Significance
The legume/rhizobia symbiosis involves a root-based exchange of bacterial fixed nitrogen for plant-derived photosynthetic carbon. The exchange takes place within the legume root nodule, which is a specialized root tissue that develops in response to plant and bacterial signal exchange. The bacteria reside within plant cells inside the nodule. In this study, we explore the activity of a membrane-bound soybean transcription factor, Glycine max basic–helix-loop–helix membrane 1, which is important for soybean nodule growth and is linked to the activity of a unique class of ammonium channels and to signaling cascades influencing a nodule circadian clock.
Keywords: nitrogen fixation, legume, nitrogen transport
Abstract
Glycine max symbiotic ammonium transporter 1 was first documented as a putative ammonium (NH4+) channel localized to the symbiosome membrane of soybean root nodules. We show that Glycine max symbiotic ammonium transporter 1 is actually a membrane-localized basic helix–loop–helix (bHLH) DNA-binding transcription factor now renamed Glycine max bHLH membrane 1 (GmbHLHm1). In yeast, GmbHLHm1 enters the nucleus and transcriptionally activates a unique plasma membrane NH4+ channel Saccharomyces cerevisiae ammonium facilitator 1. Ammonium facilitator 1 homologs are present in soybean and other plant species, where they often share chromosomal microsynteny with bHLHm1 loci. GmbHLHm1 is important to the soybean rhizobium symbiosis because loss of activity results in a reduction of nodule fitness and growth. Transcriptional changes in nodules highlight downstream signaling pathways involving circadian clock regulation, nutrient transport, hormone signaling, and cell wall modification. Collectively, these results show that GmbHLHm1 influences nodule development and activity and is linked to a novel mechanism for NH4+ transport common to both yeast and plants.
Legumes can form symbiotic interactions with soil-borne N2-fixing rhizobium bacteria. The symbiosis results in the formation of root nodules where infected nodule cells house differentiated bacteria termed bacteroids surrounded by a plant-derived symbiosome membrane (SM), forming a facultative organelle called the symbiosome (1). The symbiosis results in the exchange of carbon (C) from the plant for NH4+ produced through bacteroid nitrogenase activity (N2 fixation). Legumes promote this relationship through the expression of specific symbiotic-enhanced nodule genes (2) that allow development of the symbiosis and importantly the exchange of nutrients between symbionts.
We previously identified Glycine max basic helix–loop–helix membrane 1 (GmbHLHm1) (formerly GmSAT1) in a yeast complementation screen where it rescued growth of an NH4+ transport mutant 26972c and enhanced NH4+, methylammonium (MA), and K+ transport across the yeast plasma membrane (PM) (3). At high concentrations of MA, GmbHLHm1 made yeast cells sensitive to this toxic NH4+ analog (3). GmbHLHm1 was found localized to the PM-enriched fraction in yeast and the SM of infected cells of soybean nodules (3). The involvement of GmbHLHm1 in monovalent cation transport was controversial based on its unique structure, which contains a conserved basic helix–loop–helix (bHLH) DNA-binding motif and a single C-terminal hydrophobic transmembrane domain (TD). Subsequently, Marini et al. (4) demonstrated that GmbHLHm1 (SAT1) was not capable of NH4+ transport when expressed in the yeast NH4+ transport mutant (31019b) and suggested the role of GmbHLHm1 (SAT1) in NH4+ transport was indirect, possibly associated with changes in abundance of the NH4+ transport protein Mep3p.
In this study, we used multiple approaches to demonstrate that GmbHLHm1 (SAT1), in contrast to results reported in Kaiser et al. (3) and in accordance with Marini et al. (4), does not encode an NH4+ transporter but encodes a bHLH transcription factor (TF) that undergoes posttranslational modification for its delivery to the nucleus. In yeast, NH4+ transport is activated through overexpression of MEP3 and a unique low-affinity NH4+ transport protein common to both yeast and plants. The activity of GmbHLHm1 in soybean nodules indicates an important role in nodule development and growth that is linked ultimately to an effective N2-fixing symbiosis.
Results
GmbHLHm1 Is a Membrane-Localized DNA-Binding bHLH TF.
Sequence analysis shows that GmbHLHm1 belongs to a subset of the superfamily of plant bHLH TFs (5). GmbHLHm1-like proteins are predicted to be 28–42 kDa, contain a conserved bHLH DNA-binding domain, and a predicted C-terminal TD (SI Appendix, Fig. S1). The bHLH domain contains a conserved H-E-R amino acid motif (SI Appendix, Fig. S1) that recognizes the palindromic CANNTG promoter “E-box” element (6) commonly found in most bHLH-regulated genes. GmbHLHm1 homologs are found as multigene families in both dicot and monocot plant species (SI Appendix, Fig. S2).
