Significance
The Middle East respiratory syndrome (MERS)-coronavirus, a newly identified pathogen, causes severe pneumonia in humans, with a mortality of nearly 44%. Human-to-human spread has been demonstrated, raising the possibility that the infection could become pandemic. Mice and other small laboratory animals are not susceptible to infection. Here, we describe the development of a small-animal model for MERS, in which we use an adenovirus expressing the human host-cell receptor to sensitize mice for infection. We show that these mice are useful for determining immune responses and for evaluation of an anti-MERS vaccine and an antiviral therapy. This approach will be generally useful for the rapid (2–3 wk) development of relevant mouse and other animal models for emerging viral infections.
Keywords: emerging pathogen, interferon, SARS
Abstract
In this era of continued emergence of zoonotic virus infections, the rapid development of rodent models represents a critical barrier to public health preparedness, including the testing of antivirus therapy and vaccines. The Middle East respiratory syndrome coronavirus (MERS-CoV) was recently identified as the causative agent of a severe pneumonia. Given the ability of coronavirus to rapidly adapt to new hosts, a major public health concern is that MERS-CoV will further adapt to replication in humans, triggering a pandemic. No small-animal model for this infection is currently available, but studies suggest that virus entry factors can confer virus susceptibility. Here, we show that mice were sensitized to MERS-CoV infection by prior transduction with adenoviral vectors expressing the human host-cell receptor dipeptidyl peptidase 4. Mice developed a pneumonia characterized by extensive inflammatory-cell infiltration with virus clearance occurring 6–8 d after infection. Clinical disease and histopathological changes were more severe in the absence of type-I IFN signaling whereas the T-cell response was required for virus clearance. Using these mice, we demonstrated the efficacy of a therapeutic intervention (poly I:C) and a potential vaccine [Venezuelan equine encephalitis replicon particles expressing MERS-CoV spike protein]. We also found little protective cross-reactivity between MERS-CoV and the severe acute respiratory syndrome-CoV. Our results demonstrate that this system will be useful for MERS-CoV studies and for the rapid development of relevant animal models for emerging respiratory viral infections.
The spread of the severe acute respiratory syndrome (SARS)-coronavirus (CoV) in 2002/2003 and of the Middle East respiratory syndrome (MERS)-CoV in 2012 indicated that coronaviruses could cause severe pneumonia in humans (1, 2). As of February 7, 2014, 182 patients had been infected with MERS-CoV, with a 43.4% mortality rate. Human-to-human spread has been documented (3). A majority of patients with severe disease were elderly and had preexisting illnesses such as diabetes or renal failure whereas immunocompetent patients mostly developed mild disease (4). The pathogenesis of the infection is not well understood, in part because no autopsy information is available. Experimental infection has been demonstrated only in macaques (5–7). The expense and limited availability of these animals makes it imperative to generate a small-animal model for MERS for development of vaccines and antiviral therapies. Mice and hamsters are not infectable, and, because virus entry factors can confer virus susceptibility (7, 8), we postulated that exogenous expression of human host-cell receptor dipeptidyl peptidase 4 (hDPP4) would render mice susceptible (9).
Here, we describe a novel approach to developing a mouse model for MERS by transducing mice with a recombinant, nonreplicating adenovirus expressing the hDPP4 receptor. After infection with MERS-CoV, mice develop an interstitial pneumonia. We show that these transduced, infected mice can be used to determine antivirus immune responses and to evaluate anti–MERS-CoV vaccines and therapies.
Results
Development of Mice Susceptible to MERS-CoV Infection.
