 3). Assuming that the length of the triplex will not affect the binding site size, 1.1 neomycin/triplex was picked as the binding site size for both sites in the ITC data fits. Using the resulting values of Ka, the binding free energy (DG) were obtained using the following standard relationship:
| (1) |
Fig. 5.
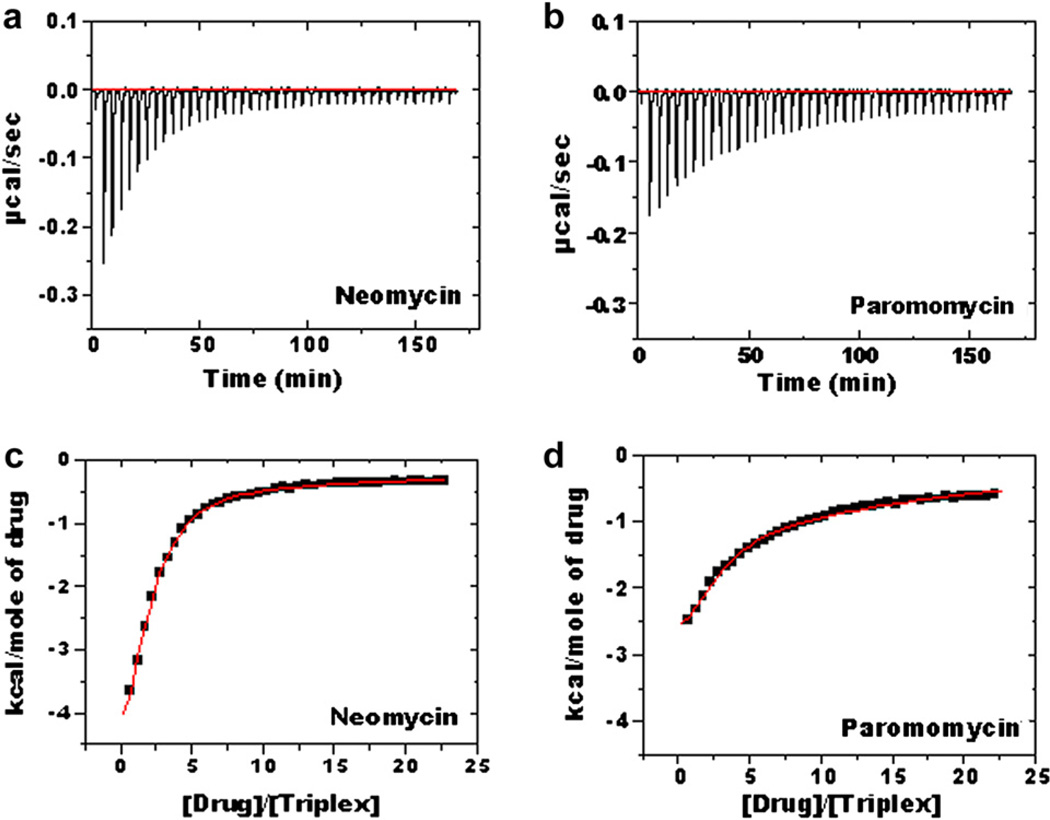
ITC studies of intramolecular triplex 5′-dA12-x-dT12-x-dT12-3′ with neomycin (a) and paromomycin (b) in 10 mM cacodylate, 0.5 mM EDTA, 150 mM KCl, pH 6.8; T= 10 °C [DNA] = 4 µM/strand, [drug] = 600 µM (c–d) Corrected injection heat as a function of [drug]/[triplex] ratio. The data points represent the experimental injection heat and the solid lines correspond to the calculated fits of the data by using a model with two binding sites (Origin 5.0).
The thermodynamic parameters (ΔHobs, ΔG, Kobs) are summarized in Table 1a–b.
From Table 1a–b, the following conclusions can be drawn: First, in both buffers, the observed binding enthalpy (ΔHobs) of neomycin with triplex is primarily exothermic. Specifically, the binding enthalpies for neomycin and paromomycin are −5.9 and −4.8 kcal/ mol in cacodylate buffer and −4.4 and −2.7 kcal/mol in MOPS buffer, respectively. At pH 6.8, the observed enthalpy includes contributions from the intrinsic binding enthalpy, drug protonation enthalpy and buffer ionization heat.
Second, at pH 6.8 in both buffers, the neomycin-intramolecular triplex interaction is enthalpy-driven. Approximately 80% of the driving force for binding results from enthalpy, with only 20% due to entropy. For paromomycin, the enthalpic contribution, smaller than for neomycin, is approximately 75% in cacodylate buffer and 44% in MOPS buffer. Given the fact that the intrinsic binding is entropy-driven at pH 5.5, the enthalpically favorable binding at pH 6.8 suggests that a non-negligible drug protonation effect is clearly involved during binding [69,73–75].
Third, low values of the Wiseman parameter “c”, which denotes the product of macromolecular concentration with the binding site size and the association constant, are found in a number of binding events. This parameter determines the shape of ITC titration curve. For example, the lack of a lower baseline is due to the low c value [76]. Previously the recommended experimental window of c values was 5–1000, with optimal values ranging from 10 to 500. For those systems with c < 5 and c > 1000, the data fits are believed to contain considerable uncertainties even when appropriate models are applied. However, this conclusion has been revised based on a study conducted by Turnbull and Daranas who have discussed in detail the application of ITC with low c values (0.01 < c < 10), especially for low affinity systems utilizing carbohydrates [76]. In their study, they proposed four requirements in the design of ITC experiments involving low c value: (1) binding stoichiometry; (2) sufficient binding isotherm used for curve fitting; (3) concentration of both macromolecule and ligand known to be accurate; and (4) adequate level of signal-to-noise in the binding data. Meeting these four requirements supports the validity of ITC model fitting for determining the association constant Ka. However, great caution is recommended in interpreting the ΔH value. The c values shown in Table 1a–b fall in the low range (all smaller than 5), with highest value of 2.1 for neomycin. However, it is not feasible to bring the c value to the socalled experimental window 10 < c < 500 because then the ligand concentrations become too high. Neomycin and triplex complexation at high concentrations causes precipitation (at 1:1 or higher drug: triplex ratios) hence a full ITC titration cannot be performed. Therefore, after obtaining the binding stoichiometry from CD titration and fulfilling all the other requirements listed above, a reliable Ka can be determined for neomycin and paromomycin. Ribostamycin binding was simply too weak to evaluate using ITC. An ITC excess site titration is also conducted to obtain a few uniform heat bursts to calculate a reliable binding enthalpy (see supporting information Figs. 4 and 5). This method uses a high concentration of nucleic acid to obtain several uniform binding heat bursts. However, for excess site titrations, the drug to triplex ratio is much lower and thus no precipitation takes place, allowing for high c values to be used in excess site titrations leading to a more accurate estimate of the binding enthalpies. A correction of the heat per injection signal with respect to the total number of moles of ligand added per injection yields a reliable independent estimate of binding enthalpy [82,83]. These results indicate that there is very little discrepancy (∼7%) in neomycin binding enthalpies obtained from ITC model fitting and ITC excess site titration (Table 1a and supporting information Table 1).
Fourth, it can be seen that all the binding enthalpies in the cacodylate buffer are greater than those in the MOPS buffer. This difference is due to the different ionization heat of the two buffers (−0.19 kcal/mol for cacodylate buffer and +5.07 kcal/mol for MOPS buffer at 10 °C) [77]. At pH 6.8, however, the drug–triplex interaction also involves a contribution from drug protonation. The number of protons linked to the drug–triplex complexation can be determined by solving the following two equations simultaneously [78]:
| (2) |
| (3) |
where ΔHint represents the intrinsic binding enthalpy (a value that excludes the enthalpic contribution from the buffer ionization), Δ Hion the heat of buffer ionization, the numerical subscripts the different buffers, and ΔHobs the observed binding enthalpies obtained from the data fitting of ITC profiles. A positive value of Δn indicates a net uptake of protons while a negative value indicates the net release of protons. Using the data listed in Table 1a–b, ΔHint and Δn for drug binding with an intramolecular triplex are calculated (Table 2).
Table 2.