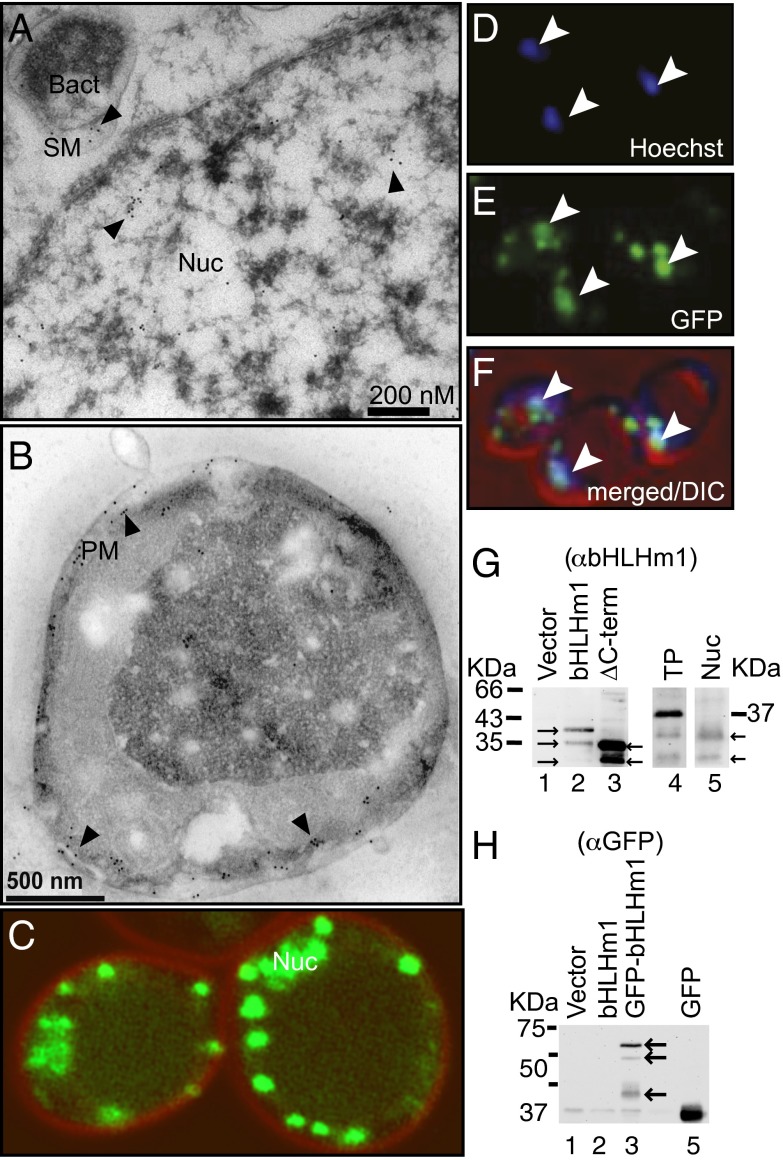
TFs located outside the nucleus are rare (7, 8) and presumably positioned to respond to cellular signaling pathways (9). To verify its location, we used immunogold labeling with a polyclonal anti-GmbHLHm1 antibody (α-bHLHm1) (3). We identified GmbHLHm1 in various membranes [PM, endoplasmic reticulum (ER), Golgi, SM] (SI Appendix, Fig. S3A) and in the nucleus of infected nodule cells (Fig. 1A). In GmbHLHm1-containing 26972c yeast cells, labeling was observed predominantly at the PM (Fig. 1B). Attachment of a N-terminal green fluorescent protein (GFP) tag to GmbHLHm1 in yeast resulted in GFP signal in peripheral punctate bodies but also in the nucleus (Fig. 1 C–F), where nuclear localization was confirmed by cross-reacting with the vital DNA stain, Hoechst 33342 (Fig. 1 D–F). Punctate and nuclear localization of GmbHLHm1 was also observed in plants when full-length GFP:GmbHLHm1 is transiently expressed in either Allium cepa (SI Appendix, Fig. S3 B–D) or Nicotiana benthamiana epidermal leaf cells (SI Appendix, Fig. S3 E–H). Loss of the C-terminal TD (S294-V347) resulted in GFP signal only in the nucleus of N. benthamiana cells (SI Appendix, Fig. S3 J–M). To understand the delivery of GmbHLHm1 to the nucleus, we used a modified split-ubiquitin assay involving the fusion of the artificial TF, LexA:VP16-Cub, to the N (LexA:VP16-Cub:GmbHLHm1) and C terminus (GmbHLHm1:LexA:VP16-Cub) of GmbHLHm1. When positioned at the N but not the C terminus, LexA:VP16-Cub complemented two nuclear reporter genes (HIS3 and lacZ) in the yeast strain DSY1 (SI Appendix, Fig. S3N). This indicated the N terminus of GmbHLHm1 is accessible to the cell cytosol, whereas the C terminus is not. To better understand the mechanism of release, we then introduced selected mutations (L277A and L277I) at a predicted subtilisin site 1 proteolytic recognition site (RXXL: L277; SI Appendix, Fig. S1) positioned upstream of the C-terminal TD in both LexA:VP16-Cub:GmbHLHm1 and native GmbHLHm1. The mutations disrupted LexA:VP16 complementation of DSY1 and reduced MA toxicity in yeast strain 26972c (SI Appendix, Fig. S3 O and P). We interrogated total yeast protein from GmbHLHm1 or N-terminal GFP-tagged GmbHLHm1 cells with α-bHLHm1 and α-GFP antibodies, respectively. With both antibodies, we identified similar banding patterns (∼37, 30, and 27 kDa: α-bHLHm1) (Fig. 1G, lane 2) and (∼66, 58, and 32 kDa: α-GFP) (Fig. 1H, lane 3). Based on the predicted size of GmbHLHm1 (3) without GFP, we believe the ∼37-kDa band represents the native full-length protein, because we only identified the 30- and 27-kDa peptides in nuclear enriched fractions from native GmbHLHm1 yeast cells or when the C-terminal TD of GmbHLHm1 was deleted (Fig. 1G, lanes 3 and 5, respectively). We believe the smaller 27-kDa protein is derived from a second uncharacterized proteolytic event at or after entry into the nucleus. Using A. cepa epidermal leaf cells, we profiled a full-length GFP:GmbHLHm1 construct against one where the N-terminal 21 aa of GmbHLHm1 was removed (∼4 kDa). The N-terminal deletion altered GFP localization, where GFP:GmbHLHm1 was located in the nucleus, whereas full-length N-terminal GFP-tagged protein resided in the outer nuclear envelope (SI Appendix, Fig. S3 B–D).
Fig. 1.