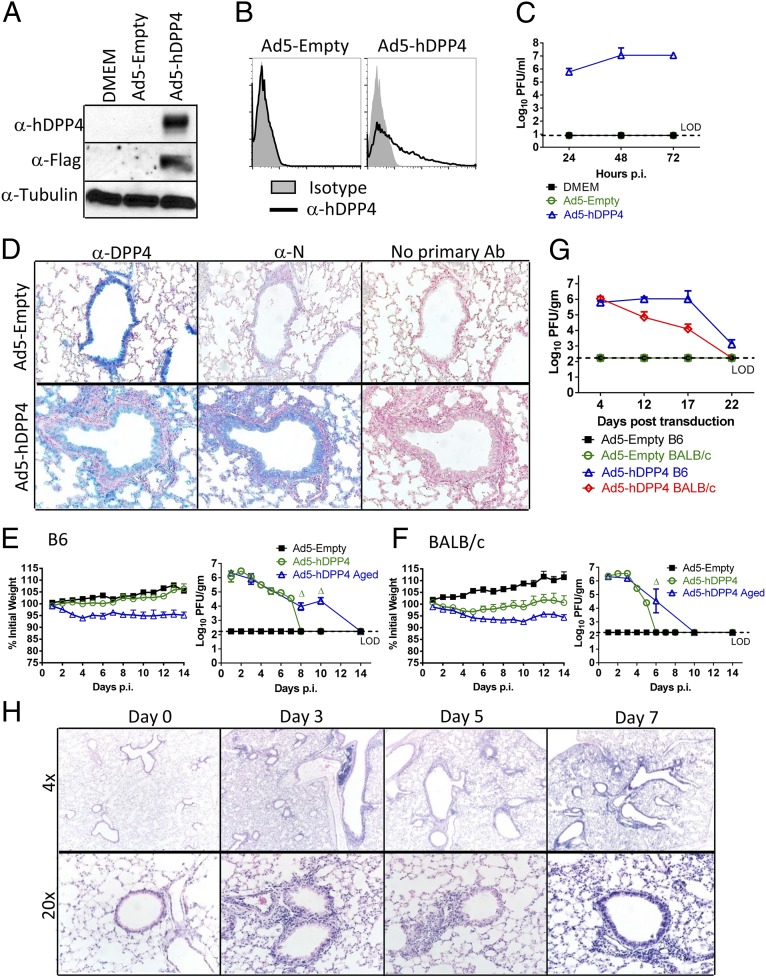
Adenoviral vectors have been used for gene therapy and to sensitize mice to systemic infection (10–14). However, their ability to render mice susceptible to mucosal infections, including those of the respiratory tract, has not been examined previously. Second-generation E1/E3-deleted Ad5 vectors are configured to minimize outgrowth of wild-type (WT) virus and to reduce immunogenicity (15). To develop adenoviruses for expressing hDPP4 in mouse lungs, we cloned a FLAG and myc-tagged cDNA into a replication-deficient adenovirus (Ad5-hDPP4). hDPP4 expression was validated by transducing MLE15 cells, a mouse alveolar type-II cell line, with Ad5-hDPP4 and analyzing cell lysates for hDPP4 expression. hDDP4 was detected using anti-hDPP4 or anti-FLAG antibodies (Fig. 1A). Surface expression of hDPP4, required to enable virus entry, was demonstrated by flow cytometry (Fig. 1B). To determine whether hDPP4 rendered cells susceptible to MERS-CoV, we infected Ad5 control (Ad5-Empty) and Ad5-hDPP4–transduced cells with MERS-CoV. Control cells were resistant to MERS-CoV whereas MERS-CoV replicated to high titers in Ad5-hDPP4–transduced cells (Fig. 1C). To identify hDPP4 expression in mice, we transduced BALB/c mice with 2.5 × 108 pfu of Ad5-hDPP4 or Ad5-Empty and detected DPP4 with an antibody that recognized human and mouse DPP4. After Ad5-Empty transduction, only endogenous mouse DPP4 expression was detected whereas, after Ad5-hDPP4 transduction, there was widespread hDPP4 expression in airway and alveolar epithelial cells (Fig. 1D). Next, we infected 6- to 12-wk-old and 18- to 22-mo-old C57BL/6 and BALB/c mice with MERS-CoV 5 d after Ad5-hDPP4 transduction (Fig. 1 E and F). We expected that innate responses triggered by the Ad vector would be largely dissipated by 5 d after transduction (16). Consistent with this, histological examination revealed no evidence for inflammatory-cell infiltration after Ad5-hDPP4 transduction but before MERS-CoV infection (Fig. 1H and Fig. S1, day 0). After MERS-CoV infection, there was no mortality, but young BALB/c mice failed to gain weight and aged mice of both strains lost weight. Transduced hDPP4 was required for virus replication (Fig. 1D), which reached ∼107 pfu/g lung tissue by 2–3 days postinfection (d.p.i.), (Fig. 1 E and F). After MERS-CoV infection, virus was then cleared by days 6–8 in young mice and days 10–14 in 18- to 22-mo-old mice. Clearance did not reflect loss of hDPP4 expression because mice could be infected as late as days 17–22 posttransduction, dependent upon strain (Fig. 1G). As expected, viral antigen was detected in the lungs of mice transduced with Ad5-hDPP4 but not control vector (Fig. 1D). After MERS-CoV infection, we identified perivascular and peribronchial lymphoid infiltration initially, with progression to an interstitial pneumonia at later times postinfection (p.i.) (Fig. 1H and Fig. S1).
Fig. 1.
