Abstract
Bardet-Biedl syndrome (BBS) is an autosomal recessive disease characterized by retinal dystrophy, obesity, postaxial polydactyly, learning disabilities, renal involvement, and male hypogenitalism. BBS is genetically heterogeneous, and to date 18 genes (BBS1-18) have been described. Mutations in known BBS genes account for approximately 70-80% of cases, and triallelic inheritance has been suggested in about 5%. Many minor features can be helpful in making the clinical diagnosis. Recently, the use of next-generation sequencing technologies has accelerated the identification of novel genes and causative disease mutations in known genes. This report presents a concise overview of the current knowledge on clinical data in BBS and the progress in molecular genetics research. A future objective will be the development of BBS diagnosis kits in order to offer genetic counseling for families at risk.
Key Words: Bardet-Biedl syndrome, Molecular diagnosis, Next-generation sequencing
Bardet-Biedl syndrome (BBS, OMIM 209900) is a pleiotropic genetic disorder characterized by a wide spectrum of clinical signs including progressive retinal degeneration, postaxial polydactyly, obesity, learning difficulties, and renal tract and genital anomalies as well as other minor frequent features such as anosmia, ataxia, or Hirschsprung disease. Clinical diagnosis is established if at least 4 major features are present in a patient [Beales et al., 1999]. The phenotypic spectrum and timing of the onset of BBS-associated symptoms are highly variable; some manifestations can appear during childhood. BBS is considered a rare disorder: its prevalence in Tunisia has been estimated at 1:156,000 [M'hamdi et al., 2011], and the current prevalence in North American and European populations ranges from 1:140,000-160,000 live births. Populations with a high rate of consanguinity or from isolated regions have been characterized with a higher frequency of BBS such as for Kuwait (1:17,000) and Newfoundland (1:18,000) [Farag and Teebi, 1989; Moore et al., 2005].
BBS is a genetically heterogeneous disorder. To date, 18 genes have been described (BBS1-18) [Scheidecker et al., 2013], and 7 BBS proteins (BBS1, 2, 4, 5, 7, 8, and 9) form a stable complex ‘BBSome’ [Nachury et al., 2007; Lechtreck et al., 2009]. BBS is considered an autosomal recessive disease. Oligogenic inheritance has been shown in some BBS families [Katsanis et al., 2001; Leitch et al., 2008; Zaghloul et al., 2010]. Mutations in BBS1-18 account for 70-80% of affected BBS families [Zaghloul and Katsanis, 2009; Muller et al., 2010; M'hamdi et al., 2013]. Founder mutations were described in Tunisian BBS families [Smaoui et al., 2006], in the Hutterite population [Innes et al., 2010], and in the Faroe Islands [Hjortshoj et al., 2009]. Recently, the advent of next-generation sequencing technologies has accelerated the identification of novel BBS genes and causative disease mutations in known genes [Otto et al., 2010; Marion et al., 2012; Redin et al., 2012; Ajmal et al., 2013; M'hamdi et al., 2013]. This report presents a concise overview on the current knowledge of BBS including clinical and molecular data as well as a discussion of the future research directions aimed at the management of molecular diagnosis.
Bardet-Biedl Syndrome: Clinical Summary
Bardet-Biedl syndrome is diagnosed if at least 4 of the main manifestations are present in a patient. Clinical evaluation during early infancy remains difficult as not all of the main manifestations are congenital but may occur later during childhood (table 1).
Table 1.