Intercellular localization of GmbHLHm1 in soybean nodule infected cells and yeast. (A and B) Transmission electron microscopy (TEM) analysis of cross-sections of wild-type soybean nodule cells (A) and 26972c yeast expressing GmbHLHm1 (B) after incubation with anti-GmbHLHm1 antiserum (α-bHLHm1, 1:500), followed by 10-nm colloidal gold conjugated with anti-rabbit IgG (1:40). Bact, bacteroid; Nuc, nucleus. (C–F) Confocal images of punctate and nuclear localized GFP::GmbHLHm1 in 26972c cells counterstained (red) with the membrane marker FM4-64. (D) Cells with Hoechst vital DNA staining. (E) N-terminal GFP-tagged GmbHLHm1. (F) Merged images of D and E with DIC image; arrows indicate nuclear localization of GFP signal and Hoechst DNA stain. Nuc, nucleus. (G and H) Identification of full-length and truncated GmbHLHm1 in whole-cell protein fractions of GmbHLHm1-transformed (α-bHLHm1 antiserum) (G) or N-terminal GFP::GmbHLHm1-transformed (using anti-GFP antiserum, α-GFP) 26972c cells (H). In G, the data in lanes 1–3, 4, and 5 represent three individual experiments, respectively. ∆C-term, deletion of the C-terminal TD of GmbHLHm1; Nuc, nuclear enriched fraction; TP, total protein.
GmbHLHm1 Is Important for Soybean Nodule Development.
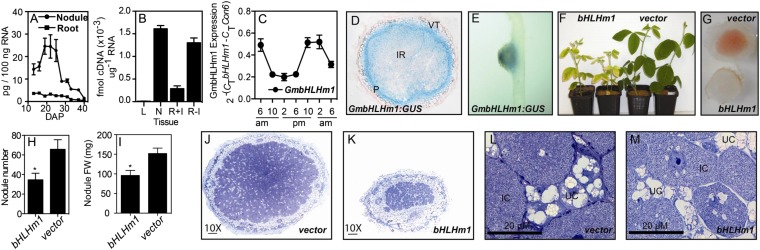
GmbHLHm1 expression is higher in nodules over nodule-detached roots (Fig. 2 A and B). When grown without rhizobium (minus N), GmbHLHm1 root expression is derepressed (Fig. 2B). In nodules, GmbHLHm1 expression increases with the onset of N2 fixation (15 d after rhizobium inoculation) and then decreases as the nodules mature (Fig. 2A). We examined the diel expression of GmbHLHm1 across a 24-h period and found a nocturnal expression pattern (Fig. 2C). GmbHLHm1 is primarily expressed in the encircling nodule parenchyma, even though a fainter signal could be detected in the bacterial infected region of the nodule (Fig. 2 D and E). Targeted RNAi silencing of the 3′-UTR of GmbHLHm1 (bhlhm1) using hairy root transformation reduced nodule numbers and nodule fresh weight (Fig. 2 H and I) (P < 0.05). In nodules that developed, N2 fixation and/or N export was compromised, resulting in shoot chlorosis (Fig. 2 F–I). bhlhm1 nodules displayed reduced leghemoglobin (Fig. 2G) and a small-infection zone (Fig. 2 J–M), where infected cells were small with variable symbiosome development, whereas uninfected cells were often vacuolated (Fig. 2 L and M and SI Appendix, Fig. S4 F and G).
Fig. 2.
Gene expression and loss of function of GmbHLHm1 in soybean nodules. (A) Developmental expression (DAP, days after planting) of GmbHLHm1 in N2-fixing soybean nodules and nodule-detached roots. (B) GmbHLHm1 expression in leaves (L), nodules (N), and nodule-detached roots with (R +I) or without (R -I) rhizobia from 32-d-old plants grown without nitrogen. (C) Diel expression of GmbHLHm1 in nodules. (D and E) Cellular expression identified using a GmbHLHm1 promoter:GUS fusion in both infected and uninfected nodule cells (D) and a nodulated root (E). IR, infected region; P, parenchymal cells; VT, vascular trace. (F) Chlorotic shoots of plants grown solely on sat1-nodulated roots supplied with nutrient solution containing no nitrogen. (G) Reduction in leghemoglobin in infected cells of sat1 nodules. (H and I) Reduction in nodule number (n = 19; P = 0.0125) and nodule fresh weight (n = 11; P = 0.0013) in bhlhm1 transgenic events. Data represent means ± SE. *P < 0.05. (J and K) Toludine blue-stained fresh tissue cross-sections of vector and bHLHm1-transformed nodules highlighting small infected cells in bHLHm1 nodules. (L and M) TEM analysis of cross-sections of infected and uninfected cells from vector- and sat1-transformed nodules. IC, bacteroid-infected cell; UC, uninfected cell.
