Abstract
Smith-Magenis syndrome (SMS) is an intellectual disability syndrome with sleep disturbance, self-injurious behaviors and dysmorphic features. It is estimated to occur in 1/25,000 births, and in 90% of cases it is associated with interstitial deletions of chromosome 17p11.2. RAI1 (retinoic acid induced 1; OMIM 607642) mutations are the second most frequent molecular etiology, with this gene being located in the SMS locus at 17p11.2. Here, we report 9 new RAI1-truncating mutations in nonrelated individuals referred for molecular analysis due to a possible SMS diagnosis. None of these patients carried a 17p11.2 deletion. The 9 mutations include 2 nonsense mutations and 7 heterozygous frameshift mutations leading to protein truncation. All mutations map in exon 3 of RAI1 which codes for more than 98% of the protein. RAI1 regulates gene transcription, and its targets are themselves involved in transcriptional regulation, cell growth and cell cycle regulation, bone and skeletal development, lipid and glucide metabolisms, neurological development, behavioral functions, and circadian activity. We report the clinical features of the patients carrying these deleterious mutations in comparison with those of patients carrying 17p11.2 deletions.
Key Words: Mutation, 17p11.2, RAI1, Smith-Magenis syndrome
Smith-Magenis syndrome (SMS; MIM 182290) involves multiple congenital anomalies and intellectual disability usually associated with an interstitial deletion of chromosome 17p11.2 [Smith et al., 1986; Greenberg et al., 1996; Dykens and Smith, 1998; Potocki et al., 2000; De Leersnyder et al., 2001; Edelman et al., 2007; Gropman et al., 2007; Elsea and Girirajan, 2008]. The common features include behavioral abnormalities with sleep disturbances, self-injurious and/or aggressive behavior, particular craniofacial and skeletal anomalies, and speech delay. The facial appearance is usually characterized by a broad square-shaped face, a tented (inverted V-shaped) upper lip with downturned corners of the mouth and upslanting palpebral fissures. Obesity is frequent.
The incidence of this syndrome has been estimated to be 1 in 25,000 births [Greenberg et al., 1991]. About 90% of SMS patients present a heterozygous interstitial deletion of chromosome 17p11.2: most (70% of SMS patients) have a common ∼3.7-Mb deletion [Greenberg et al., 1991; Juyal et al., 1996; Gropman et al., 2007] mediated by nonallelic homologous recombination (NAHR) [Chen et al., 1997; Park et al., 2002]; about 20% of SMS patients harbor atypical deletions of ∼1.5-9 Mb mediated by an end-joining mechanism [Trask et al., 1996; Bi et al., 2002; Stankiewicz et al., 2003; Vlangos et al., 2003; Shaw and Lupski, 2005]. More recently, point mutations in the RAI1 (retinoic acid induced 1) gene have been identified in less than 10% of SMS patients [Slager et al., 2003; Bi et al., 2004, 2006; Girirajan et al., 2005, 2006; Vilboux et al., 2011; Vieira et al., 2012]. This gene maps in the SMS critical region [Bi et al., 2002; Vlangos et al., 2003]. About 36 mutations have been described in RAI1 to date, and more than 50% of them are truncating mutations; the deleterious role of the missense mutations is not yet clearly established. The RAI1 gene encodes a potential transcriptional regulator and consists of 6 exons that span over 120 kb; the third exon contains more than 98% of the coding sequence [Bi et al., 2004, 2005]. The RAI1 protein contains polyglutamine and polyserine tracts, bipartite nuclear localization signals and a plant homeo/zinc finger domain at the C-terminus [Toulouse et al., 2003]. Murine Rai1 (originally reported as GT1) is upregulated by retinoic acid treatment in mouse embryonal carcinoma cells concomitantly with neuronal differentiation [Imai et al., 1995].
We report 9 new RAI1-truncating mutations in SMS patients without 17p11.2 deletions, describe associated phenotypes and compare our results with published findings.
Methods
Patients and Samples
Patients presenting with clinical symptoms of SMS, but without a 17p11.2 deletion, were referred from various clinical genetic centers in France through the network of the reference centers for developmental anomalies and malformation syndromes. All patient samples were collected after obtaining informed consent according to the guidelines of the relevant institutional review boards.
