Significance
This work describes an advance in the understanding of how the important circadian neuropeptide PDF contributes to the function of the molecular clock in Drosophila neurons. The famous perS allele of period significantly improves the rhythmicity of flies missing PDF. The perS gene product degrades rapidly, suggesting that PDF-mediated cAMP affects PER stability. Indeed, increasing cAMP levels and cAMP-mediated protein kinase A (PKA) activity stabilizes PER, in tissue culture cells and in circadian neurons. PDF addition to fly brains in vitro has a similar effect. Our observations taken together indicate that PDF contributes to clock neuron function by activating PKA, stabilizing PER, and thereby slowing the pace of clock neurons that respond to PDF.
Keywords: PDF signaling, synchronization, PDF neurons, molecular clock regulation
Abstract
The neuropeptide PDF is important for Drosophila circadian rhythms: pdf01 (pdf-null) animals are mostly arrhythmic or short period in constant darkness and have an advanced activity peak in light–dark conditions. PDF contributes to the amplitude, synchrony, as well as the pace of circadian rhythms within clock neurons. PDF is known to increase cAMP levels in PDR receptor (PDFR)-containing neurons. However, there is no known connection of PDF or of cAMP with the Drosophila molecular clockworks. We discovered that the mutant period gene perS ameliorates the phenotypes of pdf-null flies. The period protein (PER) is a well-studied repressor of clock gene transcription, and the perS protein (PERS) has a markedly short half-life. The result therefore suggests that the PDF-mediated increase in cAMP might lengthen circadian period by directly enhancing PER stability. Indeed, increasing cAMP levels and cAMP-mediated protein kinase A (PKA) activity stabilizes PER, in S2 tissue culture cells and in fly circadian neurons. Adding PDF to fly brains in vitro has a similar effect. Consistent with these relationships, a light pulse causes more prominent PER degradation in pdf01 circadian neurons than in wild-type neurons. The results indicate that PDF contributes to clock neuron synchrony by increasing cAMP and PKA, which enhance PER stability and decrease clock speed in intrinsically fast-paced PDFR-containing clock neurons. We further suggest that the more rapid degradation of PERS bypasses PKA regulation and makes the pace of clock neurons more uniform, allowing them to avoid much of the asynchrony caused by the absence of PDF.
A molecular circadian clock controls the physiology and behavior of most eukaryotes and even some prokaryotes. Transcriptional feedback loops are important features of most circadian systems, and animal molecular clocks rely on many conserved proteins controlling the cycling expression and activity of key clock genes. In Drosophila, they include period (per) and timeless (tim), which are expressed in about 150 neurons in the fly brain. per and tim are activated by a heterodimer of two basic-helix–loop–helix (bHLH) transcription factors, CLOCK (CLK) and CYCLE (CYC). After their mRNA levels peak in the early night, per and tim protein (PER and TIM, respectively) levels peak in the late night (1, 2). PER and TIM then inhibit the transcriptional activity of CLK:CYC, which decreases their own expression (3). In the absence of synthesis, PER and TIM levels decrease until CLK:CYC is liberated and per and tim mRNA synthesis begins anew. This negative-feedback loop persists with ∼24-h periodicity in constant darkness (DD), i.e., without any resetting clues from the environment. However, internal resetting clues likely contribute to the maintenance of oscillator amplitude or the synchrony of different neuronal oscillators.
The neuropeptide pigment-dispersing factor (PDF) is secreted by the ventrolateral subgroup of Drosophila clock neurons (LNvs) and makes a critical contribution to synchronizing and resetting circadian neurons (4, 5). This conclusion is based in large part on features of homozygous pdf01 flies. They have no PDF and show a 1–2 h advanced evening activity peak in light–dark (LD) as well as a short period in DD; most animals become arrhythmic within a few days (4). The DD arrhythmicity of pdf01 flies is caused at least in part by weak and asynchronous oscillators (6). The asynchrony reflects the fact that some clock neurons run faster than normal without PDF, whereas others may run more slowly (6, 7). Given the advanced evening peak in LD and the short period in DD, the fast neurons appear to dominate the behavioral profile of pdf01 flies.
The PDF receptor gene (pdfr) responds to PDF and encodes a G-protein–coupled receptor (PDFR). It is expressed in a broad subset of clock neurons, and a pdfr deletion is almost identical to the pdf01 phenotype, including the advanced evening peak and fast pace in DD (8–10). Because PDF increases cAMP levels in at least some PDFR-expressing neurons (8, 9, 11, 12), enhanced cAMP levels may slow the intrinsically fast pace of some PDFR-containing clock neurons. cAMP may then affect clock speed only indirectly or by directly impacting the clock machinery.
A clue to this relationship came from our serendipitous observation that the period-short (perS) gene substantially improves the arrhythmicity of pdf01 flies. perS flies have a very short circadian period (∼19 h vs. ∼24 h for wild-type flies) (13), and PERS has a markedly shorter half-life than PER (14–18). This suggests that the function of PDF and enhanced cAMP levels is to slow the pace of PER degradation in PDFR-expressing clock neurons; the intrinsically fast degradation of PERS can bypass this regulatory step.
To examine this possibility, we mimicked the effects of PDF by manipulating cAMP levels and PKA activity in cell culture as well as in fly brains. PER degradation was inhibited by increasing cAMP levels, and cAMP-mediated PKA activation also stabilized PER. Importantly, PERS was less sensitive than PER to increases in cAMP levels. Consistent with a stabilizing role of PDF, a light pulse caused more prominent PER degradation in pdf-null mutant flies than in wild-type flies. These observations indicate that PDF contributes to clock neuron synchronization by up-regulating cAMP levels, activating PKA, and enhancing PER stability in target neurons. The stronger and more rapid degradation of PERS may allow perS neurons to maintain better synchrony and rhythmicity without PDF.
