Abstract
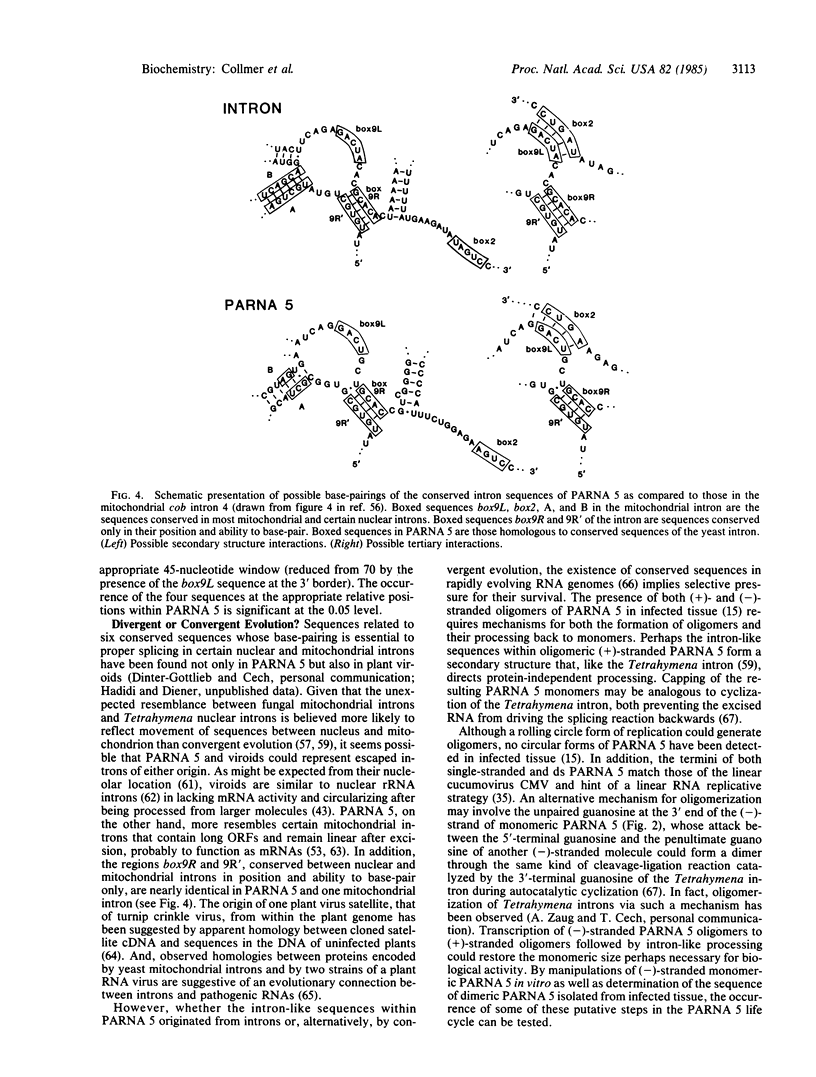
Peanut stunt virus-associated RNA 5 (PARNA 5), the satellite of a plant cucumovirus, is a linear RNA of 393 nucleotides with a 5' cap and a 3' hydroxyl group. Determination of its nucleotide sequence has revealed two consecutive open reading frames that together extend most of its length. Sequences at the 5' and 3' ends are homologous with those of the satellite of the related cucumber mosaic virus, and the double-stranded forms of both satellites contain an unpaired guanosine at the 3' end of the minus strand. However, little other homology exists between the two satellites. In contrast, PARNA 5 has several regions of 90% sequence homology with various plant viroids, including sequences of the conserved central region of most viroids. Such homologies suggest a common origin with viroids coupled with specific adaptation as a linear RNA. The presence within PARNA 5 of conserved intron sequences essential to proper RNA processing suggests a possible origin from plant introns and/or involvement of such sequences in the processing of PARNA 5 multimers to monomers at some stage of replication.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branch A. D., Robertson H. D. A replication cycle for viroids and other small infectious RNA's. Science. 1984 Feb 3;223(4635):450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R. RNA splicing: three themes with variations. Cell. 1983 Oct;34(3):713–716. doi: 10.1016/0092-8674(83)90527-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Split genes and RNA splicing. Science. 1979 Apr 20;204(4390):264–271. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- Dasgupta R., Harada F., Kaesberg P. Blocked 5' termini in brome mosaic virus RNA. J Virol. 1976 Apr;18(1):260–267. doi: 10.1128/jvi.18.1.260-267.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz J. R., Kaper J. M. Cucumber mosaic virus-associated RNA 5. III. Little nucleotide sequence homology between CARNA 5 and helper RNA. Virology. 1977 Jul 1;80(1):204–213. doi: 10.1016/0042-6822(77)90393-2. [DOI] [PubMed] [Google Scholar]
- Dickson E. A model for the involvement of viroids in RNA splicing. Virology. 1981 Nov;115(1):216–221. doi: 10.1016/0042-6822(81)90104-5. [DOI] [PubMed] [Google Scholar]
- Diener T. O. Are viroids escaped introns? Proc Natl Acad Sci U S A. 1981 Aug;78(8):5014–5015. doi: 10.1073/pnas.78.8.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener T. O. Viroids. Adv Virus Res. 1983;28:241–283. doi: 10.1016/s0065-3527(08)60725-3. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in Neurospora mitochondria. The large rRNA intron contains a noncoded, 5'-terminal guanosine residue. J Biol Chem. 1983 Dec 25;258(24):14745–14748. [PubMed] [Google Scholar]
- Gordon K. H., Symons R. H. Satellite RNA of cucumber mosaic virus forms a secondary structure with partial 3'-terminal homology to genomal RNAs. Nucleic Acids Res. 1983 Feb 25;11(4):947–960. doi: 10.1093/nar/11.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A. R., Palukaitis P., Symons R. H., Mossop D. W. Characterization of a satellite RNA associated with cucumber mosaic virus. Virology. 1978 Feb;84(2):443–455. doi: 10.1016/0042-6822(78)90261-1. [DOI] [PubMed] [Google Scholar]
- Gross H. J., Krupp G., Domdey H., Raba M., Jank P., Lossow C., Alberty H., Ramm K., Sänger H. L. Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur J Biochem. 1982 Jan;121(2):249–257. doi: 10.1111/j.1432-1033.1982.tb05779.x. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Huang A. S. Defective interfering viruses. Annu Rev Microbiol. 1973;27:101–117. doi: 10.1146/annurev.mi.27.100173.000533. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E. Cucumber mosaic virus-associating RNA 5. I. Role of host plant and helper strain in determining amount of associated RNA 5 with virions. Virology. 1977 Jul 1;80(1):186–195. doi: 10.1016/0042-6822(77)90391-9. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E., Diaz-Ruiz J. R., Tolin S. A. Peanut stunt virus-associated RNA 5: second tripartite genome virus with an associated satellite-like replicating RNA. Virology. 1978 Jul 1;88(1):166–170. doi: 10.1016/0042-6822(78)90119-8. [DOI] [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E., Lot H. A low molecular weight replicating RNA associated with a divided genome plant virus: defective or satellite RNA? Biochem Biophys Res Commun. 1976 Oct 18;72(4):1237–1243. [PubMed] [Google Scholar]