PCR and Sequencing
Patient DNA was amplified by PCR with overlapping primers covering exons 2-6 of the RAI1 gene and using the conditions described in table 1. PCR products were purified and then analyzed by direct sequencing using the BigDye terminator v3.1 Cycle Sequencing Kit and the ABI Prism 3130 Genetic Analyzer (all from Applied Biosystems, Carlsbad, Calif., USA). The gene mutation nomenclature used in this article follows the recommendations of den Dunnen and Antonarakis [2001]. When material from the parents was available, a family study was performed to prove that the mutation occurred de novo.
Table 1.
RAI1 PCR primers and conditions
| Exon | Forward primer (5′→3′) | Reverse primer (5′→3′) | Product size, bp | Annealing temp., °C |
|---|---|---|---|---|
| 2 | GGGGCTGCTAAGAAGCAAAC | CGCCGAAAACACAAAGCTC | 306 | 56 |
| 3.1 | GGTGGGTGGGAGGGTGCTTT | TGCTGTCCTGGACGCCGTAG | 489 | 57 |
| 3.2 | AGTACCACCGAGGCAGCA | TCGGTGCTGTGCAACTCTTA | 498 | 57 |
| 3.3 | CAGGGTACCCACTTTCCTCA | AGTTCTCCAGGTTTGGCATC | 496 | 57 |
| 3.4 | CAGGAAACCCTCCATTACCA | GCTGGCTTTTATGCTGCTCT | 541 | 57 |
| 3.5 | CTCCCTGAGAACCTGCTGTC | CCACGAACTTGGAGAAGCTG | 502 | 57 |
| 3.6 | AGTCCGTGTCCACCTGTTCT | GTCCGGGAAACAGTCAAAAG | 542 | 57 |
| 3.7 | TCTCGCTGGAGAACCACAG | CACCTGCCTCCTCCTTCAC | 549 | 57 |
| 3.8 | CTCAGATGGCATCAGCAAAG | TCAAACCAGCTCTGGACCTT | 504 | 57 |
| 3.9 | CACTCATCTGCACCAAGGAG | GCCGTGAGAGAACGAGTACA | 499 | 57 |
| 3.10 | AAGACCTATTGCTCCCTGAA | TTGTCGAGGAGCTTCTTGGT | 475 | 57 |
| 3.11 | GAGACAGACTCACCCAGCAC | CAGCACCTTGGTCTTCATGG | 570 | 57 |
| 3.12 | AGTGGCAATGGGGGAGAT | GCCTCGGTTTTGAAACAGTC | 530 | 57 |
| 3.13 | GGCCTCTCCTGGTAATCCTC | TGTTTCTGTGCCCTTGTCTG | 525 | 57 |
| 3.14 | GGCCAGTGAGGACAACTCTG | AGGAGAACGAGGACGAGGAT | 467 | 57 |
| 3.15 | GACGCGTTCACCACCATA | CATCACAGCAGTAGCAGCTC | 550 | 57 |
| 3.16 | ACACTGCCTCCCCAAAAAG | GTCCTTTGGAACCCCACAC | 487 | 57 |
| 4 | CCAGCCTGTAAAGCTTGAGG | CTCCAATGCCCTCCTGTTC | 197 | 56 |
| 5 | TCCTGGCTGCAGACAAAAC | GGGGACTGTGAAGGAGGTG | 207 | 56 |
| 6.1 | ACTGTGAAGTCCGAGGTCGT | GAAAGTGTCCCCAGGACAAA | 420 | 58 |
| 6.2 | GGGAGGAAAACCCGTTCC | AGAGGCCAAAGGGTGTCC | 549 | 58 |
| 6.3 | GGAGCCTTTGGAACAAACC | GTCGCGGTAGCCCCTTAC | 489 | 58 |
| 6.4 | GGAACATGCTCGCTTCTCC | CAGCTGAATCTGGTCGGAAT | 545 | 62 |
Results
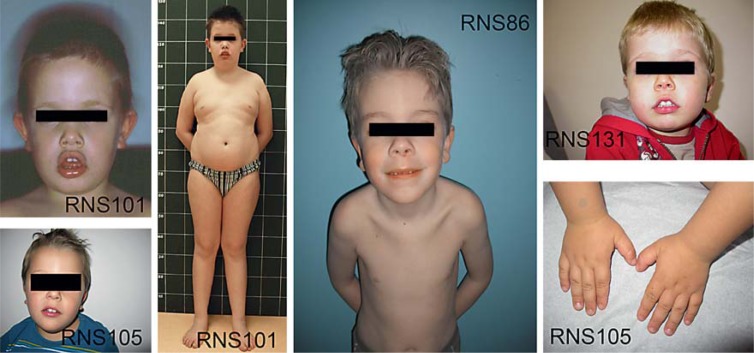
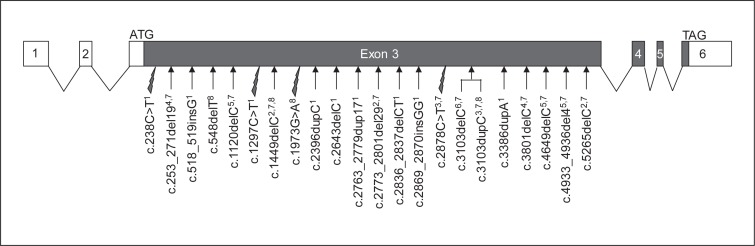
Patients presenting with clinical symptoms of SMS (fig. 1; table 2), but without 17p11.2 deletion, were referred to us, and we screened them for mutations in the RAI1 gene. We report here 9 novel mutations, all in exon 3 of the gene (fig. 2), that support a diagnosis of SMS in these cases.