Buffer dependence of the observed binding enthalpies, intrinsic binding enthalpies and the number of linked protons for the neomycin and paromomycin binding with 5′-dA12-x-dT12-x-dT12-3v triplex in 10 mM buffer, 0.5 mM EDTA and 150 mM KCl; pH 6.8, T= 10 °C.
| Drug | Buffer |
ΔHion (kcal/mol) |
ΔHobs (kcal/mol) |
ΔHint (kcal/mol) |
Δn (per drug) |
|---|---|---|---|---|---|
| Neomycin | cacodylate | −0.19 | −5.9 ± 0.2 | −5.8 ± 0.2 | 0.29 ± 0.10 |
| Neomycin | MOPS | +5.07 | −4.4 ± 0.3 | −5.8 ± 0.2 | 0.29 ± 0.10 |
| Paromomycin | cacodylate | −0.19 | −4.8 ± 0.2 | −4.7 ± 0.2 | 0.40 ±0.19 |
| Paromomycin | MOPS | +5.07 | −2.7 ± 0.8 | −4.7 ± 0.2 | 0.40 ±0.19 |
ΔHion (ionization heat) for the cacodylate buffer and MOPS buffer are obtained from Fukada and Takahashi (1998) [77]. ΔHobs are derived directly from the ITC experiments.Delta;Hint (intrinsic binding enthalpy) and Δn (the number of binding-linked protons) are calculated from the equations (2) and (3).
Table 2 shows positive values for Δn, suggesting that amino-glycoside interacting with the intramolecular triplex is coupled to the uptake of protons. The Δn values are 0.29 and 0.40 for intramolecular triplex binding to neomycin and paromomycin at 10 °C, respectively. The fractional protonation number implies the shift of pKa of amine group upon binding. Notice that the pKa of 3-amine group on the ring I is 5.74, while the remaining amine groups on both neomycin and paromomycin have the pKa values all above 7.55 [79]. Thus, it can be determined that the 3-amine group on the ring I is induced a pKa shift upon drug-DNA complexation at pH 6.8. With the Δn values of 0.29 and 0.40 observed for neomycin and paromomycin, the pKa value of 3-amine group is calculated to be 6.56 for neomycin and 6.77 with the relationship pKa = pH – log [(A−)/HA], implying a pKa shift of 0.82 for neomycin and 1.02 for paromomycin, respectively. To observe the effect of temperature on protonation, similar experiments were also conducted at 20 °C (data not shown), yielding a Δn value of 0.38 and 0.29 for neomycin and paromomycin, respectively. Comparison of the Δn value at 10 °C and 20 °C reveals that the binding-linked protonation is not affected significantly by temperature, suggesting that at physiological temperature, the intramolecular triplex–aminoglycoside interaction may have the same extent of protonation as reported here. It has been shown that a large number of binding-linked protons are involved in aminoglycosides A-site RNA interaction at pH 7.0 (Δn values are 1.42, 1.55, 1.11 for neomycin, paromomycin, and ribostamycin, respectively) [78]. It was revealed previously that the aminoglycoside (in particular neomycin) likely binds to the W–H DNA triplex groove, with rings I, II and IV as the key components involved in this process [51]. The 3′-amino group on ring II has the lowest pKa value for both neomycin and paromomycin [68].
3.5.4. Poly(dA) .2poly(dT) triplex
For the study reported here, polynucleotide DNA triplex poly (dA)-2poly(dT) was studied for comparison with intramolecular triplex. A ΔTm-based approach, derived by McGhee [80], was used to obtain binding affinities. The following equation was used to calculate the association constants at the corresponding melting temperatures where the triplex was complexed with drug:
| (4) |
where Tm0 is the melting temperature of the drug-free DNA triplex; Tm the melting temperature of drug-bound DNA triplex; ΔHHS the enthalpy change corresponding to melting enthalpy of Hoogsteen bonds in the absence of drug, determined from the DSC measurement (Fig. 6c); L is the free drug concentration at Tm (estimated by one-half of the total drug concentration), and N the apparent binding site size determined through CD titration of drug into the DNA triplex. After obtaining the association constants at Tm, the integrated van't Hoff equation (equation (5)) was used to calculate the association constants at 10 °C [81]:
| (5) |
where ΔHobs is the observed binding enthalpy of the drug to the triplex as derived from ITC excess nucleic acid binding experiments conducted at 10 °C; R the gas constant, and ΔCp the heat capacity change, determined from equation (6) by using binding enthalpies at various temperatures (Fig. 6b). The calculated binding constants are shown in Table 4.
Fig. 6.
UV melting profiles of poly(dA).2poly(dT) triplex in the absence (1) and presence of neomycin (2) with rbd 8.8. (b) A plot of ITC derived binding enthalpies vs. corresponding temperatures. The slope reflects the heat capacity change ΔCp. (c) DSC melting profile of poly(dA).2poly(dT). Buffer condition: 10 mM sodium cacodylate, 0.5 mM EDTA, 100 mM KCl, and pH 5.5.
Table 4.
Thermodynamic profiles of poly(dA) . 2poly(dT) and 5v-dA12-x-dT12-x-dT12-3′ interaction with aminoglycosides in10 mM sodium cacodylate, 0.5 mM EDTA, pH 6.8 (unless indicated otherwise) and various KCl concentrations. T= 10 °C.
| Drug | [KCl] (mM) | ΔHHS (kcal/mol base triplet) |
Tm0 (°C ) | Tm (°C ) | ΔCp (cal/mol K) | Kobs(10°C)(M −1) |
|---|---|---|---|---|---|---|
| poly(dA) 2poly(dT) | ||||||
| Neomycin | 100 | 1.20 | 22.7 | 54.2 | −218 ±15 | NA |
| Neomycin | 150 | 1.59 | 33.9 | 46.5 | −116 ± 34 | NA |
| Neomycin | 200 | 2.00 | 41.0 | 43.9 | − 76 ± 13 | NA |
| Neomycin | 100 (pH 5.5)a | 1.16 | 22.4 | 28.6 | −98 ±13 | (6.2 ± 0.5) × 105 |
| Neomycin | 120 (pH 5.5)a | 1.50 | 27.5 | 32.4 | −90 ±18 | (4.0 ±0.1) × 105 |
| Neomycin | 140 (pH 5.5)a | 1.70 | 31.4 | 34.4 | −87 ± 4 | (2.4 ±0.1) × 105 |
| Paromomycin | 100 | 1.20 | 22.7 | 32.2 | −83 ± 15 | NA |
| Paromomycin | 150 | 1.59 | 33.9 | 36.3 | −29 ±9 | NA |
| Paromomycin | 200 | 2.00 | 41.0 | 42.5 | −23 ±11 | NA |
| Ribostamycin | 100 | 1.20 | 22.7 | 26.2 | −30 ±8 | NA |
| 5′-dA12-x-dT12-x-dT12-3′ | ||||||
| Neomycin | 100 | 1.33 | 30.0 | 36.5 | −310 ± 43 | (1.4 ± 0.2) × 106 |
| Neomycin | 150 | 1.59 | 34.8 | 37.0 | −158 ± 30 | (2.5 ± 0.3) × 105 |
| Neomycin | 200 | 1.78 | 37.3 | 38.7 | −84 ±12 | (1.4 ± 0.1) × 105 |
| Neomycin | 250 | 1.81 | 42.4 | 43.7 | − 17 ± 22 | (9.3 ± 0.1) × 104 |
Experiments were done at pH 5.5.