Clinical diagnosis features in Bardet-Biedl syndrome and their frequencies
| Clinical feature | Frequency, % |
|---|---|
| Major feature | |
| Rod-cone dystrophy | 90–100 |
| Obesity | 72–92 |
| Polydactyly | 63–81 |
| Genital anomalies | 59–98 |
| Learning difficulties | 50–61 |
| Renal anomalies | 20–53 |
| Minor feature | |
| Speech delay | 54–81 |
| Developmental delay | 50–91 |
| Diabetes mellitus | 6–48 |
| Dental anomalies | 51 |
| Congenital heart disease | 7 |
| Brachydactyly/syndactyly | 46–100 |
| Ataxia/poor coordination | 40–86 |
| Cardiopathy | 10 |
| Deafness | 11–12 |
| Anosmia/hyposmia | 60 |
Retinal Degeneration
BBS is recognized as one of the major causes of syndromic retinal dystrophy which leads to a severe visual handicap before adulthood in BBS patients, and total blindness usually occurs before the second decade of life [Mockel et al., 2011]. Different forms of retinal dystrophy have been described, including a cone-rod dystrophy or rod-cone dystrophy, choroidal dystrophy, and so-called ‘global severe retinal dystrophy’. Cone-rod dystrophy is defined as a progressive retinal degeneration with initial decreased visual acuity, impaired color vision, and electroretinogram abnormalities with cone functions affected at an early stage prior to the rods. Furthermore, rod-cone dystrophies are characterized by initial rod involvement with subsequent cone alteration. Molecular genetics analysis has revealed unclear retinal genotype-phenotype correlations [Riise, 1987; Beales et al., 1999; Riise et al., 2002; Hamel, 2007; Gerth et al., 2008].
Obesity
Obesity is the second major feature in BBS patients; the current incidence of obesity in the BBS cohort has been estimated to be 72-92% [Riise et al., 1997; Beales et al., 1999; Moore et al., 2005; M'hamdi et al., 2013]. Usually beginning in early childhood and becoming severe with age, obesity appears to be widespread and diffuse [Forsythe and Beales, 2013]. Some BBS patients develop type 2 diabetes which can be related to the degree of obesity. The origin of obesity in BBS patients seems to be both central and peripheral as described by molecular and physiological studies [Mykytyn et al., 2001; Davis et al., 2007; Zhang et al., 2011]. BBS mouse models show leptin resistance; BBS proteins are required for leptin receptor localization in the hypothalamus. Moreover, primary cilium and BBS proteins play a key role in the differentiation of adipocytes, suggesting that a defect of adipogenesis contributes to the pathogenesis of obesity in BBS patients [Rahmouni et al., 2008; Marion et al., 2009; Seo et al., 2009].
Polydactyly Limb Anomalies
Polydactyly-type limb anomalies are the third major feature in BBS and may be the only clinical sign present at birth. Classically, polydactyly is postaxial (63-81%) in BBS patients. Other limb defects such as brachydactyly or syndactyly are frequently reported for both hands and feet. Limb malformations in BBS have been associated with a dysregulation of the Sonic hedgehog pathway which is a key developmental pathway implicated in limb development and left/right symmetry [McGlinn and Tabin, 2006; Bimonte et al., 2011; Mockel et al., 2011].
Hypogonadism and Genital Anomalies
Hypogonadism may manifest as delayed puberty or hypogenitalism in males and genital abnormalities in females [Beales et al., 1999]. It may include hypoplastic fallopian tubes and uterus, vaginal atresia, and hydrometrocolpos. In some cases, the vaginal malformation has been reported as leading to lethal abdominal tumors in neonates [Stoler et al., 1995]. Some BBS patients were reported to have given birth to healthy children [Klein and Amman, 1969; Beales et al., 1999].
Cognitive Impairment
Neuropsychiatric manifestations have been described in BBS patients, including intellectual disability, learning difficulties, speech deficits, and behavioral problems such as autistic traits and psychosis [Beales et al., 1999]. The primary cilium is one of the important organelle in human brain cells and is necessary for neurogenesis signaling and hippocampal development [Han et al., 2008]. A recent report showed a reduction of the volume of the hippocampus in BBS patients [Baker et al., 2011].
Renal Abnormalities
Renal failure is one of the primary features and a major cause of morbidity and mortality in BBS patients [Imhoff et al., 2010; Sowjanya et al., 2011]. The renal abnormalities are variable but classically manifest with cystic tubular disease and anatomical malformations. Most BBS patients have been characterized as having urinary concentration defects with normal renal function and no major cysts [Putoux et al., 2012; Marion et al., 2011]. The primary cilium is necessary for water absorption in the kidney [Marion et al., 2011]. In addition, a recent study reported that the vasopressin receptor AV2R is located on the primary cilium and plays a chemosensory role in renal epithelial cells [Raychowdhury et al., 2009].