Fig. 1.
Photographs of faces and extremities of some patients carrying an RAI1 mutation.
Table 2.
Clinical characteristics of patients with RAI1 mutations
| RNS78 | RNS59 | RNS101 | RNS103 | RNS86 | RNS105 | RNS131 | RNS165 | RNS208 | |
|---|---|---|---|---|---|---|---|---|---|
| RAI1 mutation | c.238C>T | c.3386dupA | c.2869_2870insGG | c.518_519insG | c.2643delC | C.1297C>T | c.2763_2779dup17 | c.2396dupC | c.2836_2837delCT |
| p.Arg80* | p.Glu1130Glyfs*36 | p.Asp957Glyfs*108 | p.Glnl74Profs*64 | p.Glu882Serfs*68 | p.Gln433* | p.Leu927Glnfs*29 | p.Gly800Trpfs*36 | p.Leu946Valfs*7 | |
| Age at time of report | 8 years | 19 years | 14 years | 29 years | 7.5 years | 5.5 years | 6 years | 9 years | 9 years |
| Sex | f | m | m | m | m | m | m | m | f |
| Growth measures, percentiles | |||||||||
| weight | 92nd | 73rd | >97th | >97th | 95th | 90th | 75th | 90th | 55th |
| height | 50th | 25th | 75th | 20th | 70th | 12th | 10th | 24th | 3rd |
| HC | 80th | <3rd | 90th | 95th | 10th | 50th | 80th | 30th | 30th |
| Overweight | + | + | + | + | + (moderate) | + | + | + | + |
| Developmental delay | + | + speech delay | + speech delay | + speech delay | + | + speech delay | + | + | + |
| Facial dysmorphism | + egg-shaped face, horizontal eyebrows, synophrys, short and upturned nose | + square face, large mouth, thick lips with tented upper lip and everted lower lip, short philtrum | + square face, high forehead, short philtrum, thick lips with tented upper lip and everted lower lip | + tented upper lip, short philtrum, uplifted ear lobes | + upslanting palpebral fissures, anteverted nostrils, long philtrum, tented upper lip | + short philtrum | + tented upper lip, brachycephaly | + synophrys, short philtrum | + anteverted nostrils, long philtrum, tented upper lip |
| Short extremities | + bilateral clinodactyly of the fifth finger | + | + | + | + | + | − | + | + bilateral clinodactyly of the fifth finger |
| Sleep disturbance | + early morning awakening (before 5 a.m.) and early sleeping in the evening | + inverted circadian rhythm for melatonin | moderate | + early morning awakening (5:30 a.m.) | + early morning awakening (4 or 5 a.m.) | + early morning awakening and early sleeping in the evening | + | + early morning awakening (5 a.m.) and inverted circadian rhythm for melatonin | + early morning awakening (4 a.m.) |
| Behavioral problems | + aggressiveness, overeating | + impulsiveness, aggressiveness, self-injurious behavior | + rage attacks, hyperactivity, self-injurious behavior, head banging, aggressiveness, overeating (compulsive behavior) | + self-injurious behavior | + self-injurious behavior, aggressiveness | + | + overeating | + | + impulsiveness, aggressiveness, overeating |
HC = Head circumference; + = present; – = absent.
Fig. 2.