3.5.5. At pH 5.5
The protonation of aminoglycosides contributes to the observed binding enthalpy, the heat capacity and the binding constant calculated at pH 6.8. However, the appropriate application of Equation (6) requires the thermodynamic parameters of binding enthalpy and heat capacity change from the intrinsic binding, both of which were obtained from the interaction at pH 5.5, in which binding induced drug protonation is minimized. Observation of the thermal stability of poly(dA).2poly(dT) at both pH 5.5 (Fig. 6a) and 6.8 (Fig. 1c) indicates that the formation of DNA triplex is not affected by pH. However, the heat capacity change (−98 cal/mol K) (Fig. 6b) at pH 5.5 was much lower than the value (−218 cal/mol K) at pH 6.8 (see supporting information Fig. 9). When studying the thermal stability increase of triplex by neomycin at saturated rbd ratio of 8.8 (see supporting information Fig. 10), Tm of triplex was increased only 6 °C by neomycin at pH 5.5 (rbd: 8.8) (Fig. 6a). In addition to the triplex transition, two transitions were observed, one at 67 °C corresponding to 2 → 1 transition and a second one at 80 °C due to the 3 → 1 transition. This phenomenon was also observed in the poly(dA)poly(dT) duplex melting profile, in which the two strands of duplex disproportionate after denaturation to form a triplex in the presence of neomycin, directly melting to a single strand at 80 °C. The 3 → 1 transition (82 °C) of poly(dA). 2poly(dT) has also been previously reported in the presence of coralyne [84]. In this case, two triplex transitions were observed, one at 29 °C (3 → 2) and the other at 80 °C (3 → 1). The former reflects neomycin binding to the preexising triplex, while the second (3 → 1) reflects a neomycin-induced structure at low pH. Thus, this result suggests that the neomycin binding to polynucleotide triplex is not as favorable as the intramolecular triplex at low pH, confirmed by the lower binding constant of (6.17 ±0.1) × 105 M−1 derived from the Equations (4) and (5) (Table 4). It was observed that the binding enthalpy of neomycin with DNA triplex at pH 5.5 is only approximately −1.0 kcal/mol at 10 °C. Entropy was calculated to be −6.50 kcal/mol K using Equations (1) and (2), contributing 86% of driving force to the binding of neomycin with poly(dA) . 2poly(dT) triplex. This significant entropic contribution to the binding force is consistent with that observed for the 5′-dA12-x-dT12-x-dT12-3′ at low pH.
3.5.6. At pH 6.8
However, the binding affinities of the aminoglycosides for the DNA triplex cannot be obtained accurately using this method at pH 6.8. As indicated previously, the binding enthalpy observed at pH 6.8 includes the drug protonation heat. As a consequence, the heat capacity change calculated from binding enthalpies is not intrinsic as well. Thus, the binding affinities calculated using Equations (4) and (5) at pH 6.8 will be overestimated due to the overestimation of both binding enthalpies and heat capacity changes.
The binding enthalpy at pH 6.8 still can be used to calculate the protonation number of neomycin upon complexation with poly (dA).2poly(dT) triplex. The binding enthalpies of neomycin-poly (dA.2poly(dT) interaction were obtained under both cacodylate and MOPS buffer. A positive value of Δn 0.23 (Table 3) was calculated using Equations (2) and (3). This number is consistent with the protonation number calculated for neomycin interacting with 5′-dA12-x-dT12-x-dT12-3′ (0.29), suggesting that the binding-linked drug protonation is not affected by the length of the nucleic acid.
Table 3.
Buffer dependence of the observed binding enthalpies, intrinsic binding enthalpies and the number of linked protons for the neomycin binding with poly(dA).2poly (dT) triplex in 10 mM buffer, 0.5 mM EDTA and 100 mM KCl; pH 6.8, T= 10 °C.
| Drug | Buffer | ΔHobs (kcal/mol) |
ΔHion (kcal/mol) |
ΔHint (kcal/mol) |
Δn (per drug) |
|---|---|---|---|---|---|
| Neomycin | cacodylate | − 7.4 ±0.1 | −0.19 | − 7.3 ±0.1 | 0.23 ± 0.06 |
| Neomycin | MOPS | −6.2 ±0.1 | +5.07 | − 7.3 ±0.1 | 0.23 ± 0.06 |
3.6. A negative heat capacity change is associated with neomycin and paromomycin binding to 5′ -dA12-x-dT12-x-dT12-3′ and poly(dA)-2poly(dT) triplex
To determine the heat capacity of drug binding with the DNA triplex, parallel ITC experiments at 5 °C, 10 °C and 15 °C were conducted to obtain the observed binding enthalpies (ΔHobs) (see supporting information Table 1). Theoretically, one can obtain the binding enthalpies by fitting the ITC profiles as described above. However, it has been found that when the heat signal is not significant, model-dependent data fitting is sensitive to any small change in data processing, for example, for paromomycin binding within the triplex. As shown in Table 1a–b, the uncertainty of the binding constant for paromomycin is much larger than for neomycin. As a result, ΔHobs could reflect significant errors. The model-independent protocol, the ITC excess site binding experiments as discussed above, was used in this case to obtain reliable binding enthalpies (see sample ITC excess titrations in supporting information Figs. 4 and 5) [85]. The ΔHobs obtained from several different temperatures facilitates determination of the heat capacity change related to the drug interaction with the intramolecular triplex by applying the following equation:
| (6) |
3.6.1. 5′-dA12-x-dT12-x-dT12-3′ triplex
Our calculations reveal that the magnitude of ΔCp value decreases with increasing salt concentration at pH 6.8 for 5′-dA12-x-dT12-x-dT12-3′ (Table 4). The ΔCp value for the neomycin interaction with the triplex decreased in magnitude from −310 cal/ (mol K) to −47 cal/(mol K) while for paromomycin it decreased from −229 cal/(mol K) to −13 cal/(mol K) as the salt concentration was increased from 100 mM to 250 mM KCl. For ribostamycin, the ΔCp was only −78 cal/(mol K), even in 100 mM KCl. Although observed heat capacity changes are overestimated to some small extent due to the drug protonation, the intrinsic heat capacity changes are expected to be still negative. Recall that ΔCp values for other minor groove binding drugs, such as netropsin, propamidine, berenil, Hoechst 33,258 and distamycin, have been found to fall in the range of −150 to −350 cal/(mol K) upon binding the A3T3 duplex [78,86], while furan derivatives, such as DB244, DB75 and DB226, have ΔCp values between −95 and −180 cal/(mol K) [87]. Intercalators including ethidium, propidium, daunomycin, and adriamycin have been observed to exhibit ΔCp values of −140 to −160 cal/(mol K) with calf thymus DNA, a smaller range than that of the groove binders [88,89]. A positive ΔCp value of+10 cal/(mol K) for ethidium binding to calf thymus was also observed using van't Hoff analysis [89]. However, to date, all groove binder-DNA interactions have been found to have a negative ΔCp.
Negative ΔCp was observed for neomycin-nucleic acid interactions at both pH 5.5 and 6.8 (Table 4). The negative sign has been suggested to be due to the removal of large amounts of nonpolar surface from water upon complex formation [90,91]. Solvent-accessible surface area change (ΔSASA) also has an impact on the value of ΔCp [78, 86,92]. The removal of a nonpolar surface causes the ΔCp value to be more negative while the removal of the polar surface leads to a more positive ΔCp. In addition to the removal of nonpolar solvent accessible surface area, changes of vibrational modes of macromolecules and water molecules [93], and conformational effects of macromolecules[94,95] can contribute to the ΔCp value. However, in our system, the conformational effects are likely minimal in the temperature range studied since those temperatures were carefully determined based on the thermal denaturation profiles from either UV or DSC and CD spectra show little change upon drug binding. Binding coupled protonation of drug molecule contributes to the observed ΔCp value at pH 6.8 since it has been found that drug protonation is associated with a more negative ΔCp [96]. At pH 5.5, the contribution of drug protonation to ΔCp should be negligible. Recently, the effect of anions to the ΔCp value in the protein-nucleic acid interaction has been reported, with the magnitude of ΔCp being larger when salt type switches from NaF, NaCl to NaBr [97]. The effect of anions such as Cl− and SO42− to the ΔCp may also exist in the small molecule-nucleic acid interaction system by interacting weakly with the positively charged neomycin, but not significant given that such interactions are likely to be weak in solution. The heat capacity change, thus, not only is linked to the changes between nonpolar and polar surface area change upon complexation, but also is probably dependent on the solution conditions.
We observe a decrease of ΔCp in magnitude as the salt concentration increases. The salt effect of observed ΔCp here, similar to those reported for protein-DNA interaction, could be explained by the disruption of salt bridges formed between positive and negative residues. Based on this model, the salt bridges formed at low salt condition are easily disrupted upon another molecule binding. This disruption of salt bridge contributes to the large and negative ΔCp. When salt concentration increases, the shielding of the ammonium groups in neomycin by excess salt leads to lesser disruption of the salt bridge and thereby leading to a smaller ΔCp.