To profile transcriptional changes in bhlhm1 nodules, we used quantitative (q) PCR and microarray analysis on isolated RNA from bhlhm1 and empty vector (vector) nodules (SI Appendix, Table S1 and Fig. S4 A–D). Although nodule development is impaired in bhlhm1 roots, we observed no change in expression of the bacteroid N2-fixation genes NifH and FixU (10) (SI Appendix, Fig. S4A). Using microarrays, we then profiled gene expression in 24-d-old bhlhm1 and vector nodules (SI Appendix, Fig. S4 B–D). Microarray analysis highlighted functional classes of genes where transcript abundance changed (SI Appendix, Table S1 and Fig. S4C). Among these were a number of down-regulated (P < 0.05) acyl-acid amido synthetase genes (GH3/BRU6) commonly involved in auxin/jasmonate amino acid conjugation (11) and a homolog of the ABC IBA transporter, ABCG37 (12). Other down-regulated genes (P < 0.05; fold-change, >2.5) included a zinc transporter (Zip1-like) (13) and multiple genes encoding xyloglucan endotransglucosylases (XTH/TCH4) (14). Interestingly, genes involved in the circadian clock, including the evening [GIGANTEA (GI)] (15) and morning expressed [PSEUDO-RESPONSE REGULATOR (PRR5, PRR7)] (16) genes were down-regulated in bhlhm1 nodules (SI Appendix, Table S1 and Fig. S4D). Legume N2 fixation has previously been shown to be responsive to changes in light and temperature, where rates of N2 fixation are strongly influenced by variations in day/night and related temperature cycles, whereas reduced N delivery from the nodule to the plant appears less variable (17–19). Because GmbHLHm1 expression is nocturnal (Fig. 2C), we profiled the expression of GmbHLHm1 against a selection of clock and nodule genes across a 24-h diel cycle. We found that GmGI1, GmGI2, GmPRR5, and GmPRR7 were expressed at their highest levels in the early evening (SI Appendix, Fig. S5 B–E). In contrast, the morning-loop regulator GmLHY1 showed increased expression in the morning (SI Appendix, Fig. S5F). Nodule-expressed N assimilatory genes [Glutamine synthetase (GmGS1γ2, GmGS1γ1, GmGS1β1)], the sucrose catabolic gene sucrose synthase (GmSS1), and the bacteroid N2-fixation genes (NifH and FixU) all showed positive responses to the daily light period, whereas the ureide biosynthetic enzyme urate oxidase (GmUO1) was relatively stable (SI Appendix, Fig. S5M). Because legume nodule growth, symbiotic N2 fixation, and NH4+ assimilation are all intimately linked to photosynthetically supplied C (20), we propose that a nodule circadian clock could be an important mechanism to integrate symbiotic C requirements across the day/night cycle.
GmbHLHm1 Activity Identifies a Unique Class of NH4+ Transport Proteins in Yeast and Plants.
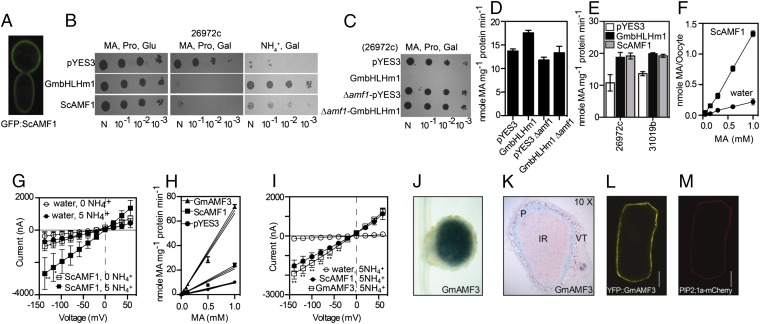
NH4+ uptake in yeast involves three PM-localized NH4+ transport proteins (Mep1-3p) belonging to the Mep/Amt/Rh superfamily (21). Using the NH4+ transport-deficient yeast mutant 26972c, GmbHLHm1 (SAT1) was originally linked to NH4+ and MA transport (3); 26972c is an EMS-derived mutant that contains a deletion of the MEP2 locus (mep2-1) and a substitution (G413D) in the C terminus of MEP1 (mep1-1) that transinactivates the functional low-affinity Mep3p through direct mep1-1p/Mep3p oligomerization (4, 22). Deleting mep1-1 or alternatively increasing MEP3 expression restores NH4+ transport activity in 26972c (4). To help explain how GmbHLHm1 complemented 26972c, we tested its influence on mep1-1 and MEP3 expression. Overexpression of GmbHLHm1 resulted in a significant increase in MEP3 (∼2.0-fold over pYES3-transformed controls; P = 0.0002) but no change in mep1-1 expression (SI Appendix, Fig. S6A). This indicates GmbHLHm1 influences MEP3 expression, an activity most likely contributing to its ability to restore growth on 1 mM NH4+, while also supporting the observation that GmbHLHm1 enhances Mep3p in 26972c (4). However, expression of GmbHLHm1 continued to enhance MA uptake (Fig. 3E) and toxicity in strains where MEP3 was deleted (26972c ∆mep3) or in a MEP1-3 deficient strain (31019b) (SI Appendix, Fig. S6B) (23). This result suggested that GmbHLHm1 enhances a MEP-independent MA transport pathway in yeast. To identify this transport mechanism, we used microarray analysis on RNA extracted from 26972c cells expressing GmbHLHm1 (SI Appendix, Table S2). We identified a gene encoding a major facilitator transport protein (YOR379w) that was significantly up-regulated (P = 8.6 × e−14) in cells expressing GmbHLHm1 (SI Appendix, Table S2). We observed that YOR378w is located on the PM when fused to GFP (Fig. 3A). YOR378w displays limited sequence similarity (SI Appendix, Fig. S7) to members of the DHA2 family of H+/drug antiporters (ATR1: YML116W, 38.7% identity; ATR2: YMR279c, 36.5% identity) involved in boron efflux and tolerance, respectively (24, 25), and animal spinster-like proteins (SPIN1) predicted to be involved in sugar transport (26). Overexpression of YOR378w (hereafter referred to as AMF1 for Ammonium Facilitator 1) resulted in MA toxicity in either 26972c (Fig. 3B) or 31019b cells (SI Appendix, Fig. S6C). Deletion of AMF1 (∆amf1) in 26972c eliminated GmbHLHm1-dependent MA transport and toxicity (Fig. 3 C and D) and reduced its NH4+ complementation capacity (SI Appendix, Fig. S6D). Introduced mutations in the GmbHLHm1 bHLH DNA-binding domain (R180K, a region involved in hydrogen bonding with phosphate in the DNA backbone) or the dimerization domains (L191V and L207V, two regions important for homo- and/or heterodimerization of bHLH proteins to bind DNA (SI Appendix, Fig. S1) (27)] disrupted [14C]MA transport, MA toxicity, and expression of ScAMF1 (SI Appendix, Fig. S6 E–G). Subsequent analysis of the ScAMF1 promoter identified eight predicted E-box binding domains (SI Appendix, Fig. S6H). Electromobility-shift analysis demonstrated that soluble GmbHLHm1 (128–270 aa) directly binds the ScAMF1 promoter (SI Appendix, Fig. S6H).