Diagram showing the structure of RAI1 in the genome with 6 exons and summarizing truncating mutations associated with SMS. All truncating mutations reported to date are indicated, including those described in this study1 and in Slager et al. [2003]2, Bi et al. [2004]3, Girirajan et al. [2005]4, Girirajan et al. [2006]5, Bi et al. [2006]6, Truong et al. [2010]7, and Vilboux et al. [2011]8: 4 nonsense (indicated by lightning flashes) and 17 frameshift (indicated by arrows) mutations. The numbering of nucleotides is based on GenBank NM_030665.3. The filled boxes represent the RAI1 coding region, and the hollow boxes represent noncoding regions.
RNS78
RNS78 is the third child of healthy unrelated parents. Pregnancy was normal, but she had major reflux problems in the neonatal period. She developed overgrowth at ∼5 months of age and is bulimic. In addition to facial dysmorphism and extremity anomalies, she suffers significant sleep disturbance including early awakening, early sleeping in the evening and multiple napping. She presents learning difficulties and an oppositional behavior with aggressiveness; she is, however, sociable.
Laboratory findings included a normal karyotype and normal FISH for del(17)(p11.2). RNS78 has a heterozygous cytosine to thymine transition at nucleotide 238 (c.238C>T) in the RAI1 gene, producing a nonsense substitution (p.Arg80*): the reading frame is interrupted by a premature stop codon.
RNS59
RNS59 presents with developmental delay, significant language retardation, behavioral problems, and major sleeping disturbance. He spoke his first word at the age of 3 years and his first sentence at 5. He displays impulsiveness and self- and heteroaggressiveness. A study of the rhythm of melatonin release gave a flat trace with an inversion of the circadian rhythm as early as at the age of 15 months. This patient is awake for long periods through the night and frequently naps during the day, with a shorter than usual total sleep time.
Pharmacogenetic analysis of cytochrome P450 2D6 revealed an ultrarapid metabolism.
RNS59 carries a duplication of a single adenine in exon 3 at nucleotide position 3,386 on one RAI1 allele (c.3386dupA). This duplication results in a frameshift starting at amino acid 1130, leading to misincorporation of 35 amino acids and a downstream stop codon. Neither parent carried this p.Glu1130Glyfs*36 in DNA.
RNS101
RNS101 is the second son of healthy unrelated parents. Pregnancy was normal. Early problems included frequent bronchiolitis. Growth was steady over the 2 first years and then tended to obesity. He displays developmental delay, significant language delay, behavioral problems including fits of anger, and moderate sleeping disturbance with night awakenings. His hands are short, and his feet are broad. He presents joint hyperlaxity.
Laboratory findings included a normal karyotype, negative fragile X and subtelomeric microrearrangement results, normal FISH for 17p11.2 deletion, and normal pangenomic array CGH. RNS101 has a heterozygous insertion of 2 guanines after nucleotide position 2,869 (c.2869_2870insGG) in the RAI1 gene, resulting in a frameshift starting at amino acid residue 957, a misincorporation of 107 amino acids and a downstream stop codon (p.Asp957Glyfs*108).
RNS103
RNS103 is a 29-year-old man presenting syndromic obesity. There is no family history of mental handicap or congenital malformations, and the pregnancy was normal. Birth weight was 3,340 g, and there were no feeding difficulties during the neonatal period. He presented subsequently with developmental delay including walking and language delay, but he learned to read and write and now works in an ‘Etablissement et Service d'Aide par le Travail’ (French working environment adapted to those with difficulties). Behavioral problems include significant sleep disturbance with frequently interrupted sleeping and early waking (5:30 a.m.) and self-aggressiveness.
He has short fingers and toes and one ‘café au lait’ spot on the trunk.
Laboratory findings include a normal karyotype, normal methylation of the chromosome 15, negative test results for fragile X, normal FISH for del(17)(p11.2), and no deleterious microrearrangement observed on array CGH.
RNS103 carries an insertion of one guanine after nucleotide 518 on one allele of RAI1 (c.518_519insG). This insertion results in a frameshift starting at amino acid 174, leading to misincorporation of 63 amino acids and a downstream stop codon (p.Gln174Profs*64).
RNS86
RNS86 presents developmental delay and major behavioral problems including self- and heteroaggressiveness. He has significant sleep disturbance and facial dysmorphisms as well as astigmatism and hypermetropia. His hands are short with a transverse palmar crease on the left hand, and he presents hyperlaxity. Over the past 2 years, his weight has increased substantially.