Results
perS Improves the Rhythmicity of pdf01 Flies.
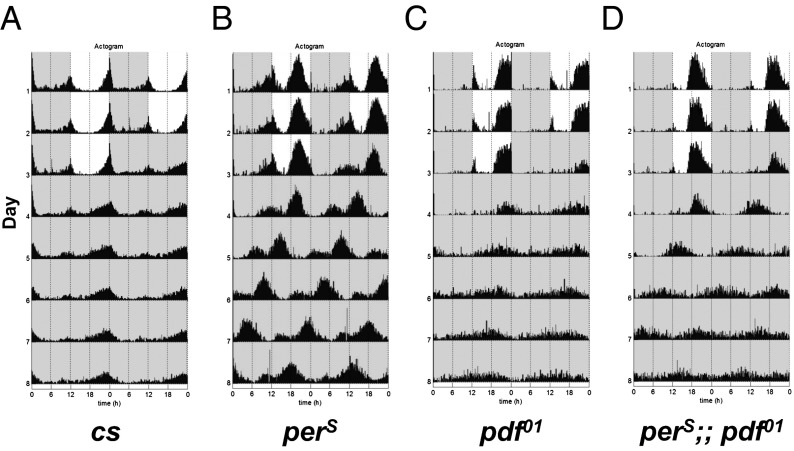
As described above, pdf01 flies show a ∼1–2 h advanced evening peak in LD and a comparably short period in DD before they become arrhythmic. This short period connects with our recent work showing that perS flies have a short-lived per protein, i.e., PERS degrades more rapidly than PER. We considered that if all circadian oscillators had the same very short ∼19-h period of perS, this might compensate for the oscillator asynchrony feature of pdf01 flies. To test this notion, we created double-mutant perS;;pdf01 flies, which indeed maintain a more robust short period in DD; in other words, the perS mutation partially rescues the arrhythmicity of the pdf01 mutation (Fig. 1 and Table 1; the actograms of representative individual flies are shown in Fig. S1). We note that the behavioral rhythms of perS flies as well as perS;;pdf01 flies also appear more robust than those of flies containing only a wild-type per gene (Fig. 1). Moreover, the period of the double-mutant flies is only marginally different (∼0.5 h shorter) than that of perS flies.
Fig. 1.
perS ameliorates the loss of PDF. Flies were entrained in 12:12-h light–dark (LD) cycles at 25 °C for at least 3 d, and then moved to constant-dark conditions (DD) for 6 d. The data were collected and double plotted on an actogram. Actograms are presenting the locomotive activity of following genotypes: (A) canton-S (cs); (B) perS; (C) rhythmic flies of pdf01; (D) rhythmic flies of perS;;pdf01. The mean of the period for the first 5 d in DD, the total number of animals, and the percentages of rhythmic animals are shown in Table 1. The actograms of several single flies are shown in Fig. S1.
Table 1.
Activity rhythm phenotypes
| Genotype | Period | SEM | n | R, % |
| pdf01 | 22.7*** | 0.2 | 48 | 45.8 |
| cs | 23.7 | 0.1 | 42 | 97.6 |
| perS;;pdf01 | 18.5*** | 0.1 | 60 | 78.3 |
| perS | 19.1 | 0.1 | 33 | 97.0 |
Period, the average period in 5 d of DD rhythmic animals; n, total animal numbers from three individual experiments; R, %, percentage of rhythmic animals. ***Significantly different from their control flies that do not have pdf01 mutation, P < 0.001, determined by Student t test.
One interpretation of this partial rescue is that there is less oscillator asynchrony in the perS background and therefore less need for PDF to slow the pace of some fast-running oscillators to maintain rhythmicity in DD (Discussion). The results also have a mechanistic implication for wild-type flies: they suggest that PDF slows the pace of circadian rhythms by enhancing PER stability. The rapid turnover of PERS may bypass this regulatory step.
Partial Rescue of PER Oscillations in pdf01 Neurons by perS.
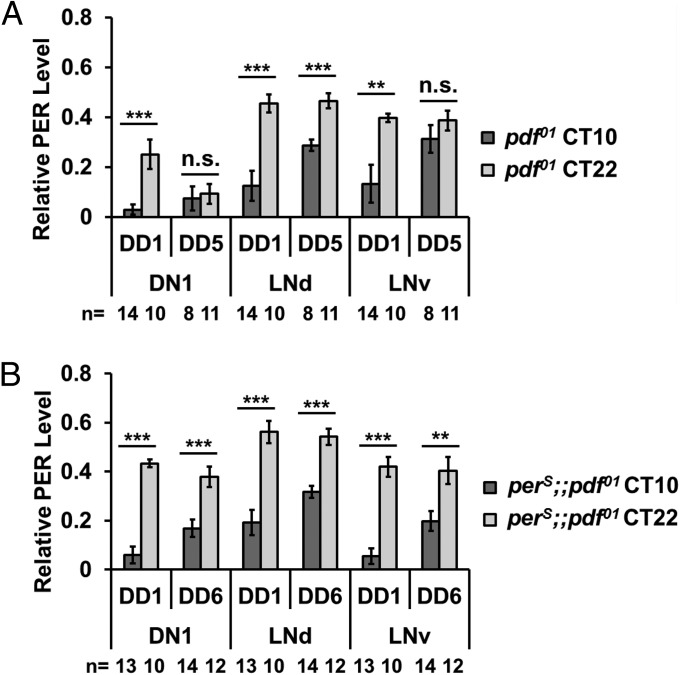
Although the rhythmicity of pdf01 flies is improved by the perS mutation (Fig. 1), it is still unclear the extent to which individual circadian neurons are rescued at the molecular level. To address this question, pdf01 and perS;;pdf01 flies were collected at circadian time 10 (CT10) and CT22, and brains were dissected and immunostained with an anti-PER antibody. These times were chosen because PER levels are normally low and high at CT10 and CT22, respectively. Flies were collected during DD1 as well as DD5. (DD1 is the first cycle in DD, whereas DD5 is the fifth, and robust PER cycling persists within many clock neuron groups for at least 5 d.) To accommodate the different period lengths of the two genotypes (Table 1), the real time points were converted as described by Yoshii et al. (7). In brief, periods were divided by 24 to get one “hour unit”: CT10 is the 10th hour unit and CT22 is the 22nd hour unit of a cycle. Because the period of the perS;;pdf01 genotype is about 4.5 h shorter than that of the pdf01 genotype, six perS;;pdf01 cycles is roughly equal to five pdf01 cycles. For simplicity, we examined and compared PER staining within three groups of clock neurons: dorsal neurons 1 (DN1s), dorsal-lateral neurons (LNds), and LNvs (includes large and small LNvs).