Development of mice susceptible to MERS-CoV infection. To assess hDPP4 expression (A) and surface localization (B), MLE15 cells were transduced with Ad5-hDPP4 or Ad5-Empty at an MOI of 20 at 37 °C for 4 h. hDDP4 expression was monitored by Western blot assay (A) or flow cytometry (B). (C) Ad5-hDPP4–transduced cells were infected with MERS-CoV at an MOI of 1 at 48 h posttransduction, and virus titers were determined by plaque assay. Five days after transduction with 2.5 × 108 pfu of Ad5-hDPP4 or Ad5-Empty in 75 μL of DMEM intranasally, mice were intranasally infected with 1 × 105 pfu of MERS-CoV in 50 μL of DMEM. (D) Lungs were harvested from BALB/c mice at day 3 after MERS-CoV infection, fixed in zinc formalin, and embedded in paraffin. Sections were stained with an anti-hDPP4 or with an anti–MERS-CoV nucleocapsid antibody (blue signal). (Original magnification, 20×.) (E and F) Weight changes in 6- to 12-wk-old (young) and 18- to 22-mo-old (aged) B6 (E) and BALB/c (F) mice were monitored daily. For B6 mice, n = 8 in Ad5-Empty group; 12 in Ad5-hDPP4 group; 8 in Ad5-hDPP4 aged group. For BALB/c mice, n = 8 in Ad5-Empty group; 12 in Ad5-hDPP4 group; 8 in Ad5-hDPP4 aged group. To obtain virus titers, lungs were homogenized at the indicated time points and titered on Vero 81 cells. Titers are expressed as pfu/g tissue (n = 4–8 mice per group per time point). Data are representative of two independent experiments. Δ, P < 0.05 when Ad5-hDPP4 aged were compared with Ad5-hDPP4 and Ad4-Empty. (G) To evaluate the length of time that Ad5-hDPP4–transduced mice could be infected with MERS-CoV, Ad5-hDPP4–transduced mice were infected with MERS-CoV at the indicated times. Lungs were harvested for titers at 2 d.p.i. (n = 4 mice per group per time point. (H) Lungs from B6 mice were removed at the indicated time points p.i., fixed in zinc formalin, and embedded in paraffin. Sections were stained with hematoxylin/eosin.
Requirements for Type-I IFN Induction and Signaling in MERS-CoV Clearance.
There are roles for both the innate and adaptive immune responses for protection from coronavirus infection (17). Initially we showed that depletion of natural killer (NK) cells, a cellular component of the innate immune response, did not change clinical disease or the kinetics of virus clearance (Fig. S2). MERS-CoV does not induce significant amounts of IFNα/β expression in vitro (18–20), but the role of type-I IFN induction and signaling in vivo is unknown. IFN is induced via RIG-I–like receptors (RLRs) and Toll-like receptors (TLRs) in coronavirus infections (21). To determine the role of each in MERS-CoV–infected mice, Ad5-hDPP4–transduced mice impaired in RLR [mitochondrial antiviral signaling protein−/− (MAVS−/−)] or TLR [myeloid differentiation primary response gene 88−/− (MyD88−/−)] signaling were infected with MERS-CoV (Fig. 2A). Infection of MyD88−/− but not MAVS−/− mice resulted in up to 20% weight loss. Without type-I IFN signaling (IFNAR−/−), infection was even more severe than in MyD88−/− mice, with weight loss beginning 2 d earlier p.i. Virus was cleared with the same kinetics in MAVS−/− and WT mice, but clearance was delayed in both MyD88−/− and IFNAR−/− mice (Fig. 2B), suggesting that TLR-dependent and IFN signaling pathways were required for MERS-CoV control. Consistent with these results, we observed increased vascular congestion and inflammation on gross pathological lung specimens from infected MyD88−/− and IFNAR−/− mice (Fig. 2C). Histological examination of Ad5-hDPP4–transduced, MERS-CoV–infected IFNAR−/− compared with B6 mice revealed earlier onset of peribronchial, perivascular, and interstitial infiltrates, relative to B6 mice (Figs. 1H and 2D). Alveolar thickening and edema and increased infiltration of granulocytes, especially eosinophils, were observed only in infected IFNAR−/− lungs (Fig. 2E). To further address the role of IFN induction and signaling in MERS-CoV protection, we transduced B6 mice with Ad5-hDPP4 and, 5 d later, treated them with poly I:C, a TLR3 agonist that signals through the MyD88-dependent pathway, or with IFN-β or IFN-γ. We then infected mice 6 h later with MERS-CoV. The treatments, particularly poly I:C and IFN-β, accelerated virus clearance (Fig. 2F) without appreciably affecting weight or extent of inflammatory-cell infiltration (Fig. S3). Poly I:C delivered 6 h after infection also accelerated virus clearance, although to a lesser extent than if delivered before infection (Fig. S4). Of note, administration of IFN-α2b (with ribavirin) to MERS-CoV–infected macaques improved clinical, radiological, and virological parameters of infection (22).