Genetics of Bardet-Biedl Syndrome
BBS is a genetic heterogeneous disease; up to date 18 genes have been described (BBS1-18) accounting for 70-80% of the BBS cases (table 2). Within the last decade, the introduction of robust genomics analysis technologies such as homozygosity mapping using SNPs arrays and high-throughput sequencing technologies have accelerated the discovery of novel BBS genes and mutations in known causative disease genes [Smaoui et al., 2006; Abu Safieh et al., 2010; Marion et al., 2012; Redin et al., 2012; M'hamdi et al., 2013, Scheidecker et al., 2013].
Table 2.
List of the locus position, the OMIM reference, and the product function, if known, of the Bardet-Biedl syndrome genes cited in this review
| Gene | Locus | Function | Gene/MIM number |
|---|---|---|---|
| BBS1 | 11q13 | BBSome protein | 209901 |
| BBS2 | 16q21 | BBSome protein | 606151 |
| BBS3/ARL6 | 3p12p13 | GTPase | 608845 |
| BBS4 | 15q22.3q23 | BBSome protein | 600374 |
| BBS5 | 2q31 | BBSome protein | 603650 |
| BBS6/MKKS | 20p12 | part of chaperonin complex/BBSome assembly | 604896 |
| BBS7 | 4q27 | BBSome protein | 607590 |
| BBS8/TTC8 | 14q32.1 | BBSome protein | 608132 |
| BBS9 | 7p14 | BBSome protein | 607968 |
| BBS10 | 12q21.2 | part of chaperonin complex | 610148 |
| BBS11/TRIM32 | 9q31q34.1 | E3 ubiquitin ligase | 602290 |
| BBS12 | 4q27 | part of chaperonin complex | 610683 |
| BBS13/MKS1 | 17q23 | basal body/centriole migration | 609883 |
| BBS14/CEP290/NPHP6 | 12q21.3 | basal body: RPGR interaction | 610142 |
| BBS15/WDPCP | 2p15 | basal body: regulation of septins localization and ciliogenesis | 613580 |
| BBS16/SDCCAG8 | 1q43 | basal body/interaction with OFD1 | 613615 |
| BBS17/LZTFL1 | 3p21.3 | BBSome: regulation of BBSome ciliary trafficking and Shh signaling | 606568 |
| BBS18/BBIP1 | 10q25.2 | BBSome protein | 613605 |
BBS Genes Epidemiology
All described BBS genes have been shown to be related to cilium biogenesis and/or function [Mockel et al., 2011]. The BBS mutation spectrum is divergent between populations. In European and Caucasian populations, the most commonly mutated BBS genes are BBS1 and BBS10, together accounting for about 21-30% of the BBS cases in those populations [Badano et al., 2003; Janssen et al., 2011]. In the Tunisian population, the pathogenic mutations have been most frequently found in BBS1, BBS2, and BBS8 [Smaoui et al., 2006; M'hamdi et al., 2013], while BBS1, BBS3, and BBS4 are frequently mutated in the population of Saudi Arabia [Abu Safieh et al., 2010, 2012], contributing to 33, 17, and 17% of disease-associated mutations, respectively. In north European BBS patients, 2 recurrent mutations are most common and predicted to result in the protein change p.M390R (BBS1) (50% of BBS1 cases) and p.C91Lfs*5 (BBS10) [Kim et al., 2010]. In isolated and highly consanguineous populations, founder mutations in BBS genes have been reported as well; in Tunisia, 2 founder mutations have been described: p.R189* in BBS2 and c.459 + 1G>A in BBS8 [Smaoui et al., 2006; Chen et al., 2011; M'hamdi et al., 2013]. In the Faroe Islands, 1 splice mutation c.1091 + 3G>C in BBS1 was reported, and in the Hutterite population, 1 other founder mutation, c.472 – 2A>G in BBS2, was described [Hjortshoj et al., 2009; Innes et al., 2010]. Several BBS genes have been described in other ciliopathies: BBS15 and BBS13 have been reported in Meckel syndrome [Otto et al., 2010]. Similarly, mutations in SDCCAG8 were reported in Meckel syndrome [Schaefer et al., 2011; Billingsley et al., 2012].