Among the three aminoglycosides studied here, a much less negative ΔCp was observed for the binding of ribostamycin than for neomycin and paromomycin. This comparatively less negative ΔCp value for the binding of ribostamycin than neomycin and paromomycin arises from the absence of ring IV in ribostamycin, which, in turn could reduce the amount of nonpolar surface that can be buried upon DNA triplex binding [78].
3.7. The salt dependence of neomycin and paromomycin binding for 5′-dA12-x-dT12-x-dT12-3′ and poly(dA).2poly(dT) triplex
The observed binding constants for neomycin with poly(dA). 2poly(dT) triplex calculated using ΔTm method at 10 °C over a range of KCl concentrations at pH 5.5 are summarized in Table 4. As this table shows, the Kobs value decreases as KCl concentration increases, indicating that electrostatic interactions play an important role in the drug-triplex interaction. The apparent number of drug NH [ineq] groups participating in electrostatic interactions with the triplex can be estimated from plots of log (Kobs) vs. log([KCl]). The observed linear dependencies have been described by the following relationship [98,99]: where Kobs is the observed binding constant, ‘m′ the number of ion pairs formed between the drug and the DNA triplex, ‘Ψ′the thermodynamic counterion binding parameter for the DNA triplex, and K the equilibrium constant for the drug–DNA interaction at 25 °C.
A plot of log (Kobs) vs. log ([KCl]) yields a slope of – mΨ, giving the smallest number of ion pairs formed between the drug and DNA triplex. As seen in Fig. 7, the linear dependence of log (Kobs) vs. log [KCl] yields a slope of 2.8 for the neomycin-DNA triplex complexation, suggesting that there are about 3 ion pairs formed upon binding at pH 5.5. Another aminoglycoside, tobramycin, using calorimetric methods, has been found to bind to poly(rI)poly(rC) as a trication (3 out of 5 NH[ineq] participating in electrostatic interaction) [99]. Neomycin and paromomycin contribute at least 3 and 3.6 amino groups, respectively, when forming an electrostatic interaction with A-site 16S rRNA [78]. Berenil has been shown to bind to poly(dA)poly(dT), poly(dA-dT)2, poly(dI-dC)2 and poly (rA)poly(rU) with m values of 2.3, 1.4, 1.9 and 2.3, respectively [100]. The result reported here suggests a strong electrostatic contribution to the drug-triplex complexation, which is comparable to aminoglycosides' binding to other RNA structures.
Fig. 7.
Salt dependence of the neomycin binding with poly(dA)-2poly(dT) triplex in 10 mM sodium cacodylate, 0.5 mM EDTA and pH 5.5. T= 10 °C. The experimental data were fit with linear regression and the solid line reflects the resulting curve fit.
At pH 6.8, however, the salt dependence study of the binding affinities of neomycin to poly(dA).2poly(dT) cannot be determined accurately. The observed binding affinities calculated with ΔTm method at this pH are overestimated due to the overestimation of binding enthalpy arising from drug protonation.
3.8. Models of neomycin vs. paromomycin docked in the W–H groove: the difference a charge makes
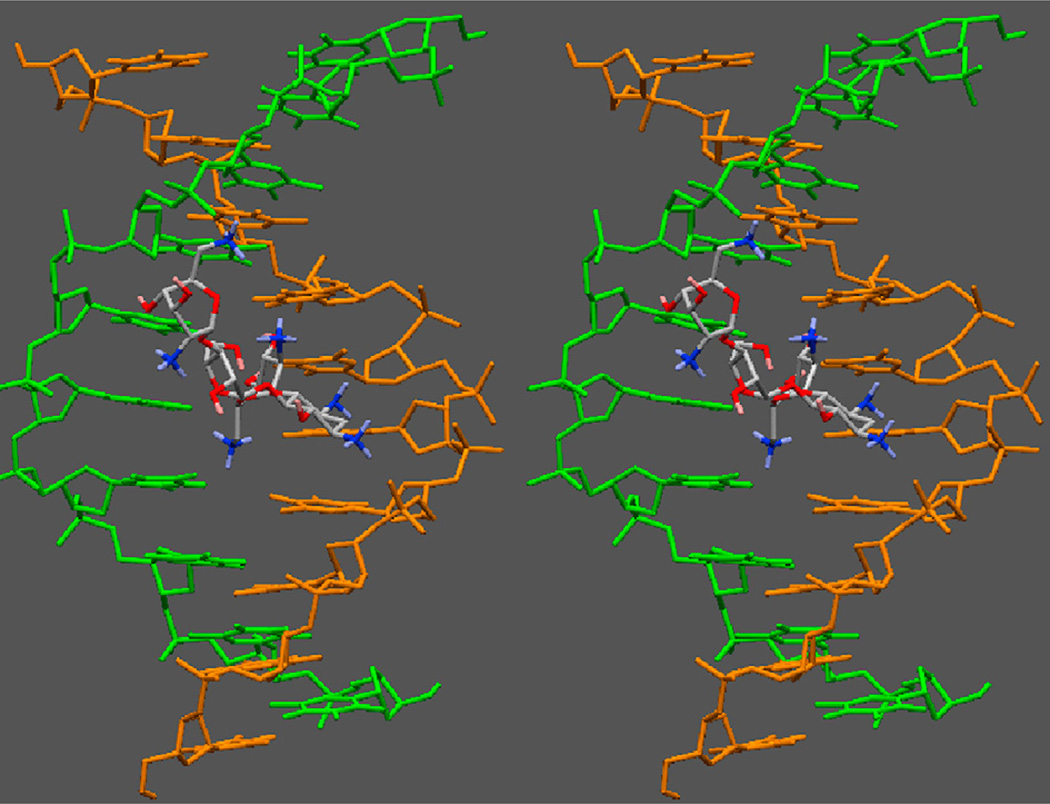
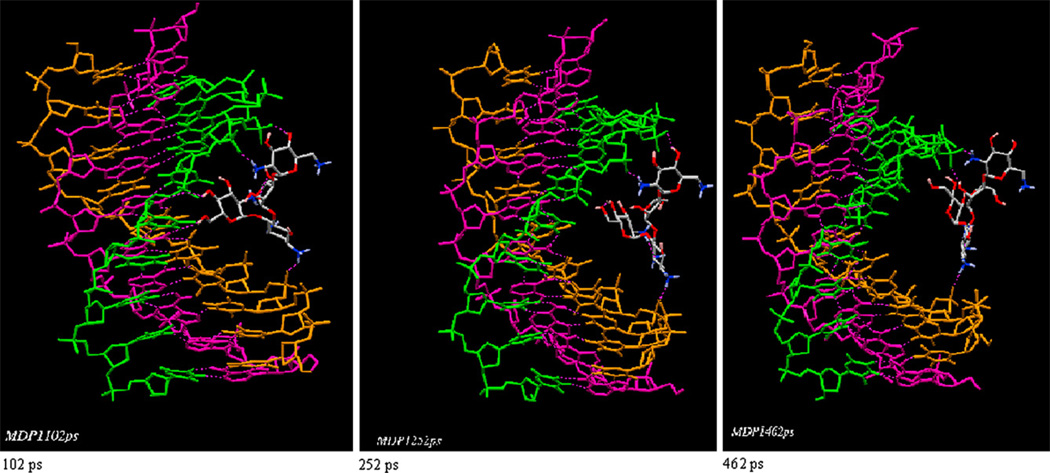
We have previously proposed a model for neomycin binding to the W-H groove of the DNA triplex [51]. The modeling studies suggest that the most likely mode of neomycin binding involves the primary amine in neomycin ring I that occupies the center of the groove, while the amines on ring II and IV help bridge the two pyrimidine strands together. A stereo view of this model is shown in Fig. 8. The salt dependent ITC studies further substantiate this model, with three ion pairs being formed between the neomycin amines and the TAT triplex groove, as shown in Fig. 8. In addition to the charge complementarity, the shape complementarity of neomycin and the W–H groove has also been suggested as a key factor in the recognition of the DNA triplex by neomycin. A molecular dynamics simulation (up to 1 ps) shows this model of neomycin to the DNA triplex to be relatively stable. On the other hand, a different picture emerges if paromomycin is docked into the W–H groove in a similar orientation (Fig. 9). The replacement of 6′-amine group on neomycin ring I with an 6′-OH in paromomycin destabilizes the complex such that in about 500 ps, paromomycin is released from the groove, and as a result, the 6′-OH group cannot realize efficient electrostatic interaction or H-bonding within W–H groove. Only the 2′-amino group can form this interaction, but it may be relatively weak due to its inflexibility and its long distance from the DNA backbone. Paromomycin perhaps binds to the W–H groove in a different orientation from neomycin (for example, ring II or ring IV being docked into the W–H groove). This difference in stability is also borne out by the lower binding affinity of paromomycin vs. neomycin (Table 1a–d), as well as the difference in DNA triplex stability induced by the two drugs (Fig. 2), showing how a single charge can have a dramatic effect on the recognition of biomolecules.