Fig. 2.
Requirements for type-I IFN induction and signaling in MERS-CoV clearance. (A) Five days after transduction with 2.5 × 108 pfu of Ad5-hDPP4, mice were intranasally infected with 1 × 105 pfu of MERS-CoV. Weight changes were monitored daily (n = 8 in B6 group; 14 in IFNAR−/− group; and 13 in MyD88−/− group; n= 9 in MAVS−/− group). (B) Virus titers in the lungs were measured at the indicated time points. Titers are expressed as pfu/g tissue (n = 3–4 mice per group per time point). Data are representative of two independent experiments. *, P < 0.05 compared with B6 group; Δ, P values of <0.05 compared with IFNAR−/− group; #, P values of <0.05 compared with MyD88−/− group. (C) Photographs of gross pathological lung specimens isolated from infected mice at day 7 p.i. (D) Sections of paraffin-embedded lungs from Ad5-hDPP4–transduced, infected IFNAR−/− mice were stained with hematoxylin/eosin. (E) Edema (asterisks) and infiltrating eosinophils (arrows) are indicated. (F) Ad5-hDPP4-–transduced mice were treated with 20 μg of poly I:C, 2,000 units of IFN-β, 200 ng of IFN-γ, or PBS in 50 μL of DMEM 6 h before intranasal infection with 1 × 105 pfu of MERS-CoV. Viral titers in lungs were measured at the indicated time points (n = 4 mice per group per time point. Data are representative of three independent experiments. *, P < 0.05 compared with PBS group; Δ, P values of <0.05 compared with poly I:C group; #, P values of <0.05 compared with IFN-β group.
Requirements for CD8 T Cells and Antibodies for MERS-CoV Clearance and Protection from Subsequent Challenge.
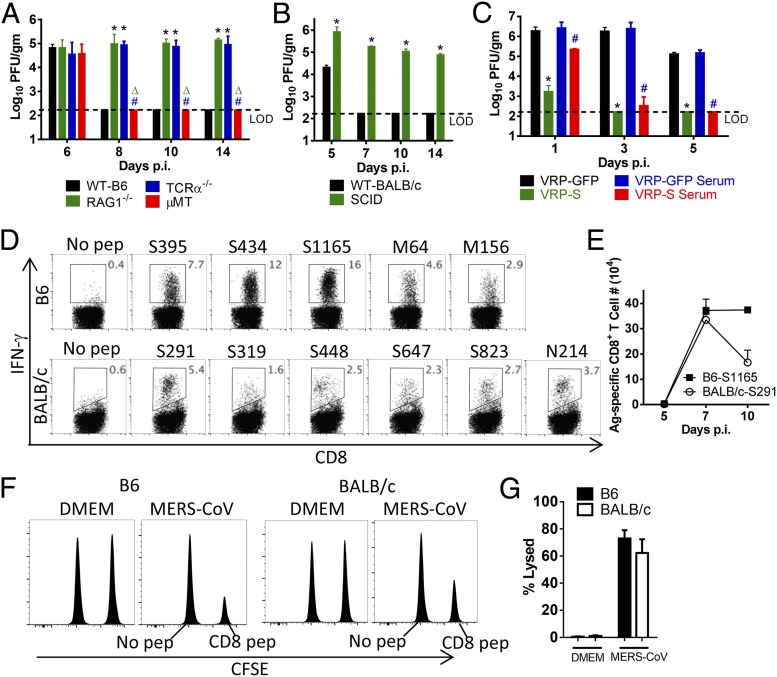
To examine the role of T- and B-cell responses in protection against MERS-CoV, we infected Ad5-hDPP4–transduced mice deficient in T cells [T-cell receptor α−/− (TCRα−/−)], B cells (μMT), or T and B cells [recombination activating gene 1−/− (RAG1−/−) severe combined immunodeficiency (SCID)] and their corresponding controls (Fig. 3 A and B). Virus was not cleared in mice lacking T cells (TCRα−/−, RAG1−/−), or in SCID mice but was cleared in μMT mice. Despite the persistent infections, none of these mice lost weight (Fig. S5). Although T cells are important for acute virus clearance, protection against subsequent challenge is generally antibody-mediated. To generate a protective antibody response, we engineered Venezuelan equine encephalitis replicon particles (VRPs) expressing the MERS-CoV spike protein as previously described (VRP-S) (6). Immunization with VRP-S using a prime-boost regimen reduced MERS-CoV titers to nearly undetectable levels by day 1 p.i. Anti-S antibody, which blocked virus attachment, was largely sufficient for this effect because transfer of sera from VRP-S–immunized mice also accelerated the kinetics of virus clearance (Fig. 3C). To analyze CD8 T-cell activity in vivo, we identified several epitopes using peptides selected for consensus binding to the MHC class I antigen (Table S1) and used them in intracellular IFN-γ–staining assays (Fig. 3D). The immunodominant epitopes recognized in both B6 and BALB/c mice were located in the S protein. The CD8 T-cell response to these epitopes in Ad5-hDPP4–transduced B6 and BALB/c mice peaked at days 7–10 p.i. (Fig. 3E). Specific killing was confirmed in vivo because target cells coated with virus-specific CD8 T-cell peptides were efficiently lysed (Fig. 3 F and G).