Triallelism and Modifier Alleles in Bardet-Biedl Syndrome
Triallelic inheritance in BBS was reported in 2001 in an affected BBS family with unaffected BBS siblings carrying 2 mutations in the BBS2 gene, whereas the affected child was found to have a third mutation in either BBS1 or BBS6. This was interpreted to mean that 3 mutations were necessary to cause the disease in these patients [Katsanis et al., 2001]. Several studies reported few BBS families for whom the third allele correlated with a more severe phenotype, suggesting the possible effect of modifier alleles. In a reported BBS family, 2 affected patients shared the homozygous mutation p.M390R (BBS1); the first patient, carrying an additional heterozygous missense mutation (BBS6), had an earlier onset of obesity and more severe mental retardation than her sister who carried only the homozygous p.M390R (BBS1) mutation. In addition, a BBS family has been described with p.M390R (BBS1) and an additional heterozygous mutation found in BBS2 associated with a higher body mass index and a more severe retinal phenotype [Badano et al., 2003]. The frequency of identified triallelism in BBS has been overall low, and several studies suggest the absence of evidence of triallelism in BBS families [Badano et al., 2006; Smaoui et al., 2006; Abu Safieh et al., 2012]. Moreover, the CCDC28B gene (synonym MGC1203) has been reported to contribute epistatic alleles modifying the BBS phenotype [Badano et al., 2006]. Interestingly, other ciliopathy genes have been described to exhibit epistatic effects on BBS gene mutations, such as MKS1, MKS3, CEP290, and AHI1 [Nachury et al., 2007; Pawlik et al., 2010; Zaghloul et al., 2010].
Genotype-Phenotype Correlation
For the majority of identified mutations, no clear-cut correlation could be established between the genotype and clinical expression of BBS. Several studies have suggested a milder phenotype in association with some BBS gene mutations [Riise et al., 2002; Hjortshoj et al., 2010; Pawlik et al., 2010]. For instance, a milder phenotype has been associated with the recurrent mutation p.M390R (BBS1) [Hjortshoj et al., 2010]. Further, in a recent study, ocular phenotype evaluation for 37 BBS patients revealed that patients with BBS1 mutations had a milder phenotype than patients with mutations in other BBS genes [Daniels et al., 2012]. Other reports suggested the association between mutations in BBS1, BBS2, BBS3, and BBS4 and specific ocular phenotypes and digital malformations [Heon et al., 2005].