- Kaper J. M., Tousignant M. E. Separation of the complementary strands of double-stranded cucumber mosaic virus-associated RNA 5 and peanut stunt virus-associated RNA 5. Biochem Biophys Res Commun. 1983 Nov 15;116(3):1168–1175. doi: 10.1016/s0006-291x(83)80265-4. [DOI] [PubMed] [Google Scholar]
- Kiefer M. C., Owens R. A., Diener T. O. Structural similarities between viroids and transposable genetic elements. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6234–6238. doi: 10.1073/pnas.80.20.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Tyc K., Filipowicz W., Sänger H. L., Gross H. J. Circularization of linear viroid RNA via 2'-phosphomonoester, 3', 5'-phosphodiester bonds by a novel type of RNA ligase from wheat germ and Chlamydomonas. Nucleic Acids Res. 1982 Dec 11;10(23):7521–7529. doi: 10.1093/nar/10.23.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Linthorst H. J., Kaper J. M. Replication of peanut stunt virus and its associated RNA 5 in cowpea protoplasts. Virology. 1984 Dec;139(2):317–329. doi: 10.1016/0042-6822(84)90377-5. [DOI] [PubMed] [Google Scholar]
- Lot H., Jonard G., Richards K. Cucumber mosaic virus RNA 5. Partial characterization and evidence for no large sequence homologies with genomic RNAs. FEBS Lett. 1977 Aug 15;80(2):395–400. doi: 10.1016/0014-5793(77)80484-5. [DOI] [PubMed] [Google Scholar]
- Marcu K. B., Schibler U., Perry R. P. The 5'-terminal sequences of immunoglobulin messenger RNAs of a mouse myeloma. J Mol Biol. 1978 Apr 15;120(3):381–400. doi: 10.1016/0022-2836(78)90426-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Monroy G., Spencer E., Hurwitz J. Characteristics of reactions catalyzed by purified guanylyltransferase from vaccinia virus. J Biol Chem. 1978 Jun 25;253(12):4490–4498. [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- Netter P., Jacq C., Carignani G., Slonimski P. P. Critical sequences within mitochondrial introns: cis-dominant mutations of the "cytochrome-b-like" intron of the oxidase gene. Cell. 1982 Apr;28(4):733–738. doi: 10.1016/0092-8674(82)90052-6. [DOI] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Intervening sequences excised in vitro. Nature. 1978 Aug 10;274(5671):530–530. doi: 10.1038/274530a0. [DOI] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J., Sänger H. L., Riesner D. Subcellular localization of viroids in highly purified nuclei from tomato leaf tissue. EMBO J. 1983;2(9):1549–1555. doi: 10.1002/j.1460-2075.1983.tb01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Cucumber mosaic virus RNA contains 7-methyl guanosine at the 5'-terminus of all four RNA species. Mol Biol Rep. 1975 Dec;2(4):277–285. doi: 10.1007/BF00357014. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Holl J., Schweyen R. J., Rödel G., Kaudewitz F. Processing of yeast mitochondrial RNA: involvement of intramolecular hybrids in splicing of cob intron 4 RNA by mutation and reversion. Cell. 1983 May;33(1):195–202. doi: 10.1016/0092-8674(83)90348-3. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grabowski P. J., Cech T. R. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature. 1983 Feb 17;301(5901):578–583. doi: 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- Zimmern D. Homologous proteins encoded by yeast mitochondrial introns and by a group of RNA viruses from plants. J Mol Biol. 1983 Dec 15;171(3):345–352. doi: 10.1016/0022-2836(83)90098-0. [DOI] [PubMed] [Google Scholar]