Fig. 3.
NH4+ transport through interactions between GmbHLHm1 and the membrane transporter AMF1. (A) Confocal image of N-terminal GFP:ScAMF1 localized to the PM in yeast. (B) 26972c cell growth (GmbHLHm1, ScAMF1, or pYES3) on minimal media containing 2% (wt/vol) galactose or glucose and either 0.1% l-proline (Pro), 0.1% l-proline plus 0.1 M MA (MA), or 1 mM NH4+ (NH4+). (C) Cell growth (26972c or 26972c:∆amf1) with GmbHLHm1 or pYES3 on minimal media containing 2% (wt/vol) galactose and 0.1% l-proline plus 0.1 M MA (MA). (D) Uptake of 1 mM [14C]MA by 26972c or 26972c:∆amf1 yeast cells (n = 6). (E) Uptake of 1 mM [14C]MA by 26972c or 31019b cells containing GmbHLHm1 or ScAMF1 (n = 6). (F) Concentration dependence of MA uptake into X. laevis oocytes injected with ScAMF1 cRNA or water (control) (n = 10). (G) Current–voltage curves of X. laevis oocytes injected with cRNA-ScAMF1 (squares) or water (circles). Injected oocytes were measured in 200 mM mannitol, 1 mM MgCl2, 0.2 mM CaCl2, 10 mM Hepes, and pH 7.5 adjusted with Tris base with or without NH4Cl as indicated (n = 3). (H) Uptake of [14C]MA by 26972c cells containing pYES3, ScAMF1, or GmAMF3 (Glyma08g06880) (n = 6). Data points are means ± SE. (I) Current–voltage curves of X. laevis oocytes injected with cRNA-ScAMF1 (solid circles), cRNA-GmAMF3 (squares), or water (open circles). *P < 0.05; **P < 0.001 (Tukey’s test). In H, data points have a nonlinear fit curve fit, and the 95% confidence intervals are presented. (J and K) Light images of a GmAMF3 promoter–GUS fusion in A. rhizogenes (K599)-mediated hairy roots inoculated with Bradyrhizobium japonicum USDA 110. (L and M) Confocal images of YFP-GmAMF3 (L) and AtPIP2;1a-mCherry (M) in bombarded onion epidermal cells viewed under a epifluorescence microscope. (Scale bar, 40 µm.)
We examined the activity of ScAMF1 in both yeast and Xenopus laevis oocytes. In yeast, ScAMF1 increased [14C]MA uptake and MA toxicity (0.1 M) in 26972c (Fig. 3 B and E). Many high-affinity NH4+ transport proteins, including Mep3p (4) and AMT1 (28), are capable of rescuing growth of 26972c on low (1 mM) NH4+. ScAMF1 behaved differently; it failed to complement growth at 1 mM NH4+ (SI Appendix, Fig. S6C), suggesting an activity more suited to higher NH4+ concentrations. To test further, we injected ScAMF1 cRNA into X. laevis oocytes. ScAMF1 increased [14C]MA uptake in a concentration-dependent manner (Fig. 3F) (P < 0.05; n = 10 oocytes). Using a two-electrode voltage clamp, increasing negative voltages across the oocyte PM resulted in a consistent NH4+ inducible inward current not present in the water-injected controls (Fig. 3 G and I).
Through sequence homology, we showed that AMF1 homologs are common in plants, including soybean, Medicago, Arabidopsis, and maize (SI Appendix, Figs. S7 and S8); interestingly, AMF1 homologs often display chromosomal microsynteny (29) with GmbHLHm1 genomic loci (SI Appendix, Fig. S9). We identified five AMF1 homologs in soybean consisting of two paralogous pairs: (i) Glyma15g06660 (GmAMF1) and Glyma13g32670 (GmAMF2); and (ii) Glyma08g06880 (GmAMF3) and Glyma07g30370 (GmAMF4); as well as Glyma09g33680 (GmAMF5). Analysis of publicly available RNA-seq data from Severin et al. (30) revealed GmAMF3 was up-regulated in nodules relative to roots (SI Appendix, Fig. S4E). Over a 24-h diel cycle, GmAMF3 expression peaked mid-morning, whereas a second AMF, GmAMF5 expression was elevated in the afternoon and evening (SI Appendix, Fig. S5 G and H). We cloned a full-length GmAMF3 cDNA from 28-d-old soybean nodule mRNA and tested its functional activity relative to ScAMF1 in both yeast and Xenopus oocytes. In yeast, GmAMF3 increased [14C]MA uptake relative to the empty vector control and ScAMF1 (Fig. 3H). In Xenopus oocytes, GmAMF3 elicited an NH4+ (5 mM)-dependent inward current when increasing negative voltages were applied across the oocyte PM (Fig. 3I). Based on the chemical and electrical signatures of ScAMF1 and GmAMF3, we suggest AMF proteins behave as NH4+ permeable transport proteins. We then examined the tissue localization of GmAMF3 in transformed soybean nodules and roots using the GmAMF3 (2.0-kB) promoter fused to the β-glucuronidase reporter GUS. GUS activity was primarily localized in cells surrounding the vascular bundles and the nodule parenchyma cell layer, which sits outside the infected zone (Fig. 3 K and L). We tested the intercellular location of GmAMF3 using a transient expression assay with YFP:GmAMF3 in onion epidermal cells. YFP:GmAMF3 was identified specifically on the PM similar to the PM localized AtPIP2a (31) (Fig. 3 M and N). In bhlhm1 nodules, we found no significant change in GmAMF3 expression; however, GmAMF5 (Glyma09g33680) was significantly (P < 0.05) down-regulated (SI Appendix, Fig. S4D).