Fig. 8.
Stereo view of neomycin bound to the W–H groove of the DNA triplex. Only the two pyrimidine strands are shown for clarity. Neomycin occupies 6–7 base-pairs per triplex.
Fig. 9.
A 500 ps MD simulation of paromomycin bound to the W–H groove of the DNA triplex, with ring I inside the groove. Three snapshots show the dissociation of paromomycin with time. Dissociation of ring I drives the molecule outside the groove.
Our previous work[53]has also suggested that aminoglycoside specificity (neomycin in high nM- low µM range) may be for nucleic acid forms that show some features characteristic of an A-type conformation {RNA triplex [38,101], DNA–RNA hybrid duplex [39], RNA duplex [102], DNA triplex [38,50,103–105], A-form DNA duplex[103]and DNA tetraplex [106]}. The conformation of the triplex and of the Watson–Crick duplex within the triplexes has been debated for some time. Both A-like[107–110]and B-like [111– 113]geometry have been described in the literature. Some previous studies [67,112–116] on different types of DNA triple helices have suggested that the structure of the DNA triple helix to be closer to a B-form helix. Because DNA triplex have mixed properties, such as an axial rise similar to B-DNA, an X-displacement intermediate between the A- and B-DNA, a low helical twist (A-DNA), and S-type sugar puckers (B-DNA), terms such as Ψ-DNA and P-DNA, have been used to describe the DNA triplex conformation [117,118]. Given the data available, it can be safely said that DNA triplexes have some A-form characteristics. The lower association constant observed in 2-deoxystreptamine containing aminoglycoside–triplex interactions than in RNA triplex, RNA duplex and RNA.DNA hybrids [53,105,119–126] can then be explained by the fact that the DNA triplex lacks all the A-form characteristics that are present in RNA containing duplexes and triplexes. Based on this hypothesis, as the A-form features increase in a set of nucleic acid structures, the association constant of aminoglycoside–nucleic acid should increase. According to the available set of aminoglycoside–nucleic acid interactions, this seems to be the case [53,121–123].
4. Conlusion
The following conclusions can be drawn from above results: Among the aminoglycosides studied here, neomycin is the most effective agent in the stabilization of the 5′-dA12-x-dT12-x-dT12-3′ and polynucleotide poly(dA).2poly(dT) triplex. UV, ITC, fluorescence and DSC were used to calculate the observed binding affinities for both the 5′-dA12-x-dT12-x-dT12-3′ and the poly(dA).2poly (dT) triplex. The observed Tm(3→2) values for neomycin appear to be approximately 25 °C greater for poly(dA).2poly(dT) than for 5′-dA12-x-dT12-x-dT12-3′ at pH 6.8 but the trend is reversed at pH 5.5. Furthermore, the observed heat capacity changes are smaller in magnitude for poly(dA).2poly(dT) than 5′-dA12-x-dT12-x-dT12-3′. These observations can most likely be attributed to the differences in conformation between the poly(dA) . 2poly(dT) triplex and the intramolecular covalently linked triplex. The salt dependence studies reveal that there are approximately 3.0 ion pairs formed upon neomycin binding with poly(dA).2poly(dT). The FID and ITC based ΔTm methods provide a range of affinities for aminoglycoside binding to the DNA triplex. The work reported here gives an increased understanding of drug–triplex interactions from a thermodynamic standpoint, forming the basis for further investigation and development of triplex selective agents and therapeutics.
Supplementary Material
Acknowledgement
PI thanks NIH (R15CA125724) and NSF (CHE/MCB-0134972) for support.
Abbreviations
- ITC
Isothermal titration calorimetry
- DSC
Differential scanning calorimetry
- MOPS
3-[N-morpholino] propanesulfonic acid
- HS
Hoogsteen
- WC
Watson–Crick
- W–H
Watson–Hoogsteen
- FID
Fluorescence intercalator displacement
Appendix. Supplementary data
UV melting profiles of salt dependence of melting temperature for DNA triplex. CD titration curve of paromomycin with DNA triplex. ITC excess-site titration curves at different salt concentrations and temperatures. This material is available free of charge via the Internet at http://pubs.acs.org. Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.biochi.2010.02.004.
References
- 1.Maher LJ. Prospects for the therapeutic use of antigene oligonucleotides. Cancer Invest. 1996;14:66–82. doi: 10.3109/07357909609018437. [DOI] [PubMed] [Google Scholar]
- 2.Giovannangeli C, Helene C. Progress in developments of triplex-based strategies antisense. Nucleic Acid Drug Dev. 1997;7:413–421. doi: 10.1089/oli.1.1997.7.413. [DOI] [PubMed] [Google Scholar]
- 3.Praseuth D, Guieysse-Peugeot A, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochem. Biophys. Res. Commun. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN, Filippov SA, Frank-Kamenetskii M. DNA H form reuires a homopurine-homopyrimidine mirror repeat. Nature. 1987;330:495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- 5.Maher LJI, Wold B, Dervan PB. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989;245:725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- 6.Strobel SA, Doucette-Stamm L, Riba L, Housman DE, Dervan PB. Sitespecific cleavage of human chromosome 4 mediated by triple helix formation. Science. 1991;254:1639–1642. doi: 10.1126/science.1836279. [DOI] [PubMed] [Google Scholar]
- 7.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Chen ZW, Zhang SJ, Wilson GL, Jing K. Detection and determination of oligonucleotide tripler formation-mediated transcription-coupled DNA repair in HeLa nuclear extracts. Nucleic Acids Res. 2001;29:1801–1807. doi: 10.1093/nar/29.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francois JC, Saisonbehmoaras T, Thuong NT, Helene C. Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry. 1989;28:9617–9619. doi: 10.1021/bi00451a011. [DOI] [PubMed] [Google Scholar]
- 10.Carbone GM, Napoli S, Valentini A, Cavalli F, Watson DK, Catapano CV. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 2004;32:4358–4367. doi: 10.1093/nar/gkh744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carbone GM, McGuffie E, Napoli S, Flanagan CE, Dembech C, Negri U, Arcamone F, Capobianco ML, Catapano CV. DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene. Nucleic Acids Res. 2004;32:2396–2410. doi: 10.1093/nar/gkh527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone GM, McGuffie EM, Collier A, Catapano CV. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schleifman EB, Chin JY, Glazer PM. Triplex-mediated gene modification. Methods Mol. Biol. 2008;435:175–190. doi: 10.1007/978-1-59745-232-8_13. [DOI] [PubMed] [Google Scholar]
- 14.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. The triple helix: 50 years later the outcome. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirkin SM. Discovery of alternative DNA structures: a heroic decade (1979–1989) Front Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 17.Simon P, Cannata F, Concordet JP, Giovannangeli C. Targeting DNA with triplex-forming oligonucleotides to modify gene sequence. Biochimie. 2008;90:1109–1116. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Vekhoff P, Ceccaldi A, Polverari D, Pylouster J, Pisano C, Arimondo PB. Triplex formation on DNA targets: how to choose the oligonucleotide. Biochemistry. 2008;47:12277–12289. doi: 10.1021/bi801087g. [DOI] [PubMed] [Google Scholar]