The normally dramatic oscillation of PER signal is significantly dampened by the fifth cycle in pdf01 DN1s and LNvs, reflecting asynchrony and/or weaker oscillation amplitude in these two neuronal groups. They contrast with the LNds within which persistent strong PER cycling persists for the 5 d in DD (Fig. 2A). This result is essentially identical to that previously reported for TIM oscillations within pdf01 brains (7). In striking contrast, PER oscillations are still robust in all three groups of neurons in the perS;;pdf01 strain even in the sixth circadian day of DD (Fig. 2B). As this cycling is indistinguishable from what is observed for wild-type and perS flies in DD (Fig. S2) (6), it indicates that the behavioral change reflects an alteration of molecular oscillations within the individual clock neurons. Given the effect of pdf01 on clock pace as well as the effect of the perS mutation on PER turnover, the observations suggest that PDF may decrease the rate of PER degradation within some clock neurons.
Fig. 2.
Oscillation of PER in the different clock neurons of pdf01 and perS;;pdf01 flies in DD. Pdf01 and perS;;pdf01 flies were entrained in 12:12 light–dark (LD) cycles at 25 °C for 3 d, and then moved to DD. Relative PER levels in DN1s, LNds, and LNvs from the brains at CT10 and CT22 were compared. Because the periods of these flies did not maintain a period of 24 h in DD (Table 1), their periods were divided by 24 to designate a specific one “hour unit.” CT10 and CT22 are the 10th and 22nd “hour units,” respectively. DD1, 5, and 6 indicate the first, fifth, and sixth period cycle in DD, respectively. Because the period of a perS;;pdf01 fly was about 4.5 h shorter than a pdf01 fly, the time length of six cycles in a perS;;pdf01 fly was roughly equivalent to the length of five cycles in a pdf01 fly. The histograms represent the mean relative PER intensity in the indicated neurons in pdf01 flies (A) and perS;;pdf01 flies (B). The number of hemispheres is indicated by “n.” Error bars represent ±SEM. The triple asterisk (***) represents P < 0.001, the double asterisk (**) represents P < 0.01, and “n.s.” represents no statistical significance as determined by Student t test. The oscillations of PER in the clock neurons of WT and perS flies are shown in Fig. S2.
Increasing cAMP and PKA Activity Stabilizes PER in S2 Cells.
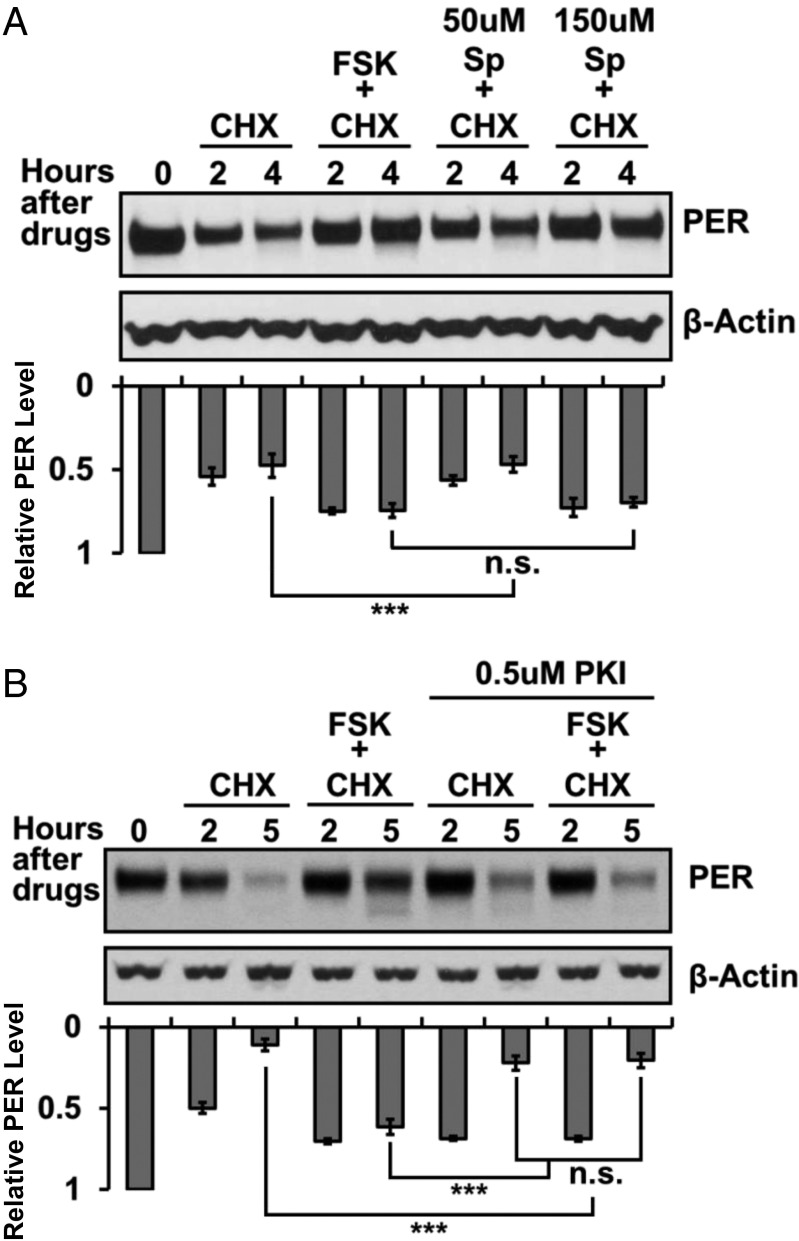
The positive effect of PDF on cAMP levels in PDFR-containing target neurons (12) suggests that an increase in cAMP levels might enhances PER stability. To test this possibility, we turned to the PER degradation S2 cell assay recently described. Following transcriptional activation of a heat shock per gene, the translation inhibitor cycloheximide (CHX) is added and then PER levels determined over a time course with anti-PER Western blots (17). This assay was combined with the addition of forskolin (FSK) or the addition of the cAMP analog Sp-adenosine-3′,5′-cyclic monophosphorothioate triethylamine (Sp-cAMPS). FSK activates adenylyl cyclase and has been previously used to phenocopy the effects of PDF in brain-imaging experiments (12). Sp-cAMPS is a specific activator of cAMP-dependent protein kinases (19). The rate of PER degradation is markedly decreased by FSK addition or by adding increasing amounts of Sp-cAMPS (Fig. 3A). Another cAMP analog, Rp-adenosine-3′,5′-cyclic monophosphorothioate triethylamine (Rp-cAMPS), is a specific competitive inhibitor of cAMP-dependent protein kinases (20). Rp-cAMPS has the opposite effect of FSK and Sp-cAMPS, and increases the rate of PER degradation (Fig. S3). The results indicate that PER stability is enhanced by activation of cAMP-dependent pathways including PKA and suggest that PDF functions in part by stabilizing PER.