Molecular Analysis of Bardet-Biedl Syndrome: Future Directions
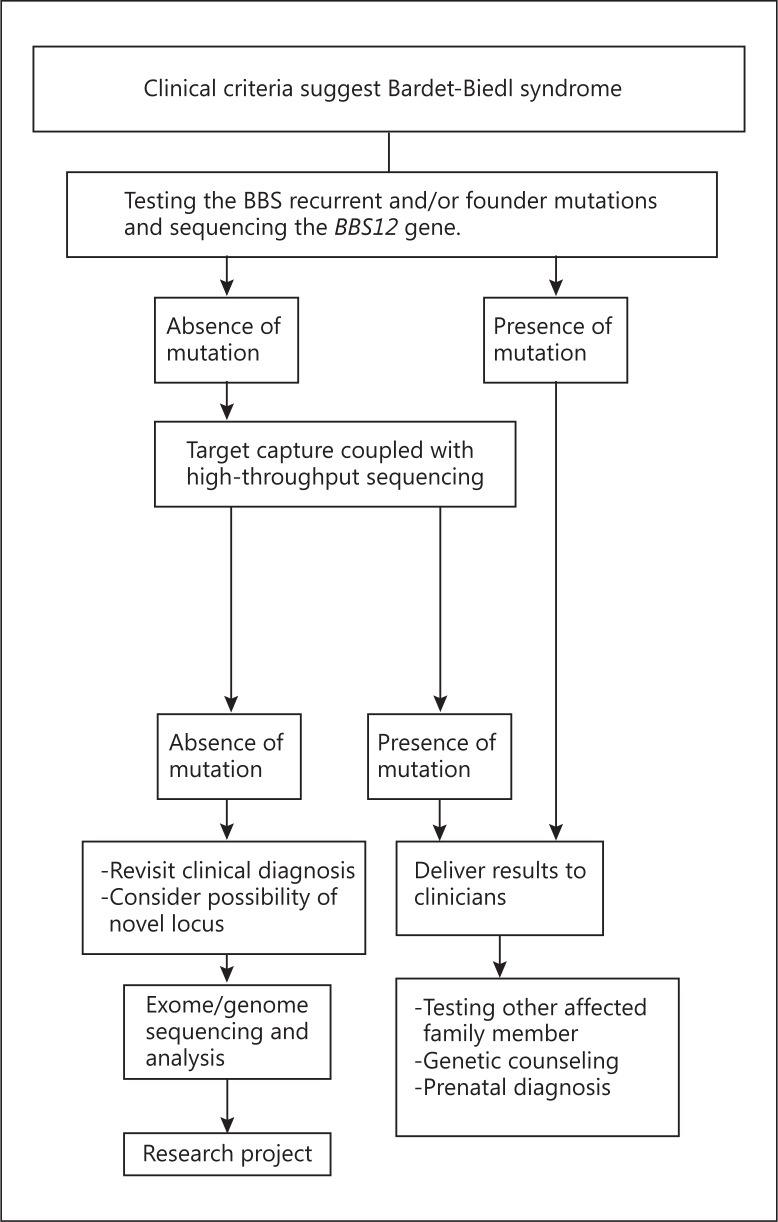
The extensive clinical and genetic heterogeneity of BBS generates difficulties for molecular diagnosis and genetic counseling. Within the last decade, many molecular strategies have been proposed to optimize the frequency of mutation detection. These strategies include homozygosity mapping using SNPs arrays in consanguineous families and screening of all BBS genes by direct sequencing [Abu Safieh et al., 2010; Billingsley et al., 2011; Janssen et al., 2011]. Recently, the implementation of next-generation sequencing has accelerated the molecular analysis of BBS patients [Choi et al., 2009; Marion et al., 2012; Redin et al., 2012; M'hamdi et al., 2013]. Targeted exon capture strategy coupled with high-throughput sequencing of 30 ciliopathy genes including 16 BBS genes, 12 nephronophthisis genes, the ALMS1 gene, and the CCDC28B gene showed high efficiency of mutation detection in BBS patients (70-80%) [Redin et al., 2012; M'hamdi et al., 2013]. Furthermore, the advent of strategies for scanning the human genome at high resolution coupled with the recognition of copy number variation has led to new methodologies in the identification of clinically relevant genes [Alkuraya, 2013; de Ligt et al., 2013]. We hope that these new molecular strategies will be adopted by medical geneticists. In figure 1 we propose a specific algorithm that could be applied to delineate the clinical and molecular diagnosis of BBS in the future.
Fig. 1.
Suggested algorithm for the molecular analysis of Bardet-Biedl syndrome patients in clinical genetics practice.
Acknowledgement
We sincerely wish to thank Prof. Alkuraya Fowzan Sami from the Department of Genetics, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia and Prof. Hejtmancik J. Fielding from the National Eye Institute for their assistance in revising the manuscript.