Metabolic balance, fragile X analysis, MRI, high resolution karyotype, and pangenomic array CGH did not reveal any anomaly.
RNS86 carries a deletion of a single cytosine in exon 3 at nucleotide 2,643 on one allele of RAI1 (c.2643delC). This deletion results in a frameshift starting at amino acid 882, leading to misincorporation of 67 amino acids and a downstream stop codon (p.Glu882Serfs*68).
RNS105
RNS105 presents with moderate developmental delay and significant language retardation. He could walk at 16 months. He displays a characteristic sleep disturbance including early sleeping in the evening, early waking and frequent napping; however, there is no aggressive behavior. He has shawl scrotum and shows hyperlaxity with bilateral sloped feet.
Additional laboratory findings include a normal karyotype, a normal pangenomic array CGH excluding an interstitial deletion of chromosome 17p11.2 and negative fragile X and FGD1 studies. RNS105 has a heterozygous cytosine to thymine transition at nucleotide 1,297 in the RAI1 gene, producing a nonsense substitution (p.Gln433*): the reading frame is interrupted by a premature stop codon.
RNS131
RNS131 presents developmental delay, hypotonia, dysmorphism, macrocephaly, behavioral problems, and overweight. He could walk at 21 months. His BMI increased excessively until he was 3 years old, and then, it declined following strict diet control. Sleep disturbances began in the second year of life with multiple waking during the night. He presents moderate hyperlaxity. He is the first of 2 children. His younger sister has a paternally inherited microrearrangement of the X chromosome, but RNS131 did not inherit this familial rearrangement.
Laboratory findings for this boy include a normal karyotype, negative fragile X and Prader-Willi studies, normal FISH for del(17)(p11.2), and normal pangenomic array CGH. The metabolic findings are normal. Sequencing of exon 3 of the RAI1 gene revealed a 17-bp duplication (c.2763_2779dup17) that results in a frameshift starting at amino acid 927 and leading to a downstream stop codon (p.Leu927Glnfs*29).
RNS165
RNS165 presents with sleep disturbances and inversion of the circadian rhythm for melatonin, developmental delay, behavioral problems, facial dysmorphism, astigmatism, hypermetropia, and brachydactyly.
RNS165 has a duplication of one cytosine (c.2396dupC) on one allele of RAI1 leading to a frameshift and a downstream stop codon at position 836 (p.Gly800Trpfs*36).
RNS208
RNS208 is the first child of healthy unrelated parents. Pregnancy was normal. She walked at the age of one year and spoke her first words at only 2. She displays impulsiveness and aggressiveness and always awakes early.
RAI1 sequencing revealed a 2-bp deletion (c.2836_ 2837delCT) that results in a frameshift starting at amino acid 946 and ending by a premature stop codon (p.Leu946Valfs*7).
Discussion
We report 9 patients with deleterious mutations in the RAI1 gene, all displaying similar features consistent with a diagnosis of SMS; in all 9 cases, a 17p11.2 deletion was excluded by FISH, MLPA or array CGH. Clinical symptoms observed in these patients correlate with previous reports of RAI1-mutated patients [Slager et al., 2003; Bi et al., 2004, 2006; Girirajan et al., 2005, 2006; Vilboux et al., 2011]. No immunologic, cardiovascular or renal features were noted.
The RAI1 gene, located on chromosome 17p11.2, comprises 6 exons generating a 7.6-kb mRNA [Toulouse et al., 2003]. This mRNA (GenBank NM_030665.3) contains a 470-bp 5′UTR (the first 2 exons and the 16 first nucleotides of exon 3) and a 1,452-bp 3′UTR (corresponding to the sixth exon). RAI1 encodes a nuclear protein containing a single zinc finger plant homeo domain, which is also present in the trithorax family of chromatin-remodeling transcriptional regulators, implicating RAI1 in the regulation of gene transcription [Bi et al., 2005, 2006]. RAI1 has 2 putative bipartite nuclear localization signals. Functional and cellular analyses of 5 mutated forms of the human RAI1 protein indicate that the N-terminal half of the protein has transcription factor activity (within the first 1,034 amino acids), and the C-terminal half is responsible for its transportation into the nucleus [Carmona-Mora et al., 2010].
All previously reported mutations map in exon 3 (5,581 bp) which codes for more than 98% of the protein. The mutations are scattered all along this third exon, and there is no evidence for a hot spot. We report 7 frameshift and 2 nonsense mutations, also all in the third exon (fig. 2). The nonsense and frameshift mutations are predicted to result in truncated proteins due to direct or downstream stop codons.