Fig. 3.
The up-regulation of cAMP in S2 cells inhibits PER degradation. PER degradation rate was assayed in vitro using Drosophila S2 cells. The expression of per under the control of a heat shock promoter was induced using a 37 °C heat shock for 30 min. (A) The effect of up-regulation of cAMP on PER stability was assayed by adding cycloheximide (CHX) with or without forskolin (FSK) and Sp-cAMPS (Sp) 2 h post induction. (B) The effect of PKA on the stability of PER was examined by adding a PKA inhibitor PKI to the cells to inhibit the activity of PKA after FSK induction. Total protein was extracted at different time points after each treatment. PER was detected using an anti-PER. β-Actin was used as loading control. The histogram below the blots represents the mean relative intensity of the blots from three independent experiments. Error bars represent ±SEM. The triple asterisk (***) represents P < 0.001, and “n.s.” represents no statistical significance as determined by one-way ANOVA test.
FSK and Sp-cAMPS activate a number of cAMP-dependent pathways including PKA (21). This was a prime candidate in part because Neurospora PKA phosphorylates and stabilizes the important transcriptional repressor FREQUENCY (FRQ) (22), which functions in Neurospora rhythms similarly to Drosophila PER. To assess the role of PKA in PER stability regulation, we combined FSK with the specific PKA inhibitor PKI(14–22). The results show that PKI addition significantly reduces the effect of FSK (Fig. 3B), indicating that PKA mediates a substantial fraction of the cAMP effect on PER degradation. There may be a slight effect of PKI alone (Fig. 3B), suggesting that PKA impacts PER stability independent of PDF-mediated up-regulation of cAMP.
Increasing cAMP Stabilizes PER in Fly Brains.
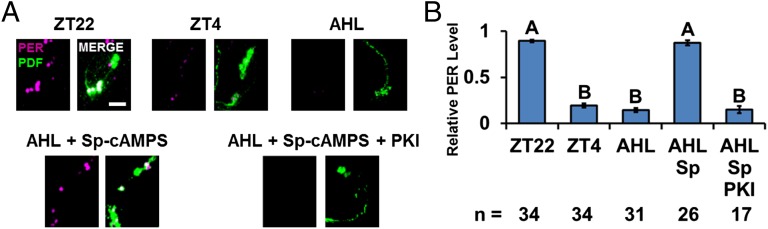
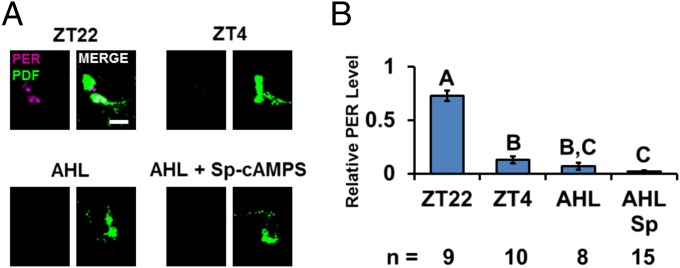
To confirm and extend this conclusion, a further PER stability assay was done in fly clock neurons. We focused on the ventrolateral group (LNvs), which controls circadian clock timing. LNvs are divided into two subgroups based on size, large-LNvs (l-LNvs) and small-LNvs (s-LNvs). Notably, all four s-LNvs and two of the four l-LNvs express PDFR (23), and all eight LNvs express PDF. Although this neuropeptide is usually an excellent marker to recognize LNvs and also to distinguish LNv cell body size, incubation in adult hemolymph-like saline (AHL) for the several hours required for this experiment caused a substantial but variable decrease in PDF signal. This made it impossible to distinguish s-LNvs from l-LNvs in some brain samples, so all PDF cells were included as a single group in this assay. To avoid the well-known and important effect of light on clock protein stability (17, 24, 25), circadian-blind cry01 flies were used in these experiments; cry01 flies manifest normal PER cycling within PDF neurons (23, 26). Flies were collected at Zeitgeber time 22 (ZT22) (10 h after lights off and 2 h before lights on) when PER is at or near peak levels (15, 17), and the brains were immediately dissected. They were incubated in AHL for 6 h, corresponding to an in vivo time of ZT4 or 4 h after lights on. PER levels at ZT4 are much lower than their peak levels at ZT22 (15, 17). As there is rather little per mRNA and therefore probably rather little PER synthesis between ZT22 and ZT4 (27), it is likely that the normal decrease in PER levels observed in vivo between ZT22 and ZT4 is due to PER degradation.