References
- 1.Abu Safieh L, Aldahmesh MA, Shamseldin H, Hashem M, Shaheen R, et al. Clinical and molecular characterization of Bardet-Biedl syndrome in consanguineous populations: The power of homozygosity mapping. J Med Genet. 2010;47:236–241. doi: 10.1136/jmg.2009.070755. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Safieh L, Al-Anazi S, Al-Abdi L, Hashem M, Alkuraya H, et al. In search of triallelism in Bardet-Biedl syndrome. Eur J Hum Genet. 2012;20:420–427. doi: 10.1038/ejhg.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajmal M, Khan MI, Neveling K, Tayyab A, Jaffar S, et al. Exome sequencing identifies a novel and a recurrent BBS1 mutation in Pakistani families with Bardet-Biedl syndrome. Mol Vis. 2013;2013:644–53. [PMC free article] [PubMed] [Google Scholar]
- 4.Alkuraya FS. The application of next-generation of sequencing in the autozygosity mapping of human recessive diseases. Hum Genet. 2013;132:1197–1211. doi: 10.1007/s00439-013-1344-x. [DOI] [PubMed] [Google Scholar]
- 5.Badano JL, Kim JC, Hoskins BE, Lewis RA, Ansley SJ, et al. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum Mol Genet. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 6.Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 7.Baker K, Norham GB, Chong WK, Banks T, Beales P, et al. Neocortical and hippocampal volume loss in a human ciliopathy: a quantitative MRI study in Bardet-Biedl syndrome. Am J Med Genet A. 2011;155A:1–8. doi: 10.1002/ajmg.a.33773. [DOI] [PubMed] [Google Scholar]
- 8.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- 9.Billingsley G, Deveault C, Héon E. BBS mutational analysis: a strategic approach. Ophthalmic Genet. 2011;32:181–187. doi: 10.3109/13816810.2011.567319. [DOI] [PubMed] [Google Scholar]
- 10.Billingsley G, Vincent A, Deveault C, Héon E. Mutational analysis of SDCCAG8 in Bardet-Biedl syndrome patients with renal involvement and absent polydactyly. Ophtalmic Genet. 2012;33:150–154. doi: 10.3109/13816810.2012.689411. [DOI] [PubMed] [Google Scholar]
- 11.Bimonte S, De Angelis A, Quagliata L, Giusti F, Tammaro R, et al. Ofd1 is required in limb bud patterning and endochondral bone development. Dev Biol. 2011;349:179–191. doi: 10.1016/j.ydbio.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Smaoui N, Hammer MB, Jiao X, Riazuddin SA, et al. Molecular analysis of Bardet-Biedl syndrome families: report of 21 novel mutations in 10 genes. Invest Ophtalmol Vis Sci. 2011;52:5317–5324. doi: 10.1167/iovs.11-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels AB, Sandberg MA, Chen J, Weigel-DiFranco C, Fielding Hejtmancic J, Berson EL. Genotype-phenotype correlations in Bardet-Biedl syndrome. Arch Ophthalmol. 2012;130:901–907. doi: 10.1001/archophthalmol.2012.89. [DOI] [PubMed] [Google Scholar]
- 15.Davis RE, Swiderski RE, Rahmouni K, Nishimura DY, Mullins RF, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Ligt J, Boone PM, Pfundt R, Vissers LE, Richmond T, et al. Detection of clinically relevant copy number variants with whole exome sequencing. Hum Mutat. 2013;34:1439–1448. doi: 10.1002/humu.22387. [DOI] [PubMed] [Google Scholar]
- 17.Farag TI, Teebi AS. High incidence of Bardet-Biedl syndrome among the Bedouin. Clin Genet. 1989;36:463–464. doi: 10.1111/j.1399-0004.1989.tb03378.x. [DOI] [PubMed] [Google Scholar]
- 18.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013;21:8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerth C, Zawadzki RJ, Werner JS, Heon E. Retinal morphology in patients with BBS1 and BBS10 related Bardet-Biedl syndrome evaluated by Fourier domain optical coherence tomography. Vis Res. 2008;48:392–399. doi: 10.1016/j.visres.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 22.Heon E, Westall C, Carmi R, Elbedour K, Panton C, et al. Ocular phenotypes of three genetic variants of Bardet-Biedl syndrome. Am J Med Genet A. 2005;132A:283–287. doi: 10.1002/ajmg.a.30466. [DOI] [PubMed] [Google Scholar]