Indeed, most deleterious mutations identified in the RAI1 gene lead to the production of truncated proteins. These mutations are unique, dominant and, in cases for which familial analysis was possible, de novo.
Antonarakis et al. [2000] suggested that single-base deletions causing frameshifts occurred in runs of identical bases, and Truong et al. [2010] postulated a preferential frameshift mutation hotspot in the RAI1 gene in a heptameric C-tract (c.3097_3103). We did not detect any mutation at this site in RAI1; however, the c.2643delC alteration predicted to cause a p.Glu882Serfs*68 frameshift mutation occurred in a mononucleotide repeat of 4 cytosines. The c.2869_2870insGG frameshift mutation maps immediately downstream from a guanine and the c.3386dupA from 2 adenines. These mutations may therefore have been the consequence of slipped mispairing [Streisinger et al., 1966; Tautz and Schlötterer, 1994]. It is noticeable, however, that guanine and cytosine runs have a higher mutation rate than adenine and thymine runs [Sagher et al., 1999; Boyer et al., 2002].
A polymorphic CAG repeat, starting from RAI1 nucleotide position 832, encodes a polyglutamine stretch in the N-terminus of the protein. Alleles of 10-16 glutamines have been described in the general population [Bi et al., 2006]. In our cohort of 80 patients with the clinical symptoms of SMS, but without 17p11.2 deletion, 44% have 14 glutamines, 52% 13 glutamines, 2% 15 glutamines, 1% 11 glutamines, and 1% 10 glutamines. Five of our 9 patients with an identified RAI1 mutation have 14 glutamines and 4 have 13 glutamines. No significant association has been reported between the length of CAG repeats and the SMS phenotype when the number of CAG trinucleotides remains in the range from 10 to 16 repeats. However, large CAG repeats could influence expressivity and severity of the SMS phenotype in patients who present a RAI1 mutation on the second allele [Seranski et al., 2001; Bi et al., 2006].
Downstream targets of the transcription factor RAI1 are involved in transcriptional regulation (RXRB, ZNF236, ZIC1, RUNX1T1, AKR7A3, FBLN1), cellular growth and cell cycle regulation (SPTBN1, POLDIP3, PPP1R14D, GLI3, KMT2A, ADD3), bone and skeletal development (PSTPIP2, ANKH), lipid biosynthesis and cholesterol metabolism (LIPE, HMGCS1, INSIG1), neurological development (ZIC1, PSEN2, RXRB, CLN8, SMA4, NF1, KMT2A), behavioral functions (SCN12A), circadian activity (NR1D2, PER2, PER3, CRY1, ARNTL), and insulin and glucose regulation (INSIG1, PIK3R1, ZNF236, LIPE) [Girirajan et al., 2009]. Interestingly, each of these biological functions is dysregulated in SMS patients: these patients present with, in particular, obesity, altered circadian rhythm, hypercholesterolemia, hypertriglyceridemia, intellectual disability, and abnormal behavior.
The role of RAI1 haploinsufficiency in the complex pathogenicity of obesity on the one hand and of sleep disturbances on the other hand has been demonstrated. First, expression analysis reveals that Bdnf (brain-derived neurotrophic factor), a gene associated with hyperphagia and obesity, is downregulated in the hypothalamus of Rai1+/- mice, and reporter studies show that RAI1 directly regulates the expression of BDNF [Burns et al., 2010]. Second, haploinsufficiency of RAI1 in SMS fibroblasts and in the mouse hypothalamus, respectively, results in the transcriptional dysregulation of the circadian clock and causes altered expression and regulation of multiple circadian genes, including PER2, PER3, CRY1, and ARNTL [Williams et al., 2012]. This study suggests that a heterozygous mutation of RAI1 alters the circadian rhythm and results in an abnormal sleep-wake cycle, which can contribute to an abnormal feeding pattern and dependent cognitive performance.
In summary, haploinsufficiency of RAI1 is the main responsible for the neurological, behavioral and craniofacial features of SMS, even if other genes or genetic background may play a role in modulating these features. Consistent with patients previously described with RAI1 mutations, the patients we report differ from patients carrying a 17p11.2 deletion: RAI1 mutations are associated with less substantial motor delay, greater inclination to overweight, no hearing loss, no seizures, and no associated systemic malformations. Therefore, one or more other genes in the 17p11.2 region are probably involved in these variable associated features more frequently observed in SMS patients with deletions [Gropman et al., 2007]. More research is required for a better understanding of the role of additional genes and to consider new treatment strategies.