PER staining intensity within LNvs decreased dramatically during the 6-h in vitro incubation in AHL (Fig. 4 A and B). Because this residual PER signal is comparable to that in LNvs of flies dissected and directly stained at ZT4, PER degradation occurs similarly in vitro and in vivo after ZT22 (Fig. 4B, group B). Although we could not use FSK in this assay (the solvent DMSO had strong effects), the addition of water-soluble Sp-cAMPS dramatically increased PER staining intensity at the end of the 6-h incubation as there was no significant reduction of PER levels within PDF neurons (Fig. 4B, group A); this effect was inhibited by the concomitant addition of PKI (Fig. 4 A and B). Interestingly, PERS stability was less sensitive than PER to increases in cAMP levels (Fig. 5), suggesting that this is why the perS flies are less sensitive to the absence of PDF (Figs. 1 and 2B).
Fig. 4.
Activation of PKA inhibits PER degradation in fly brains. Brains of light-insensitive cry01 flies were dissected and immunostained at ZT22 and ZT4 to show PER degradation during that period. As controls, fly heads collected at ZT22 and ZT4 were fixed and dissected. Another group of heads were collected and dissected at ZT22 in adult hemolymph-like media (AHL) with drugs including PKA activator Sp-cAMPS and PKA inhibitor PKI as indicated in the figure. Brains were incubated in the AHL, with or without drugs, until ZT4 before fixed. (A) PER staining (magenta) in PDF cells (green). (Scale bar: 20 µm.) (B) Quantification of PER signal in PDF cells. All results were normalized to the brain that had the strongest signal at ZT22. The number of hemispheres tested is indicated by “n.” The bars in the histogram indicate the mean relative PER intensity. Each experiment was performed two to four times, and the number of hemispheres quantified is indicated below each bar. Error bars represent ±SEM. The letters above each bar represent the statistically significant groups as determined by one-way ANOVA, Turkey post hoc test.
Fig. 5.
Degradation of PERS cannot be inhibited by PKA activation. Brains of perS;;cry01 flies were immunostained using anti-PER antibody. PER signal in PDF neurons was quantified as described in Fig. 4. (A) PER staining (magenta) in PDF cells (green). (Scale bar: 20 µm.) (B) Quantification of PER signal in PDF cells. The histogram shows the mean relative PER intensity and the number of hemispheres quantified. Error bars represent ±SEM. The letters above each bar represent the statistically significant groups as determined by one-way ANOVA, Turkey post hoc test.
In Vitro Addition of PDF Stabilizes PER in Fly Brains.
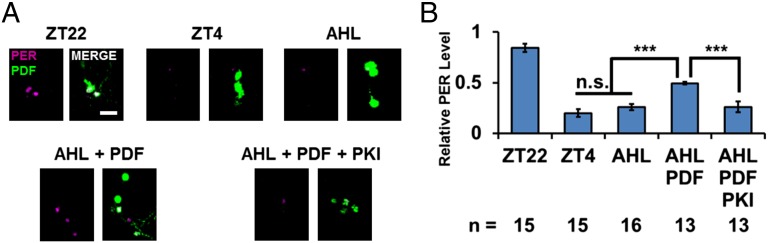
Because Sp-cAMP may exaggerate physiological levels of cAMP induced by PDF, the assay was repeated but with the application of PDF rather than Sp-cAMPS directly onto the dissected fly brains. The peptide concentration was 10 µM, which is sufficient to induce the cAMP response within circadian neurons (12).
The addition of PDF dramatically stabilized PER during the 6-h incubation (Fig. 6); this effect was also inhibited by the PKA inhibitor PKI (Fig. 6). As these clock neuron results are quite similar to those from S2 cells (Fig. 3), they further suggest that PDF increases cAMP levels, enhances PKA activity, and stabilizes PER.
Fig. 6.
In vitro addition of PDF stabilizes PER in fly brains. Same experiment was performed as in Fig. 4, except that 10 µM PDF peptide instead of Sp-cAMPS was added to the AHL. Brains were dissected at ZT22 in AHL with peptide and drug as indicated in the figure. The brains were incubated for 6 h until ZT4 before and fixed. (A) PER staining (magenta) in PDF cells (green). (Scale bar: 20 µm.) (B) PER signal in PDF cells was quantified. Data were analyzed as in Fig. 4. The bars in the histogram indicate the mean relative PER intensity. The number of brain hemispheres tested in two independent experiments is indicated by “n.” Error bars represent ±SEM. The triple asterisk (***) represents P < 0.001, and “n.s.” represents no statistical significance as determined by one-way ANOVA, Turkey post hoc test.
PDF Inhibits Light-Induced PER Degradation.