- 19.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 20.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 21.Hoyne PR, Gacy AM, McMurray CT, Maher LJ. Stabilities of intrastrand pyrimidine motif DNA and RNA triple helices. Nucleic Acids Res. 2000;28:770775. doi: 10.1093/nar/28.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyamichev VI, Mirkin SM, Frank-Kamenetskii M. Structures of homopurine-homopyrimidine tract in superhelical DNA. J. Biomol. Struct Dyn. 1986;3:667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- 23.Lyamichev VI, Mirkin SM, Frank-Kamenetskii M. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J. Biomol. Struct Dyn. 1985;3:327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- 24.Kohwi Y, Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc. Natl. Acad. Sci U.S.A. 1988;85:3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu M, Hanvey JC, Wells RD. Intramolecular DNA triplexes in supercoiled plasmids .1. Effect of loop size on formation and stability. J. Biol Chem. 1989;264:5944–5949. [PubMed] [Google Scholar]
- 26.Kato M, Shimizu N. Effect of the potential triplex DNA region on the invivo expression of bacterial beta-Lactamase gene in superhelical recombinant plasmids. J. Biochem. 1992;112:492–494. doi: 10.1093/oxfordjournals.jbchem.a123927. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar PS, Brahmachari SK. Intramolecular triplex potential sequence within a gene down regulates its expression invivo. Nucleic Acids Res. 1992;20:5713–5718. doi: 10.1093/nar/20.21.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JS, Latimer LJP, Haug BL, Pulleyblank DE, Skinner DM, Burkholder GD. Triplex DNA in plasmids and chromosomes. Gene. 1989;82:191–199. doi: 10.1016/0378-1119(89)90044-9. [DOI] [PubMed] [Google Scholar]
- 29.Parniewski P, Kwinkowski M, Wilk A, Klysik J. Dam methyltransferase sites located within the loop region of the oligopurine-oligopyrimidine sequences capable of forming H-DNA are undermethylated invivo. Nucleic Acids Res. 1990;18:605–611. doi: 10.1093/nar/18.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ussery DW, Sinden RR. Environmental-influences on the in-vivo level of intramolecular triplex DNA in Escherichia coli. Biochemistry. 1993;32:6206–6213. doi: 10.1021/bi00075a013. [DOI] [PubMed] [Google Scholar]
- 31.van Dongen MJP, Doreleijers JF, van DM, van Boom JH, Hilbers CW, Wijmenga SS. Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix. Nat. Struct. Biol. 1999;6:854–859. doi: 10.1038/12313. [DOI] [PubMed] [Google Scholar]
- 32.Postel EH, Mango SE, Flint SJ. A nuclease-hypersensitive element of the human C-Myc promoter interacts with a transcription initiation-factor. Mol. Cell. Biol. 1989;9:5123–5133. doi: 10.1128/mcb.9.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinreb A, Collier DA, Birshtein BK, Wells RD. Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J. Biol. Chem. 1990;265:1352–1359. [PubMed] [Google Scholar]
- 34.Liu QR, Chan PK. Identification of a long stretch of homopurine homopyrimidine sequence in a cluster of retroposons in the human genome. J. Mol. Biol. 1990;212:453–459. doi: 10.1016/0022-2836(90)90324-F. [DOI] [PubMed] [Google Scholar]
- 35.Horwitz EM, Maloney KA, Ley TJ. A human protein containing a cold shock domain binds specifically to H-DNA upstream from the human gammaglobin genes. J. Biol. Chem. 1994;269:14130–14139. [PubMed] [Google Scholar]
- 36.Craig ME, Crother DM, Doty P. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol. 1971;62:383–401. doi: 10.1016/0022-2836(71)90434-7. [DOI] [PubMed] [Google Scholar]
- 37.Porschke D, Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic-oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 38.Arya DP, Lane Coffee RL, Willis B, Abramovitch AI. Aminoglycosides-nucleic acid interactions: remarkable stabilization of DNA and RNA triple helices by neomycin. J. Am. Chem. Soc. 2001;123:538–5395. doi: 10.1021/ja003052x. [DOI] [PubMed] [Google Scholar]
- 39.Arya DP, Coffee RL, Jr, Charles I. Neomycin-induced hybrid triplex formation. J. Am. Chem. Soc. 2001;123:11093–11094. doi: 10.1021/ja016481j. [DOI] [PubMed] [Google Scholar]
- 40.Mergny JL, Duvalvalentin G, Nguyen CH, Perrouault L, Faucon B, Rougee M, Montenaygarestier T, Bisagni E, Helene C. Triple helix specific ligands. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 41.Ren J, Chaires JB. Preferential binding of 3,3′-diethyloxadicarbocyanine to triplex DNA. J. Am. Chem. Soc. 2000;122:424–425. [Google Scholar]
- 42.Tam VK, Liu Q, Tor Y. Extended ethidium bromide analogue as a triple helix intercalator: synthesis photophysical properties and nucleic acids binding. Chem. Commun. 2006;25:2684–2686. doi: 10.1039/b604281c. [DOI] [PubMed] [Google Scholar]
- 43.Wilson WD, Mizan S, Tanious FA, Yao S. The interaction of intercalators groove-binding agents with DNA triple-helical structures: the influence of ligand structure DNA backbone modifications and sequence. J. Mol. Recognit. 1994;7:89–98. doi: 10.1002/jmr.300070206. [DOI] [PubMed] [Google Scholar]
- 44.Cassidy SA, Strekowski L, Wilson WD, Fox KR. Effect of a triplex-binding ligand on parallel and antiparallel DNA triple helixes using short unmodified and acridine-linked oligonucleotides. Biochemistry. 1994;33:15338–15347. doi: 10.1021/bi00255a015. [DOI] [PubMed] [Google Scholar]
- 45.Choi S-D, Kim M-S, Kim SK, Lincoln P, Tuite E, Norden B. Binding mode of [ruthenium(II) (1,10-phenanthroline)2L]2+ with Poly(dT*dA-dT) triplex. Ligand size effect on third-strand stabilization. Biochemistry. 1997;36:214–223. doi: 10.1021/bi961675a. [DOI] [PubMed] [Google Scholar]
- 46.Mergny JL, Collier D, Rougee M, Montenay-Garestier T, Helene C. Intercalation of ethidium bromide into a triple-stranded oligonucleotide. Nucleic Acids Res. 1991;19:1521–1526. doi: 10.1093/nar/19.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durand M, Thuong NT, Maurizot JC. Berenil complexation with a nucleic acid triple helix. J. Biomol. Struct. Dyn. 1994;11:1191–1202. doi: 10.1080/07391102.1994.10508063. [DOI] [PubMed] [Google Scholar]
- 48.Park YW, Breslauer KJ. Drug binding to higher ordered DNA structures: netropsin complexation with a nuclear acid triple helix. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6653–6657. doi: 10.1073/pnas.89.14.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durand M, Thuong NT, Maurizot JC. Interaction of hoechst 33258 with a DNA triple helix. Biochimie. 1994;76:181–186. doi: 10.1016/0300-9084(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 50.Arya DP, Coffee RL. DNA triple helix stabilization by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett. 2000;10:1897–1899. doi: 10.1016/s0960-894x(00)00372-3. [DOI] [PubMed] [Google Scholar]
- 51.Arya DP, Micovic L, Charles I, Coffee RL, Willis B, Xue L. Neomycin binding to DNA triplex Watson-Hoogsteen (W–H) groove: a model. J. Am. Chem. Soc. 2003;125:3733–3744. doi: 10.1021/ja027765m. [DOI] [PubMed] [Google Scholar]
- 52.Arya DP, Xue L, Tennant P. Combining the best in triplex recognition: synthesis and nucleic acid binding of a BQQ-Neomycin conjugate. J. Am. Chem. Soc. 2003;125:8070–8071. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 53.Arya DP, Xue L, Willis B. Aminoglycoside (Neomycin) preference is for A-form nucleic acids ot just RNA: results from a competition dialysis study. J. Am. Chem. Soc. 2003;125:10148–10149. doi: 10.1021/ja035117c. [DOI] [PubMed] [Google Scholar]
- 54.Napoli S, Carbone GM, Catapano CV, Shaw N, Arya DP. Neomycin improves cationic lipid-mediated transfection of DNA in human cells. Bioorg. Med. Chem. Lett. 2005;15:3467–3469. doi: 10.1016/j.bmcl.2005.04.038. [DOI] [PubMed] [Google Scholar]