- 23.Hjortshoj TD, Gronskov K, Brondum-Nielsen K, Rosenberg T. Novel founder BBS1 mutation explains a unique high prevalence of Bardet-Biedl syndrome in the Faroe Islands. Br J Ophthalmol. 2009;93:409–413. doi: 10.1136/bjo.2007.131110. [DOI] [PubMed] [Google Scholar]
- 24.Hjortshoj TD, Gronskov K, Philp AR, Nishimura DY, Riise R, et al. Bardet-Biedl syndrome in Denmark – report of 13 novel sequence variations in six genes. Hum Mutat. 2010;31:429–436. doi: 10.1002/humu.21204. [DOI] [PubMed] [Google Scholar]
- 25.Imhoff O, Marion V, Stoetzel C, Durand M, Holder M, et al. Bardet-Biedl syndrome: a study of the renal and cardiovascular phenotypes in a French cohort. Clin J Am Soc Nephrol. 2010;6:22–29. doi: 10.2215/CJN.03320410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Innes AM, Boycott KM, Puffenberger EG, Redl D, MacDonald IM, et al. A founder mutation in BBS2 is responsible for Bardet-Biedl syndrome in the Hutterite population: utility of SNP arrays in genetically heterogeneous disorders. Clin Genet. 2010;78:424–431. doi: 10.1111/j.1399-0004.2010.01481.x. [DOI] [PubMed] [Google Scholar]
- 27.Janssen S, Ramaswami G, Davis EE, Hurd T, Airik R, et al. Mutations analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum Genet. 2011;129:79–90. doi: 10.1007/s00439-010-0902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 29.Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, et al. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein D, Ammann F. The syndrome of Laurence Moon, Bardet-Biedl and allied diseases in Switzerland. Clinical, genetic and epidemiological studies. J Neurol Sci. 1969;9:479–513. doi: 10.1016/0022-510x(69)90091-4. [DOI] [PubMed] [Google Scholar]
- 31.Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leitch CC, Zaghloul NA, Davis EE. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 33.Marion V, Stoetzel C, Schlicht D. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci USA. 2009;106:1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marion V, Schlicht D, Mockel A, Caillard S, Imhoff O, et al. Bardet-Biedl syndrome highlights the major role of the primary cilium in efficient water reabsorption. Kidney Int. 2011;79:1013–1025. doi: 10.1038/ki.2010.538. [DOI] [PubMed] [Google Scholar]
- 35.Marion V, Stutzmann F, Gérard M, De Melo C, Schaefer E, et al. Exome sequencing identifies mutations in LTZFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet-Biedl syndrome with situs inversus and insertional polydactyly. J Med Genet. 2012;49:317–321. doi: 10.1136/jmedgenet-2012-100737. [DOI] [PubMed] [Google Scholar]
- 36.McGlinn E, Tabin CJ. Mechanistic insight into how Shh patterns the vertebrate limb. Curr Opin Genet Dev. 2006;16:426–432. doi: 10.1016/j.gde.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 37.M'hamdi O, Ouertani I, Maazoul F, Chaabouni H. Prevalence of Bardet-Biedl syndrome in Tunisia. J Community Genet. 2011;2:97–99. doi: 10.1007/s12687-011-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M'hamdi O, Redin C, Stoetzel C, Ouertani I, Chaabouni M, et al. Clinical and genetic characterization of Bardet-Biedl syndrome in Tunisia: defining a strategy for molecular diagnosis. Clin Genet, E-pub ahead of print (2013). [DOI] [PubMed]
- 39.Mockel A, Perdomo Y, Stutzmann F, Letsch J, Marion V, Dollfus H. Retinal dystrophy in Bardet-Biedl syndrome and related syndromic ciliopathies. Prog Retin Eye Res. 2011;30:258–274. doi: 10.1016/j.preteyeres.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Moore SJ, Green JS, Fan Y, Bhogal AK, Dicks E, et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22 year prospective, population-based, cohort study. Am J Med Genet A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller J, Stoetzel C, Vincent MC, Leitch CC, Laurier V, et al. Identification of 28 novels mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet. 2010;127:583–593. doi: 10.1007/s00439-010-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 43.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, et al. A core complex of BBS proteins cooperate with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 44.Otto EA, Hurd TW, Airik R, Chaki M, Zhou W, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawlik B, Mir A, Iqbal H, Li Y, Nürnberg G, et al. A novel familial BBS12 mutation associated with a mild phenotype: implication for clinical and molecular diagnosis strategies. Mol Syndromol. 2010;1:27–34. doi: 10.1159/000276763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putoux A, Attie-Bitach T, Martinovic J, Gubler MC. Phenotypic variability of Bardet-Biedl syndrome: focusing on the kidney. Pediatr Nephrol. 2012;27:7–15. doi: 10.1007/s00467-010-1751-3. [DOI] [PubMed] [Google Scholar]