Light enhances the firing rate of PDF neurons, and morning phase shifts appear to require the l-LNvs (28, 29). Although TIM degradation is intimately connected with light-mediated phase shifts (24, 30–32), our recent study showed that PER degradation is also relevant (17). Given the likely positive relationship between PDF signaling and PER stabilization, we asked whether PER disappearance was more pronounced after a light pulse in the absence of PDF. To this end, we assayed the effect of a light pulse at ZT18 on PER staining intensity within the LNvs of pdf01 flies, which were labeled with GFP. Brains were dissected and stained at ZT19 after a standard light pulse protocol, namely10 min of light and 50 min in the dark. PER staining was compared with control brains dissected at ZT19 in the absence of a light pulse (17).
As previously described, a light pulse at ZT18 has no effect on PER signal within the LNvs of wild-type flies (Fig. 7 A and B) (17). In contrast, the same light pulse protocol dramatically reduced PER signal within LNvs of pdf01flies (Fig. 7 A and B). This is consistent with the notion that light causes PDF release, which then enhances PER stability.
Fig. 7.
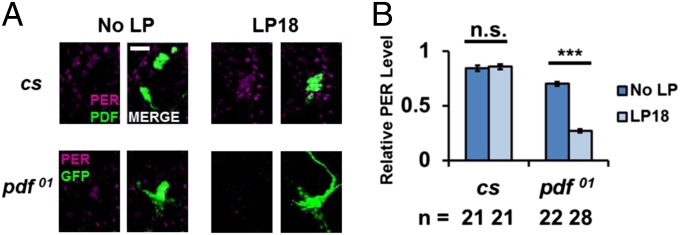
PDF protects PER from light-induced degradation at ZT18. A 10-min light pulse was given to flies at ZT18. Fly brains were dissected and stained at ZT19 and compared with flies from ZT19 with no light pulse. (A) In cs flies, PER staining (magenta) in PDF cells (green, PDF neurons immunostained with anti-PDF antibody) with or without the light pulse at ZT18. In UAS-mCD8-GFP; pdf-GAL4; pdf01 flies, PER staining (magenta) in PDF cells (green, PDF neurons were immunostained with anti-GFP) with or without a light pulse at ZT18. (Scale bar: 20 µm.) (B) PER signal in PDF cells from both genotypes with or without a light pulse was quantified. All results were normalized to the cs brain with the highest signal intensity at ZT19. The experiment was performed twice. The bars in the histogram indicate the mean relative PER intensity. The number of brain hemisphere tested is indicated by “n.” Error bars represent ±SEM. The triple asterisk (***) represents P < 0.001, and “n.s.” represents no statistical significance as determined by Student t test.
Discussion
Since the original observation that pdf01 flies have a highly reliable 1–2 h advanced activity phase in LD and short period in DD before they become arrhythmic (4, 5), it has been assumed that PDF functions at least in part to lengthen the period of at least some brain oscillators that run too fast in its absence. Indeed, there is evidence in favor of this notion (6, 7), and it is likely that the pdf01 strain arrhythmicity results from conflicts between neuronal oscillators that run too fast and others that maintain a ∼24-h pace or may even run more slowly without PDF (7). The substantial improvement of pdf01 rhythmicity by the perS gene (Figs. 1 and 2) therefore suggests that perS endows all oscillators with such a short period that they have a more uniform pace and substantially reduced oscillator asynchrony without PDF.
Although there was no information on how PDF might function to lengthen the period of the fast oscillators, the effect of perS implicates PER as a candidate molecular target. Because PERS is known to disappear rapidly in the nighttime, this further suggests that the PER degradation rate might be the biochemical target of PDF period lengthening. An even more specific version of this notion follows from the PDF-mediated increases in cAMP levels in PDFR-expressing clock neurons (12). Because PDFR is expressed in many clock neurons, including subsets of LNvs, LNds, and DN1s (23), this increase in cAMP may slow the pace of PER degradation in intrinsically fast-paced PDFR-expressing clock neurons. Indeed, the data indicate that increasing cAMP levels and PKA activity inhibits PER degradation in cell culture as well as in fly brains (Figs. 3, 4, and 6). Although these increases are probably in excess of what normally occurs in response to PDF, addition of PDF to brains in vitro has a similar effect. Because the additions of kinase inhibitors Rp-cAMPS and PKI increased the rate of PER degradation in S2 cells as well as in brains (Figs. 3, 4, and 6, and Fig. S3), we suggest that PDF-induced up-regulation of cAMP level and PKA activity likely affect PER stability.
A light pulse at night caused more prominent PER degradation in pdf01 mutant flies than in wild-type flies (Fig. 7). As nighttime light also causes premature TIM degradation and a consequent advance in PER degradation in many clock neurons (17), some of these neurons could be the intrinsically fast (22- to 23-h period) oscillators that are impacted by PDF and experience enhanced cAMP levels to slow their rate of PER degradation and clock pace. These probably include the s-LNvs and the DN1s, many of which are PDFR-positive [Fig. 2, Fig. S2, and Yoshii et al. (7)]. Based on the behavioral phenotype of pdf01 flies in LD and DD, the effect of PDF on PER degradation probably occurs in the late night–early morning in a LD cycle and at the same (subjective) time in DD. This is also the time when PER degradation is most prominent.
Interestingly, the firing rate of PDF-containing neurons, the l-LNvs as well as the s-LNvs, is also maximal near the beginning of the day (28, 33), in DD as well as LD; this is also the likely time of maximal PDF release from s-LNv dorsal projections (34). In addition, the l-LNvs promote light-mediated arousal, also mediated at least in part by PDF (23, 29, 35). Taken together with the fact that light has been shown to increase the firing rate of l-LNvs in a CRY-dependent manner (28), it is likely that lights on in the morning also potentiates the PDF-cAMP system. Note that the end of the night–beginning of the day is the time in the circadian cycle dominated by clock protein turnover, i.e., this is when there is little per or tim RNA or protein synthesis. This further supports a focus on clock protein turnover regulation at these times.