Acknowledgements
We thank all clinicians who referred SMS cases and all families for giving their consent for this study.
References
- 1.Antonarakis SE, Krawczak M, Cooper DN. Disease-causing mutations in the human genome. Eur J Pediatr 159 Suppl. 2000;3:S173–S178. doi: 10.1007/pl00014395. [DOI] [PubMed] [Google Scholar]
- 2.Bi W, Yan J, Stankiewicz P, Park SS, Walz K, et al. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 2002;12:713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi W, Saifi GM, Shaw CJ, Walz K, Fonseca P, et al. Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum Genet. 2004;115:515–524. doi: 10.1007/s00439-004-1187-6. [DOI] [PubMed] [Google Scholar]
- 4.Bi W, Ohyama T, Nakamura H, Yan J, Visvanathan J, et al. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum Mol Genet. 2005;14:983–995. doi: 10.1093/hmg/ddi085. [DOI] [PubMed] [Google Scholar]
- 5.Bi W, Saifi GM, Girirajan S, Shi X, Szomju B, et al. RAI1 point mutations, CAG repeat variation, and SNP analysis in non-deletion Smith-Magenis syndrome. Am J Med Genet A. 2006;140:2454–2463. doi: 10.1002/ajmg.a.31510. [DOI] [PubMed] [Google Scholar]
- 6.Boyer JC, Yamada NA, Roques CN, Hatch SB, Riess K, Farber RA. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum Mol Genet. 2002;11:707–713. doi: 10.1093/hmg/11.6.707. [DOI] [PubMed] [Google Scholar]
- 7.Burns B, Schmidt K, Williams SR, Kim S, Girirajan S, Elsea SH. Rai1 haploinsufficiency causes reduced Bdnf expression resulting in hyperphagia, obesity and altered fat distribution in mice and humans with no evidence of metabolic syndrome. Hum Mol Genet. 2010;19:4026–4042. doi: 10.1093/hmg/ddq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmona-Mora P, Encina CA, Canales CP, Cao L, Molina J, et al. Functional and cellular characterization of human Retinoic Acid Induced 1 (RAI1) mutations associated with Smith-Magenis Syndrome. BMC Mol Biol. 2010;11:63. doi: 10.1186/1471-2199-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, et al. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 10.De Leersnyder H, De Blois MC, Claustrat B, Romana S, Albrecht U, et al. Inversion of the circadian rhythm of melatonin in the Smith-Magenis syndrome. J Pediatr. 2001;139:111–116. doi: 10.1067/mpd.2001.115018. [DOI] [PubMed] [Google Scholar]
- 11.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 12.Dykens EM, Smith AC. Distinctiveness and correlates of maladaptive behaviour in children and adolescents with Smith-Magenis syndrome. J Intellect Disabil Res. 1998;42:481–489. doi: 10.1046/j.1365-2788.1998.4260481.x. [DOI] [PubMed] [Google Scholar]
- 13.Edelman EA, Girirajan S, Finucane B, Patel PI, Lupski JR, et al. Gender, genotype, and phenotype differences in Smith-Magenis syndrome: a meta-analysis of 105 cases. Clin Genet. 2007;71:540–550. doi: 10.1111/j.1399-0004.2007.00815.x. [DOI] [PubMed] [Google Scholar]
- 14.Elsea SH, Girirajan S. Smith-Magenis syndrome. Eur J Hum Genet. 2008;16:412–421. doi: 10.1038/sj.ejhg.5202009. [DOI] [PubMed] [Google Scholar]
- 15.Girirajan S, Elsas LJ, 2nd, Devriendt K, Elsea SH. RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet. 2005;42:820–828. doi: 10.1136/jmg.2005.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girirajan S, Vlangos CN, Szomju BB, Edelman E, Trevors CD, et al. Genotype-phenotype correlation in Smith-Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet Med. 2006;8:417–427. doi: 10.1097/01.gim.0000228215.32110.89. [DOI] [PubMed] [Google Scholar]
- 17.Girirajan S, Truong HT, Blanchard CL, Elsea SH. A functional network module for Smith-Magenis syndrome. Clin Genet. 2009;75:364–374. doi: 10.1111/j.1399-0004.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith AC, et al. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2) Am J Hum Genet. 1991;49:1207–1218. [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, et al. Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2) Am J Med Genet. 1996;62:247–254. doi: 10.1002/(SICI)1096-8628(19960329)62:3<247::AID-AJMG9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Gropman AL, Elsea S, Duncan WC, Jr, Smith AC. New developments in Smith-Magenis syndrome (del17p11.2) Curr Opin Neurol. 2007;20:125–134. doi: 10.1097/WCO.0b013e3280895dba. [DOI] [PubMed] [Google Scholar]