- 55.Charles I, Xi H, Arya DP. Sequence-Specific targeting of RNA with an oligonucleotide-neomycin conjugate. Bioconjug. Chem. 2007;18:160–169. doi: 10.1021/bc060249r. [DOI] [PubMed] [Google Scholar]
- 56.Shaw N, Xi H, Arya DP. Novel targets for aminoglycosides. In: Dev Arya P, editor. Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. John Wiley and Sons; 2007. Chapter 11. [Google Scholar]
- 57.Trent JO. Molecular modeling of drug-DNA complexes: an update. Meth. Enzymol. 2001;340:290 –326. doi: 10.1016/s0076-6879(01)40428-9. [DOI] [PubMed] [Google Scholar]
- 58.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986;7:230. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 59.McDonald DQ, Still WC. AMBER torsional parameters for the peptide backbone. Tetrahedron Lett. 1992;33:7743–7746. [Google Scholar]
- 60.Qiu D, Shenkin PS, Hollinger FP, Still WC. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate born radii. J. Phys. Chem. 1997;101:3005–3014. [Google Scholar]
- 61.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:61207–6129. [Google Scholar]
- 62.Shields GC, Laughton CA, Orozco M. Molecular dynamics simulations of the d(T.A.T) triple helix. J. Am. Chem. Soc. 1997;119:7463–7469. [Google Scholar]
- 63.Tarkoy M, Phipps AK, Schultze P, Feigon J. Solution structure of an intramolecular DNA triplex linked by hexakis (ethylene glycol) units: D[AGA-GAGAA-(EG)6-TTCTCTCT-(EG)6TCTCTCTT] Biochemistry. 1998;37:5810–5819. doi: 10.1021/bi9728102. [DOI] [PubMed] [Google Scholar]
- 64.Chang G, Guida WC, Still WC. An internal coordinate Monte-Carlo method for searching conformational space. J. Am. Chem. Soc. 1989;111:4379–4386. [Google Scholar]
- 65.Kolossvary I, Guida WC. Torsional flexing conformational searching of cyclic molecules in biased internal coordinate space. J. Comput. Chem. 1993;14:691–698. [Google Scholar]
- 66.Durand M, Peloille S, Thuong NT, Maurizot JC. Triple-helix formation by an oligonucleotide containing one (dA)12 and 2. (dT)12 sequences bridged by 2 hexaethylene glycol chains. Biochemistry. 1992;31:9197–9204. doi: 10.1021/bi00153a012. [DOI] [PubMed] [Google Scholar]
- 67.Bartley JP, Brown T, Lane AN. Solution conformation of an intramolecular DNA triplex containing a nonnucleotide linker: comparison with the DNA duplex. Biochemistry. 1997;36:14502–14511. doi: 10.1021/bi970710q. [DOI] [PubMed] [Google Scholar]
- 68.Kaul M, Barbieri CM, Kerrigan JE, Pilch DS. Coupling of drug protonation to the specific binding of aminoglycosides to the A site of 16 S rRNA: elucidation of the number of drug amino groups involved and their identities. J. Mol. Biol. 2003;326:1373–1387. doi: 10.1016/s0022-2836(02)01452-3. [DOI] [PubMed] [Google Scholar]
- 69.Singh SK, Caram-Lelham N. Thermodynamics of kappa-carrageenan-mmphiphilic drug interaction as influenced by specific counterions and temperature: a microcalorimetric and viscometric study. J. Colloid Interface Sci. 1998;203:430–446. doi: 10.1006/jcis.1998.5502. [DOI] [PubMed] [Google Scholar]
- 70.Boger DL, Fink BE, Hedrick MP. Total synthesis of distamycin A 2640 analogues: a solution-phase combinatorial approach to the discovery of new bioactive DNAbinding agents and development of a rapid high-throughput screen for determining relative DNA binding affinity or DNA binding sequence selectivity. J. Am. Chem. Soc. 2000;122:6382–6394. [Google Scholar]
- 71.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP, simple A. high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 72.Boger DL, Tse WC. Thiazole orange as the fluorescent intercalator in a high resolution FID assay for determining DNA binding affinity and sequence selectivity of small molecules. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 73.Junquera E, Aicart E. Thermodynamic analysis of the binding of a hep-atoprotectant drug thioctic acid by beta-cyclodextrin. J. Pharm. Sci. 1999;88:626–631. doi: 10.1021/js980458n. [DOI] [PubMed] [Google Scholar]
- 74.Ehtezazi T, Govender T, Stolnik S. Hydrogen bonding and electrostatic interaction contributions to the interaction of a cationic drug with poly-aspartic acid. Pharm. Res. 2000;17:871–878. doi: 10.1023/a:1007520628237. [DOI] [PubMed] [Google Scholar]
- 75.Cervoni L, Lascu I, Xu Y, Gonin P, Morr M, Merouani M, Janin J, Giartosio A. Binding of nucleotides to nucleoside diphosphate kinase: a calorimetric study. Biochemistry. 2001;40:4583–4589. doi: 10.1021/bi002432s. [DOI] [PubMed] [Google Scholar]
- 76.Turnbull WB, Daranas AH. On the value ofc: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 77.Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 78.Kaul M, Pilch DS. Thermodynamics of aminoglycoside-rRNA recognition: the binding of neomycin-class aminoglycosides to the A site of 16S rRNA. Biochemistry. 2002;41:7695–7706. doi: 10.1021/bi020130f. [DOI] [PubMed] [Google Scholar]
- 79.Botto RE, Coxon B. Nitroen-15 nuclear magnetic resonance spectroscopy of neomycin B and related aminoglycosides. J. Am. Chem. Soc. 1983;105:1021–1028. [Google Scholar]
- 80.McGhee JD. Theoretical calculations of the helix-coil transition of DNA in the presence of large cooperatively binding ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 81.Doyle ML, Brigham-Burke M, Blackburn MN, Brooks IS, Smith TM, Newman R, Reff M, Stafford WF, Sweet RW, Truneh A, Hensley P, O'Shannessy DJ. Measurement of protein interaction bioenergetics: application to structural variants of anti-sCD4 antibody. Meth. Enzymol. 2000;323:207–230. doi: 10.1016/s0076-6879(00)23368-5. [DOI] [PubMed] [Google Scholar]
- 82.Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Chaires JB. Specific binding of hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 83.Jenkins TC, Chowdhry BZ, Ren J, Chaires JB. Drug-DNA interaction protocols. Methods Mol. Biol. 1997:195–218. [Google Scholar]
- 84.Polak M, Hud NV. Complete disproportionation of duplex poly(dT)*poly (dA) into triplex poly(dT)*poly(dA)*poly(dT) and poly(dA) by coralyne. Nucleic Acids Res. 2002;30:983–992. doi: 10.1093/nar/30.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barbieri CM, Srinivasan AR, Pilch DS. Deciphering the origins of observed heat capacity changes for aminoglycoside binding to prokaryotic eukaryotic ribosomal RNA a-sites: a calorimetric computational and osmotic stress study. J. Am. Chem. Soc. 2004;126:14380–14388. doi: 10.1021/ja0457516. [DOI] [PubMed] [Google Scholar]
- 86.Haq I. Thermodyanmics of drug-DNA interactuibs. Arch. Biochem. Biophys. 2002;403:1–15. doi: 10.1016/S0003-9861(02)00202-3. [DOI] [PubMed] [Google Scholar]
- 87.Mazur S, Tanious FA, Ding DY, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A thermodynamic and structural analysis of DNA minor-groove complex formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 88.Ren J, Jenkins TC, Chaires JB. Energetics of DNA intercalation reactions. Biochemistry. 2000;39:8439–8447. doi: 10.1021/bi000474a. [DOI] [PubMed] [Google Scholar]
- 89.Hopkins HPJ, Fumero J, Wilson WD. Temperature dependence of enthalpy changes for ethidium and propidium binding to DNA: effect of alkylamine chains. Biopolymers. 1990;29:449–459. doi: 10.1002/bip.360290215. [DOI] [PubMed] [Google Scholar]