Because the mammalian neuropeptide VIP contributes to oscillator synchrony within the SCN in a manner that resembles at least superficially the contribution of PDF to oscillator synchrony within the fly brain circadian network, VIP might function similarly to PDF (36). However, VIP probably connects differently to the mammalian clock system. For example, morning light almost certainly up-regulates clock protein transcription in mammals, for example, per1 transcription (37). Therefore, VIP-mediated up-regulation of cAMP levels probably activates CREB and clock gene transcription through CRE sites in mammalian clock gene promoters rather than influencing clock protein turnover like in flies (38, 39).
The stabilization effect of PDF and cAMP on PER requires PKA activity within circadian neurons (Figs. 4 and 6). The effect could be indirect, through unknown PKA targets including other clock proteins. However, PER is known to be directly phosphorylated by multiple kinases; they include NEMO, which stabilizes PER (16). In addition, a study in Neurospora shows that PKA directly phosphorylates and stabilizes FRQ (22). Because FRQ and PER have similar roles (40, 41), protein turnover in the two clock systems may be similar beyond the shared role of the CK1 kinase (42). Based also on the S2 cell experiments (Fig. 3), we suggest that PKA directly phosphorylates PER and enhances its stability. This could occur by inhibiting a conformational switch to a less stable structure, a possibility that also applies to NEMO-mediated PER stabilization (16, 43, 44). PKA could also phosphorylate other clock proteins; this is by analogy to the known PER kinases NEMO and DOUBLETIME (DBT), which also phosphorylate CLK (45, 46).
The more rapid intrinsic degradation of PERS may at least partially bypass the effect of PKA phosphorylation and therefore PDFR stimulation. This may endow all circadian neurons with a more uniform period, which can maintain synchrony and therefore rhythmicity without PDF. The fact that PERS is less sensitive than PER to increases in cAMP levels (Fig. 5) is consistent with this interpretation, although an earlier phase of PERS degradation might also influence this result.
One further consideration is the 0.5-h period difference between the perS and the perS;;pdf01 strains. A residual period-lengthening effect of PDF suggests that perS does not endow all oscillators with the identical period, i.e., that there is still some asynchrony between different perS neurons without PDF. This may reflect an incomplete bypass of PKA by PERS or an additional effect of cAMP or PKA on other clock proteins. Nonetheless, several perS neuronal oscillators maintain a strong amplitude without PDF (Fig. 2 and Fig. S2). Although this is commonly taken to reflect an effect on synchrony, another possibility is based on data indicating that PDF normally enhances oscillator amplitude as well as synchrony; weak amplitudes may then be the more proximal cause of behavioral arrhythmicity. With this notion in mind, we suggest that PERS-containing oscillators are not only short period but also more robust, i.e., that the more rapid turnover of PERS makes the clock stronger. More robust rhythmicity is also apparent in the behavioral records of all perS-containing strains (Fig. 1). In this view, the stronger degradation “drive” of PERS makes these oscillators more cell autonomous and therefore less dependent on neuronal mechanisms like firing and PDF release, which enhance oscillator synchrony and amplitude. The general notion is that discrete differences in clock molecule properties can change the relationship of the transcriptional cycle to the circadian brain network.
Experimental Procedures
Drosophila Stocks and Plasmids.
Drosophila melanogaster were reared on standard cornmeal/agar medium supplemented with yeast. The wild-type (Canton-S) and perS flies were described by Konopka and Benzer in 1971 (13); pdf-null mutant (pdf01) was described by Renn et al. (4), and it was crossed with UAS-mCD8-GFP;pdf-GAL4 (47) flies to generate UAS-mCD8-GFP;pdf-GAL4;pdf01 flies; it was also crossed with perS flies to generate perS;;pdf01 flies; cry01 fly was described by Dolezelova et al. (48). The flies were entrained in 12:12 LD cycles at 25 °C. For the heat shock experiment in culture, the coding region of per with a V5 tag sequence was cloned into a pCaSpeR-hs vector (17).
S2 Cell Transfection and In Vitro Degradation Assay.
Drosophila Schneider 2 S2 (Drosophila Schneider 2) cells were maintained in insect tissue culture medium (HyClone) with 10% (vol/vol) FBS and 1% (vol/vol) antibiotics–antimycotics (Gibco) at 25 °C. Transfection was performed when cell confluency reached 50–70%. Cellfectin II (Invitrogen) was used according to the previously described protocol (49). The protocol of inducing PER expression was the same as previously described (17). S2 cells transfected with pCaSpeR-hs-per were heat shocked in 37 °C water bath for 30 min. Then the plates were put in a 25 °C incubator for 2 h. Then the cells were treated with 100 µg/mL CHX (10 mg/mL stock solution in water; Sigma-Aldrich), alone or with 20 µM FSK (50 mM stock solution in DMSO; Sigma), 150 µM Sp-cAMPS, 150 µM Rp-cAMPS (10 mM stock solution in water; Santa Cruz Biotechnology), and 0.5 µM PKI(12–22) (1 mM stock solution in water; Invitrogen). The cells were lysed with 300 µL of RBS buffer (45) at different time points. The lysates were denatured in SDS buffer at 100 °C for 10 min. The total proteins were resolved on 3–8% Tris-acetate gel (Invitrogen) and transferred to nitrocellulose membranes using the iBlot Dry Blotting (Invitrogen). The antibodies against PER (rabbit; 1:3,000) (50) and β-Actin (mouse; 1:3,000; Sigma) were used.
Locomotor Activity Analysis.