- 21.Imai Y, Suzuki Y, Matsui T, Tohyama M, Wanaka A, Takagi T. Cloning of a retinoic acid-induced gene, GT1, in the embryonal carcinoma cell line P19: neuron-specific expression in the mouse brain. Brain Res Mol Brain Res. 1995;31:1–9. doi: 10.1016/0169-328x(95)00020-s. [DOI] [PubMed] [Google Scholar]
- 22.Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, et al. Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet. 1996;58:998–1007. [PMC free article] [PubMed] [Google Scholar]
- 23.Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, et al. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potocki L, Glaze D, Tan DX, Park SS, Kashork CD, et al. Circadian rhythm abnormalities of melatonin in Smith-Magenis syndrome. J Med Genet. 2000;37:428–433. doi: 10.1136/jmg.37.6.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagher D, Hsu A, Strauss B. Stabilization of the intermediate in frameshift mutation. Mutat Res. 1999;423:73–77. doi: 10.1016/s0027-5107(98)00227-9. [DOI] [PubMed] [Google Scholar]
- 26.Seranski P, Hoff C, Radelof U, Hennig S, Reinhardt R, et al. RAI1 is a novel polyglutamine encoding gene that is deleted in Smith-Magenis syndrome patients. Gene. 2001;270:69–76. doi: 10.1016/s0378-1119(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 27.Shaw CJ, Lupski JR. Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet. 2005;116:1–7. doi: 10.1007/s00439-004-1204-9. [DOI] [PubMed] [Google Scholar]
- 28.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 29.Smith AC, McGavran L, Robinson J, Waldstein G, Macfarlane J, et al. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet. 1986;24:393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- 30.Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet. 2003;72:1101–1016. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, et al. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Tautz D, Schlötterer C. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 33.Toulouse A, Rochefort D, Roussel J, Joober R, Rouleau GA. Molecular cloning and characterization of human RAI1, a gene associated with schizophrenia. Genomics. 2003;82:162–171. doi: 10.1016/s0888-7543(03)00101-0. [DOI] [PubMed] [Google Scholar]
- 34.Trask BJ, Mefford H, van den Engh G, Massa HF, Juyal RC, et al. Quantification by flow cytometry of chromosome-17 deletions in Smith-Magenis syndrome patients. Hum Genet. 1996;98:710–718. doi: 10.1007/s004390050291. [DOI] [PubMed] [Google Scholar]
- 35.Truong HT, Dudding T, Blanchard CL, Elsea SH. Frameshift mutation hotspot identified in Smith-Magenis syndrome: case report and review of literature. BMC Med Genet. 2010;11:142. doi: 10.1186/1471-2350-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vieira GH, Rodriguez JD, Carmona-Mora P, Cao L, Gamba BF, et al. Detection of classical 17p11.2 deletions, an atypical deletion and RAI1 alterations in patients with features suggestive of Smith-Magenis syndrome. Eur J Hum Genet. 2012;20:148–154. doi: 10.1038/ejhg.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilboux T, Ciccone C, Blancato JK, Cox GF, Deshpande C, et al. Molecular analysis of the Retinoic Acid Induced 1 Gene (RAI1) in patients with suspected Smith-Magenis syndrome without the 17p11.2 deletion. PLoS ONE. 2011;6:e22861. doi: 10.1371/journal.pone.0022861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlangos CN, Yim DK, Elsea SH. Refinement of the Smith-Magenis syndrome critical region to approximately 950 kb and assessment of 17p11.2 deletions. Are all deletions created equally? Mol Genet Metab. 2003;79:134–141. doi: 10.1016/s1096-7192(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 39.Williams SR, Zies D, Mullegama SV, Grotewiel MS, Elsea SH. Smith-Magenis syndrome results in disruption of CLOCK gene transcription and reveals an integral role for RAI1 in the maintenance of circadian rhythmicity. Am J Hum Genet. 2012;90:941–949. doi: 10.1016/j.ajhg.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]