Discussion
GmbHLHm1 Is a Soybean Membrane-Localized DNA-Binding bHLH TF.
One of the unique features of GmbHLHm1 is its membrane association when not in the nucleus. In soybean, GmbHLHm1 is located across multiple membranes in nodule infected cells, including the SM but also the PM, Golgi, and ER. Membrane association is also observed when the TF is heterologously expressed in other systems, including yeast, onion, and N. benthamiana leaf epidermal cells. Membrane bound TFs have been identified in plants, but less than 2% of the putative 1,533 TFs in Arabidopsis have been classified as membrane tethered (32). Examples of these include an Arabidopsis ER stress-responsive bZIP protein (33) and Arabidopsis nascent polypeptide-associated complex TFs (34). Membrane tethered TFs are assumed to be located outside the nucleus as a measure to regulate activity and to respond to cellular signaling pathways. Their activity is based on the anchored TF undergoing regulated intramembrane proteolysis or regulated ubiquitin/proteasome-dependent processing involving proteolytic cleavage and release of the soluble DNA-binding domain to the nucleus (9). Our data suggest that GmbHLHm1 can be delivered to the nucleus from its membrane anchor through a yet-unidentified proteolytic event upstream of its C-terminal TD (Fig. 1 H–J) and a predicted further modification at its N terminus, which may aid entry into the nucleus (SI Appendix, Fig. S3 C and D). This is consistent with other membrane-bound TFs, where release involves cleavage by membrane-associated serine, rhomboid, and possibly calpain-like proteases (9, 33–35). We observed via sequence comparison with other membrane bound TFs (OASIS, ATF2, SREBP2), a putative subtilisin site 1 proteolytic recognition site (RXXL: L277) upstream of the C-terminal TD of GmbHLHm1 (SI Appendix, Fig. S1). Substitutions at this site (L277A, L277I) reduced MA sensitivity and prevented nuclear entry of the artificial TF LexA-VP16 (SI Appendix, Fig. S3 O and P). Transcriptional activity of GmbHLHm1 increased the expression of a select number of yeast genes involved in nutrient transport, phosphorus homeostasis and cell wall modification (SI Appendix, Table S2). In particular, we identified an uncharacterized yeast gene ScAMF1 (YOR378w) that was strongly up-regulated by GmbHLHm1 (SI Appendix, Table S2). We confirmed this regulation was dependent on the DNA-binding domain of GmbHLHm1 through site-directed mutagenesis and through electromobility-shift analysis that indicated an affinity of purified GmbHLHm1 protein to the promoter of ScAMF1 (SI Appendix, Fig. S6H).
GmbHLHm1 Activity Identifies a Unique Low-Affinity NH4+ Transport Protein in Yeast and Plants.
Discrepancies between previous interpretations of GmbHLHm1 (SAT1) acting as an independent NH4+ transport protein (3), or not (4), were resolved in this study by demonstrating that GmbHLHm1 (SAT1) is instead a bHLH TF that enhances the expression of the high-affinity NH4+ transporter, MEP3 (SI Appendix, Fig. S6A), a phenotype that explains the ability of GmbHLHm1 (SAT1) to complement growth of yeast mutants 26972c but not 31019b on low NH4+ concentrations. Because MEP3 does not present E-box binding domains (6) in its promoter region, the enhanced expression by GmbHLHm1 may be indirect. This could be mediated by altered cellular NH4+ homeostasis influenced by the GmbHLHm1-enhanced expression of the NH4+ channel ScAMF1, which our data indicate is responsible for both MA uptake and MA toxicity when GmbHLHm1 is expressed in either 26972c (3) or 31019b (Fig. 3 B–E).
The identification of ScAMF1 reveals a previously unknown mechanism by which yeast cells can manage NH4+ transport. We have shown ScAMF1 is a yeast PM protein that is capable of facilitating MA uptake (Fig. 3 B and E). We have verified its transport activity further in X. laevis oocytes using both chemical (MA uptake) and electrical (NH4+-induced ion currents) studies. Incubating oocytes with increasing concentrations of [14C]MA significantly enhanced MA uptake across both high-affinity (<250 µM) and low-affinity (>250 µM) ranges. Similarly, we observed inward electrical currents at high concentrations (5 mM) of NH4+ in ScAMF1 cRNA-injected oocytes (Fig. 3G). Collectively, the transport studies in both yeast and Xenopus oocytes suggest ScAMF1 is most likely a low-affinity NH4+ transport protein. Its role in yeast NH4+ transport/metabolism is still to be defined. Mep(1-3)p transporters are clearly involved in the high-affinity uptake of NH4+ and are required for growth at low NH4+ concentrations (23). However, the mechanisms responsible for low-affinity NH4+ transport (physiological and genetic) have until now been poorly understood. Our data suggest ScAMF1 is most likely a participant in this process.