- 47.Rahmouni K, Fath MA, Seo S, Thedens DR, Berry CJ, et al. Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J Clin Invest. 2008;118:1458–1467. doi: 10.1172/JCI32357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raychowdhury MK, Ramos AJ, Zhang P, McLaughin M, Dai XQ, et al. Vasopressin receptor-mediated functional signaling pathway in primary cilia of renal epithelial cells. Am J Physiol Renal Physiol. 2009;296:F87–F97. doi: 10.1152/ajprenal.90509.2008. [DOI] [PubMed] [Google Scholar]
- 49.Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, et al. Targeted high-throughput sequencing for molecular diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedel and Alström syndromes. J Med Genet. 2012;49:502–512. doi: 10.1136/jmedgenet-2012-100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riise R. Visual function in Laurence-Moon-Bardet-Biedl syndrome. A survey of 26 cases. Acta Ophthalmol Suppl. 1987;182:128–131. doi: 10.1111/j.1755-3768.1987.tb02610.x. [DOI] [PubMed] [Google Scholar]
- 51.Riise R, Andreasson S, Borgastrom MK, Wright AF, Tommerup N, et al. Intrafamilial variation of the phenotype in Bardet-Biedl syndrome. Br J Ophthalmol. 1997;81:378–385. doi: 10.1136/bjo.81.5.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riise R, Tornqvist K, Wright AF, Mykytyn K, Sheffield VC. The phenotype in Norwegian patients with Bardet-Biedl syndrome with mutations in the BBS4 gene. Arch Ophthalmol. 2002;120:1364–1367. doi: 10.1001/archopht.120.10.1364. [DOI] [PubMed] [Google Scholar]
- 53.Schaefer E, Zaloszyc A, Lauer J, Durand M, Stutzmann F, et al. Mutations in SDCCAG8/NPHP10 cause Bardet-Biedl syndrome and are associated with penetrant renal disease and absent polydactyly. Mol Syndromol. 2011;1:273–281. doi: 10.1159/000331268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheidecker S, Etard C, Pierce NW, Geoffroy V, Schaefer E, et al. Exome sequencing of Bardet-Biedl syndrome patient identifies a null mutation in the BBSome subunit BBIP1 (BBS18). J Med Genet, E-pub ahead of print (2013). [DOI] [PMC free article] [PubMed]
- 55.Seo S, Guo DF, Bugge K, Morgan DA, Rahmouni K, Sheffield VC. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet. 2009;18:1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smaoui N, Chaabouni M, Sergeev YV, Kallel H, Li S, Mahfoudh N, et al. Screening of the eight BBS genes in Tunisian families: no evidence of triallelism. Invest Ophthalmol Vis Sci. 2006;47:3487–3495. doi: 10.1167/iovs.05-1334. [DOI] [PubMed] [Google Scholar]
- 57.Sowjanya B, Sreenivasulu U, Naidu JN, Sivaranjani N. End stage renal disease, differential diagnosis, a rare genetic disorder: Bardet-Biedl syndrome: case report and review. Indian J Clin Biochem. 2011;26:214–216. doi: 10.1007/s12291-011-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoler JM, Herrin JT, Holmes LB. Genital abnormalities in females with Bardet-Biedl syndrome. Am J Med Genet. 1995;55:276–278. doi: 10.1002/ajmg.1320550306. [DOI] [PubMed] [Google Scholar]
- 59.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaghloul NA, Liu Y, Gerdes JM, Gascue C, Oh EC, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc Natl Acad Sci USA. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Q, Nishimura D, Seo S, Vogel T, Morgan DA, et al. Bardet-Biedl syndrome 3 (Bbs3) knockout mouse model reveals common BBS-associated phenotypes and Bbs3 unique phenotypes. Proc Natl Acad Sci USA. 2011;108:20678–20783. doi: 10.1073/pnas.1113220108. [DOI] [PMC free article] [PubMed] [Google Scholar]