Locomotor activity of individual male flies (aged 2–5 d) were measured with Trikinetics Activity Monitors for at least 3 d under 12:12 LD conditions followed by at least 5 d of DD at 25 °C. The group activity were generated and analyzed with MATLAB (MathWorks) (51).
Fly Brain Immunocytochemistry.
To test the effect of drugs on PER stability in fly brains, flies were dissected in AHL (52) at room temperature. Brains were incubated in the AHL with drugs as described above (same concentration used in S2 cell culture) or 10 µM PDF peptide (100 µM stock in AHL) and kept in incubator for 6 h. Then the brains were fixed and processed as the protocol previously described (53). For PER staining, a precleaned polyclonal rabbit anti-PER by per01 fly head extract was used at a 1:50 dilution (50). For PDF staining, a mouse anti-PDF antibody (Development Studies Hybridoma, University of Iowa) was used at a 1:20 dilution. The secondary antibodies were Alexa Fluor 622-conjugated anti-rabbit (for PER) and Alexa Fluor 488-conjugated anti-mouse (for PDF; Molecular Probes) at 1:200 dilution. The brains were viewed in 1.1-µm sections sequentially at 20× on a Leica SP2 confocal microscope. To compare the PER signals from different samples, the laser intensity and other parameters of the confocal were set at the same level during each experiment. The neuron groups in pdf01 flies were identified according to their positions in the brains. PER signals in specific neurons were quantified by with Selection Brush Tool in ImageJ.
Supplementary Material
Acknowledgments
We thank Kate Abruzzi, Jerome Menet, Weifei Luo, Sean Bradley, Kim Kimberly, and Sudeep Agarwala for their thoughtful discussion. This work was supported by National Institutes of Health Grant P01 NS44232 (to M.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402562111/-/DCSupplemental.
References
- 1.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343(6258):536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 2.Sehgal A, et al. Rhythmic expression of timeless: A basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270(5237):808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 3.Darlington TK, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280(5369):1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 4.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99(7):791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 5.Lear BC, Zhang L, Allada R. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol. 2009;7(7):e1000154. doi: 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24(36):7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshii T, et al. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29(8):2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lear BC, et al. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48(2):221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Hyun S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48(2):267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Mertens I, et al. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48(2):213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol. 2012;108(2):684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafer OT, et al. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58(2):223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91(6):2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: Evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15(24):6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145(3):357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Rosbash M. Accelerated degradation of perS protein provides insight into light-mediated phase shifting. J Biol Rhythms. 2013;28(3):171–182. doi: 10.1177/0748730413489797. [DOI] [PubMed] [Google Scholar]
- 18.Kivimäe S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6(7):e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothermel JD, Parker Botelho LH. A mechanistic and kinetic analysis of the interactions of the diastereoisomers of adenosine 3′,5′-(cyclic)phosphorothioate with purified cyclic AMP-dependent protein kinase. Biochem J. 1988;251(3):757–762. doi: 10.1042/bj2510757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botelho LH, Webster LC, Rothermel JD, Baraniak J, Stec WJ. Inhibition of cAMP-dependent protein kinase by adenosine cyclic 3′-,5′-phosphorodithioate, a second cAMP antagonist. J Biol Chem. 1988;263(11):5301–5305. [PubMed] [Google Scholar]
- 21.Meinkoth JL, et al. Signal transduction through the cAMP-dependent protein kinase. Mol Cell Biochem. 1993;127-128:179–186. doi: 10.1007/BF01076769. [DOI] [PubMed] [Google Scholar]
- 22.Huang G, et al. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 2007;21(24):3283–3295. doi: 10.1101/gad.1610207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im SH, Li W, Taghert PH. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One. 2011;6(4):e18974. doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84(5):677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 25.Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66(3):378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95(5):681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 27.Kula-Eversole E, et al. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA. 2010;107(30):13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99(2):976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105(50):19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceriani MF, et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285(5427):553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 31.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380(6570):129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 32.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271(5256):1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 33.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28(25):6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JH, et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97(7):3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheeba V, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18(20):1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol. 2007;152(2-3):165–175. doi: 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan L, Takekida S, Shigeyoshi Y, Okamura H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: Circadian profile and the compartment-specific response to light. Neuroscience. 1999;94(1):141–150. doi: 10.1016/s0306-4522(99)00223-7. [DOI] [PubMed] [Google Scholar]
- 38.Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274(25):17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 39.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320(5878):949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClung CR, Fox BA, Dunlap JC. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature. 1989;339(6225):558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- 41.Millar AJ. Circadian rhythms: PASsing time. Curr Biol. 1997;7(8):R474–R476. doi: 10.1016/s0960-9822(06)00240-5. [DOI] [PubMed] [Google Scholar]
- 42.Görl M, et al. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 2001;20(24):7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menet JS, Rosbash M. A new twist on clock protein phosphorylation: A conformational change leads to protein degradation. Mol Cell. 2011;43(5):695–697. doi: 10.1016/j.molcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Querfurth C, et al. Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell. 2011;43(5):713–722. doi: 10.1016/j.molcel.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 45.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20(6):723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu W, Houl JH, Hardin PE. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 2011;21(9):756–761. doi: 10.1016/j.cub.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagoshi E, et al. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13(1):60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177(1):329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawathean P, Menet JS, Rosbash M. Assaying the Drosophila negative feedback loop with RNA interference in S2 cells. Methods Enzymol. 2005;393:610–622. doi: 10.1016/S0076-6879(05)93032-2. [DOI] [PubMed] [Google Scholar]
- 50.Dembinska ME, Stanewsky R, Hall JC, Rosbash M. Circadian cycling of a PERIOD-beta-galactosidase fusion protein in Drosophila: Evidence for cyclical degradation. J Biol Rhythms. 1997;12(2):157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 51.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3(1):1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112(2):271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 53.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508(6):952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.