We identified AMF1 homologs across multiple plant genomes (SI Appendix, Fig. S8). Soybean contains five, where four are physically associated (within 20 Kb) with GmbHLHm1 loci. Publicly available RNA-seq expression data in soybean (30) demonstrated that GmAMF3 (Glyma08g06880) is nodule enhanced. GmAMF3 is primarily expressed in nodule parenchyma cells and the enveloping vascular tissues. Similar to ScAMF1, functional expression of GmAMF3 in both yeast and X. laevis oocytes resulted in [14C]MA and NH4+ transport activity (Fig. 3 H and I). This result suggests that a low-affinity NH4+ channel may operate in both the nodule parenchyma and vascular cells that connect the N2-fixing inner cortical cells to the root. Because the majority of fixed nitrogen (N) exported from soybean nodules are ureides (36), the requirement of an NH4+ channel in these cell types is unclear. Previous studies in Lotus japonicus have shown that PM localized high-affinity (AMT) NH4+ transporters are expressed across nodule-infected, vascular, and outer parenchyma cells (37, 38). Together, this would suggest NH4+ is present in many cell types of the nodule and that transport systems are required for efficient NH4+ recapture and assimilation or potentially release from the nodule through excessive N2 fixation (39).
GmbHLHm1 Activity Is Important to the Rhizobium/Legume Symbiosis.
GmbHLHm1 expression occurs primarily in the nodule parenchyma cells, as GmAMF3, but is also weakly detected in the infected region of the nodule and shows differential expression between roots exposed to or grown in the absence of rhizobia. GmbHLHm1 expression in nodules occurs during the night and decreases during the day. At the intercellular level, GmbHLHm1 is associated with the SM but also the PM, Golgi, ER, and the nucleus, which introduces an interesting link between membrane-based signaling and transcriptional activity in the nucleus. Apart from the association with AMF1, GmbHLHm1’s activity in legume nodules suggests an important role in the success of the symbiosis. Loss of bhlhm1 in soybean roots disrupted nodule development, resulting in reduced nodule numbers, which were generally smaller with impaired symbiosome development. These developmental changes were accompanied by a modest shift in transcriptional expression in the nodule that included down-regulated genes involved in the maintenance of auxin concentrations through amino acid conjugation (GH3/BRU6) and cell wall xyloglucan modification (XTH) and, interestingly, the regulation of the morning and evening components of the circadian clock (GI, PRR5, and PRR7).
The role of the circadian clock in N2-fixing legume nodules is unknown, but we assume it is similar to that of roots and linked to an underlying metabolic control regulating growth (15, 40). The circadian clock is also linked to the global regulation of endogenous auxin signaling involving auxin-induced genes in Arabidopsis, including homologs of the auxin-amido synthetases (41) found down-regulated in bhlhm1 nodules. How this relates to the role of bHLHm-like TFs identified in nonlegumes is less clear (SI Appendix, Fig. S5). In Arabidopsis, a bHLHm1 homolog, At2g22770 (NAI1), is involved in stress-induced synthesis of ER bodies in root and leaf tissues (42). It is possible that bHLHm1 TFs may have a role in facilitating other microbial interactions, including pathogen responses or mycorrhizal symbioses, both of which involve C supply from the plant. The identification of GmbHLHm1 presents an exciting opportunity to begin unraveling the subtle interactions that circadian rhythms may play in the establishment and maintenance of the rhizobium/legume symbiotic partnership.
In conclusion, we propose that GmbHLHm1 is an important plant regulator used for the rhizobium/legume symbiosis. Activity in yeast is linked to NH4+ transport through AMF1 activity, whereas AMF1 and bHLHm1 share chromosomal conservation across many plant species, including soybean. The link with nodule-expressed circadian clock genes suggests a role of GmbHLHm1 in mediating the underlying symbiosis-specific exchange of C from the plant for fixed N from rhizobia. Because bHLHm1 sequences are present in plants, but not in lower-order eukaryotes or prokaryotes, we suggest that bHLHm1-like proteins are likely an adaptation involved in mediating plant–microbe interactions.
Materials and Methods
Plant Materials and Growth Conditions.
Soybeans (Glycine max L. cv. Djakal) were grown in sand in either a growth chamber (28/25 °C, 16-/8-h day/night regime) under mercury halide lights (∼600 photosynthetic active radiation) or a temperature-controlled glasshouse (25/15 °C, day/night temperature) supplemented with mercury halide lights (16-/8-h day/night regime). Plants were inoculated with Bradyrhizobium japonicum US Department of Agriculture (USDA) 110 and watered with N-free nutrient solution. See SI Appendix, SI Materials and Methods for details.
Cellular Localization and Expression of GmbHLHm1.
The cellular localization of GmbHLHm1 was defined using immunogold labeling with anti-GmbHLHm1 polyclonal antibodies in wild-type nodules and yeast. Promoter-GUS fusion constructs were expressed in nodulated hairy roots generated with Agrobacterium rhizogenes. In planta GFP tagging experiments were carried out in N. benthamiana via Agrobacterium tumefacians infiltration or gold particle bombardment of A. cepa. Distribution of full-length and truncated versions of GmbHLHm1 was verified using a modified split-ubiquitin system and GFP tagging of GmbHLHm1 when expressed in Saccharomyces cerevisiae cells. Western blot analysis using anti-GmbHLHm1 and/or anti-GFP antibodies were used to investigate posttranslational modification of GmbHLHm1 in soybean and yeast protein extracts. Gene expression was quantified by qPCR from soybean and yeast total RNA. Whole-genome transcriptional profiling in yeast and soybean was conducted using Affymetrix microarray analysis. See SI Appendix, SI Materials and Methods for details.
Modification of GmbHLHm1 Activity in Soybean Nodules and Yeast Cells.
GmbHLHm1 expression in soybean roots and nodules was repressed using a 3′-UTR hairpin loop inserted into the soybean genome via A. rhizogenes root transformation (43). Modification to the putative bHLH DNA-binding and dimerization domain of GmbHLHm1 was evaluated using site-directed mutagenesis. AMF1-mediated transport activities were measured using yeast NH4+ transport mutants (26972c, 31019b) and cRNA AMF1-injected X. laevis oocytes with [14C]MA influx analysis and/or two-voltage electrode clamping. See SI Appendix, SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This work was supported by Australian Discovery Grants DP110100245 (to B.N.K., S.E.S., and T.B.) and DP0450577 (to D.A.D., M. Ludwig, S.D.T., and B.N.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE55804 and GSE55896).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312801111/-/DCSupplemental.