- 90.Spolar RS, Record MTJ. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 91.Ha JH, Spolar RS, Record MTJ. Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J. Mol. Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- 92.Spolar RS, Livingstone JR, Record MTJ. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 93.Bergqvist S, Williams MA, O'Brien R, Ladbury JE. Heat capacity effects of water molecules and ions at a protein-DNA interface. J. Mol. Biol. 2004;336:829–842. doi: 10.1016/j.jmb.2003.12.061. [DOI] [PubMed] [Google Scholar]
- 94.Ferrari ME, Lohman TM. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 95.Peters WB, Edmondson SP, Shriver JW. Thermodynamics of DNANA binding and distortion by the hyperthermophile chromatin protein Sac7d. J. Mol. Biol. 2004;343:339–360. doi: 10.1016/j.jmb.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 96.Barbieri CM, Pilch DS. Complete thermodynamic characterization of the multiple protonation equilibria of the aminoglycoside antibiotic paromo-mycin: a calorimetric and natural abundance N-15 NMR study. Biophys. J. 2006;90:1338–1349. doi: 10.1529/biophysj.105.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kozlov AG, Lohman TM. Effects of monovalent anions on a temperature-dependent heat capacity change for Escherichia coli SSB tetramer binding to single-stranded DNA. Biochemistry. 2006;45:5190–5205. doi: 10.1021/bi052543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.deHaseth PL, Lohman TM, Record MT. Nonspecific interaction of Lac reperssor with DNA: an association reaction driven by counerion release. Biochemistry. 1977;16:4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]
- 99.Jin E, Katritch V, Olson WK, Kharatisvili M, Abagyan R, Pilch DS. Ami-noglycoside binding in the major groove of duplex RNA: the thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol. 2000;298:95–110. doi: 10.1006/jmbi.2000.3639. [DOI] [PubMed] [Google Scholar]
- 100.Pilch DS, Kirolos MA, Liu XY, Plum GE, Breslauer KJ. Berenil [1,3-bis (4′-amidinophenyl)triazene] binding to DNA duplexes and to a RNA duplex - evidence for both intercalative and minor-groove binding-properties. Biochemistry. 1995;34:9962–9976. doi: 10.1021/bi00031a019. [DOI] [PubMed] [Google Scholar]
- 101.Xue L, Charles I, Arya DP. Pyrene-neomycin conjugate: dual recognition of a DNA triple helix. Chem. Commun. 2002:70–71. doi: 10.1039/b108171c. [DOI] [PubMed] [Google Scholar]
- 102.Chen Q, Shafer RH, Kuntz ID. Structure-based discovery of ligands targeted to the RNA double helix. Biochemistry. 1997;36:11402–11407. doi: 10.1021/bi970756j. [DOI] [PubMed] [Google Scholar]
- 103.Robinson H, Wang AHJ. Neomycin spermine and hexaamminecobalt(III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res. 1996;24:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arnott S, Bond PJ, Selsing E, Smith PJ. Models of triple-stranded polynucleotides with optimised stereochemistry. Nucleic Acids Res. 1976;3:2459–2470. doi: 10.1093/nar/3.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xi H, Arya DP. Recognition of triple helical nucleic acids by aminoglycosides. Curr. Med. Chem. Anticancer Agents. 2005;5:327–338. doi: 10.2174/1568011054222328. [DOI] [PubMed] [Google Scholar]
- 106.Kypr J, Fialova M, Chladkova J, Tumova M, Vorlickova M. Conserved guanine-guanine stacking in tetraplex and duplex DNA. Eur. Biophys. J. 2001;30:555–558. doi: 10.1007/s002490100174. [DOI] [PubMed] [Google Scholar]
- 107.Weerasinghe S, Smith PE, Mohan V, Cheng Y-, Pettitt BM. Nanosecond dynamics and structure of a model DNA triple helix in saltwater solution. J. Am. Chem. Soc. 1995;117:2147–2158. [Google Scholar]
- 108.Umemoto K, Sarma MH, Gupta G, Luo J, Sarma RH. Structure and stability of a DNA triplex in solution: NMR studies on d(T)6.d(A)6.d(T)6 and its complex with a minor groove binding drug. J. Am. Chem. Soc. 1990;112:4539–4545. [Google Scholar]
- 109.Arnott S, Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT) J. Mol. Biol. 1974;88:509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- 110.Betts L, Josey JA, Veal JM, Jordan SR. A nucleic-acid triple-helix formed by a peptide nucleic-acid DNA complex. Science. 1995;270:1838–1841. doi: 10.1126/science.270.5243.1838. [DOI] [PubMed] [Google Scholar]
- 111.Radhakrishnan I, Gao X, Santos CDL, Live D, Patel DJ. NMR structural studies of intramolecular (Y+)n.Cntdot. (R+)n(Y-)n DNA triplexes in solution: imino and amino proton and nitrogen markers of G.Cntdot.AT base triple formation. Biochemistry. 1991;30:9022–9030. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- 112.Macaya RF, Schultze P, Feigon J. Sugar conformations in intramolecular DNA triplexes determined by coupling constants obtained by automated simulation of P.COSY cross peaks. J. Am. Chem. Soc. 1992;114:781–783. [Google Scholar]
- 113.Koshlap KM, Schultze P, Brunar H, Dervan PB, Feigon J. Solution structure of an intramolecular DNA triplex containing an N7-glycosylated guanine which mimics a protonated cytosine. Biochemistry. 1997;36:2659–2668. doi: 10.1021/bi962438a. [DOI] [PubMed] [Google Scholar]
- 114.Dagneaux C, Liquier J, Taillandier E. Sugar conformations in DNA and RNA—DNA triple helixes determined by FTIR spectroscopy: role of backbone composition. Biochemistry. 1995;34:16618–16623. doi: 10.1021/bi00051a009. [DOI] [PubMed] [Google Scholar]
- 115.Howard FB, Miles HT, Liu K, Frazier J, Raghunathan G, Sasisekharan V. Structure of d(T)d(A)d(T): the DNA triple helix has B-form geometry with C2′-endo sugar pucker. Biochemistry. 1992;31:10671–10677. doi: 10.1021/bi00159a005. [DOI] [PubMed] [Google Scholar]
- 116.Radhakrishnan I, Patel DJ, Veal JM, Gao X. Solution conformation of a G. cntdotTA [Guanosine.cntdot.thymidine-5′-adenylic acid] triple in an intramolecular pyrimidine.cntdot.purine.cntdot.pyrimidine DNA triplex. J. Am. Chem. Soc. 1992;114:6913–6915. [Google Scholar]
- 117.Saminathan M, Antony T, Shirahata A, Sigal LH, Thomas T, Thomas TJ. Ionic structural specificity effects of natural synthetic polyamines on the aggregation resolubilization of single-, double-, and triple-stranded DNA. Biochemistry. 1999;38:3821–3830. doi: 10.1021/bi9825753. [DOI] [PubMed] [Google Scholar]
- 118.Radhakrishnan I, Patel DJ. DNA triplexes: solution structures hydration sites energetics interactions and function. Biochemistry. 1994;33:11405–11416. doi: 10.1021/bi00204a001. [DOI] [PubMed] [Google Scholar]
- 119.Shaw NN, Arya DP. Recognition of the unique structure of DNA: RNA hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 120.Shaw NN, Xi H, Arya DP. Molecular recognition of a DNA: RNA hybrid: sub-nanomolar binding by a neomycin-methidium conjugate. Bioorg. Med. Chem. Lett. 2008;18:4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 121.Arya DP. Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. Hoboken NJ: Wiley-Interscience; 2007. [Google Scholar]
- 122.Willis B, Arya DP. An expanding view of aminoglycoside-nucleic acid recognition. Adv. Carbohydr. Chem. Biochem. 2006;60:251–302. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 123.Arya DP. Aminoglycoside-nucleic acid interactions: the case for neomycin. Top. Curr. Chem. 2005;253:149–178. [Google Scholar]
- 124.Willis B, Arya DP. Triple recognition of B-DNA. Bioorg. Med. Chem. Lett. 2009;19:4974–4979. doi: 10.1016/j.bmcl.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 125.Willis B, Arya DP. Triple recognition of B-DNA by a Neomycin-Hoechst 33258-pyrene conjugate. Biochemistry. 2010;49:452–469. doi: 10.1021/bi9016796. [DOI] [PubMed] [Google Scholar]
- 126.Xi H, Gray D, Kumar S, Arya DP. Molecular recognition of single-stranded RNA: neomycin binding to poly(A) FEBS Lett. 2009;13:2269–2275. doi: 10.1016/j.febslet.2009.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.