References
- 1.Roth E, Jeon K, Stacey G. Homology in endosymbiotic systems: The term “symbiosome. In: Palacios R, Verma D, editors. Molecular Genetics of Plant-Microbe Interactions. St. Paul, MN: American Phytopathological Society; 1988. pp. 220–225. [Google Scholar]
- 2.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64(1):781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser BN, et al. Characterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science. 1998;281(5380):1202–1206. doi: 10.1126/science.281.5380.1202. [DOI] [PubMed] [Google Scholar]
- 4.Marini AM, Springael JY, Frommer WB, André B. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol Microbiol. 2000;35(2):378–385. doi: 10.1046/j.1365-2958.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 5.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 6.Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48(5):501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- 7.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu J-X, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22(3):782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoppe T, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102(5):577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 10.Sciotti MA, Chanfon A, Hennecke H, Fischer HM. Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum. J Bacteriol. 2003;185(18):5639–5642. doi: 10.1128/JB.185.18.5639-5642.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westfall CS, Herrmann J, Chen Q, Wang S, Jez JM. Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal Behav. 2010;5(12):1607–1612. doi: 10.4161/psb.5.12.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Růžička K, et al. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA. 2010;107(23):10749–10753. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreau S, et al. GmZIP1 encodes a symbiosis-specific zinc transporter in soybean. J Biol Chem. 2002;277(7):4738–4746. doi: 10.1074/jbc.M106754200. [DOI] [PubMed] [Google Scholar]
- 14.Maldonado-Mendoza IE, Dewbre GR, Blaylock L, Harrison MJ. Expression of a xyloglucan endotransglucosylase/hydrolase gene, Mt-XTH1, from Medicago truncatula is induced systemically in mycorrhizal roots. Gene. 2005;345(2):191–197. doi: 10.1016/j.gene.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Dalchau N, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA. 2011;108(12):5104–5109. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA. 2012;109(42):17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minchin FR, Pate JS. Diurnal functioning of the legume root nodule. J Exp Bot. 1974;25:295–308. [Google Scholar]
- 18.Rainbird RM, Atkins CA, Pate JS. Diurnal variation in the functioning of cowpea nodules. Plant Physiol. 1983;72(2):308–312. doi: 10.1104/pp.72.2.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisz PR, Sinclair TR. Soybean nodule gas permeability, nitrogen fixation and diurnal cycles in soil temperature. Plant Soil. 1988;109:227–234. [Google Scholar]
- 20.Vance CP, Heichel GH. Carbon in N2 fixation: Limitation or exquisite adaptation. Annu Rev Plant Physiol. 1991;42:373–392. [Google Scholar]
- 21.Pantoja O. High affinity ammonium transporters: Molecular mechanism of action. Front Plant Sci. 2012;3(34):34. doi: 10.3389/fpls.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. A cytosolic trans-activation domain essential for ammonium uptake. Nature. 2007;446(7132):195–198. doi: 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- 23.Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(8):4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaya A, Karakaya HC, Fomenko DE, Gladyshev VN, Koc A. Identification of a novel system for boron transport: Atr1 is a main boron exporter in yeast. Mol Cell Biol. 2009;29(13):3665–3674. doi: 10.1128/MCB.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozdag GO, Uluisik I, Gulculer GS, Karakaya HC, Koc A. Roles of ATR1 paralogs YMR279c and YOR378w in boron stress tolerance. Biochem Biophys Res Commun. 2011;409(4):748–751. doi: 10.1016/j.bbrc.2011.05.080. [DOI] [PubMed] [Google Scholar]
- 26.Rong Y, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci USA. 2011;108(19):7826–7831. doi: 10.1073/pnas.1013800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferré-D’Amaré AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 28.Ninnemann O, Jauniaux JC, Frommer WB. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994;13(15):3464–3471. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowers JE, et al. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc Natl Acad Sci USA. 2005;102(37):13206–13211. doi: 10.1073/pnas.0502365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Severin AJ, et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pumplin N, Zhang X, Noar RD, Harrison MJ. Polar localization of a symbiosis-specific phosphate transporter is mediated by a transient reorientation of secretion. Proc Natl Acad Sci USA. 2012;109(11):E665–E672. doi: 10.1073/pnas.1110215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S-Y, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35(1):203–213. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(42):16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y-S, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18(11):3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Pate JS, Atkins CA, White ST, Rainbird RM, Woo KC. Nitrogen nutrition and xylem transport of nitrogen in ureide-producing grain legumes. Plant Physiol. 1980;65(5):961–965. doi: 10.1104/pp.65.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Apuzzo E, et al. Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 2004;134(4):1763–1774. doi: 10.1104/pp.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogato A, et al. Tissue-specific down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus. Plant Mol Biol. 2008;68(6):585–595. doi: 10.1007/s11103-008-9394-5. [DOI] [PubMed] [Google Scholar]
- 39.Silvester WB, Parsons R, Watt PW. Direct measurement of release and assimilation of ammonia in the Gunnera–Nostoc symbiosis. New Phytol. 1996;132(4):617–625. doi: 10.1111/j.1469-8137.1996.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 40.James AB, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322(5909):1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 41.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5(8):e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell. 2004;16(6):1536–1549. doi: 10.1105/tpc.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammadi-Dehcheshmeh M, Ebrahimie E, Tyerman S, Kaiser B. A novel method based on combination of semi-in vitro and in vivo conditions in Agrobacterium rhizogenes-mediated hairy root transformation of Glycine species. In Vitro Cell Dev Biol Plant. 2013 doi: 10.1007/s11627-013-9575-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.