Fig. 3.
Requirements for CD8 T cells and antibodies for MERS-CoV clearance and protection from subsequent challenge. (A and B) Ad5-hDPP4–transduced mice were infected with 1 × 105 pfu of MERS-CoV. Virus titers in the lungs were measured at the indicated time points. Titers are expressed as pfu/g tissue (n = 3–4 mice per group per time point. Data are representative of two independent experiments. *, P values of <0.05 compared with WT group; Δ, P values of <0.05 compared with RAG1−/− group; #, P values of <0.05 compared with TCRα−/− group. (C) BALB/c mice were immunized with 1 × 105 infectious units (IU) of VRP-GFP or VRP-S in the footpad in 20 μL of PBS and boosted with the same dose 4 wk later. Mice were transduced and infected with 1 × 105 pfu of MERS-CoV 2–4 wk after the booster. For adoptive transfer of serum, sera were obtained 2–4 wk after booster. Then, 300 μL of serum was transferred into transduced mice intraperitoneally 1 d before MERS-CoV infection. *, P values of <0.05 compared with VRP-GFP group; #, P values of <0.05 compared with VRP-GFP serum group. (D) To identify MERS-CoV–specific CD8 T-cell epitopes, single-cell suspensions were prepared from the lungs of transduced/infected mice and stimulated with 1 µM peptides for 5–6 h in the presence of brefeldin A. Frequencies of MERS-CoV–specific T cells (determined by IFN-γ intracellular staining) are shown. Kinetics of immune responses to dominant CD8 T-cell epitopes in Ad5-hDPP4–transduced B6 and BALB/c mice are summarized in E. Data are representative of five independent experiments. (F and G) In vivo cytotoxicity assays were performed on day 7 (BALB/c mice; peptides S395, S434, and S1165 combined) or day 8 (B6 mice; peptides S291, S823, and N214 combined) p.i. as described in Materials and Methods (F) and summarized (G) (n = 4 mice per group). Data are representative of two independent experiments.
Low Level of Cross-Reactivity Between MERS-CoV and SARS-CoV.
A critical question is whether SARS-CoV and MERS-CoV, which both likely originate from bat sources (23, 24), elicit cross-reactive, protective immune responses. To address this question, we infected young BALB/c mice (after Ad5-hDPP4 transduction) with either MERS-CoV (Fig. 4 A–C) or a sublethal dose of SARS-CoV (Fig. 4 D–F). BALB/c mice were used in these experiments because young B6 mice are resistant to SARS-CoV (25). After 5 wk, mice were challenged with SARS-CoV or (after Ad5-hDPP4 transduction) with MERS-CoV. Mice initially infected with MERS-CoV were as fully susceptible to subsequent challenge with SARS-CoV as DMEM-treated mice (Fig. 4 A–C). Regardless of the MERS-CoV immunization regimen (no Ad5, Ad5-empty, or Ad5-hDPP4), mortality and weight loss were equivalent (Fig. 4 A and B and Fig. S6 A and B). The reciprocal initial infection with SARS-CoV resulted in a statistically significant but minor decrease in MERS-CoV titers at day 5 p.i. after challenge (Fig. 4F). This decrease in titer may reflect a cross-reactive T-cell response because no differences were detected at early times p.i. Again, the immunization regimen (no Ad5 or Ad5-empty before SARS-CoV infection) did not effect any weight changes after MERS-CoV challenge (Fig. S6 C and D). As controls, mice were treated before infection with serum obtained from SARS-CoV– or MERS-CoV–infected mice, which conferred protection (Fig. 4 C and F).
Fig. 4.
Low level of cross-reactivity between MERS-CoV and SARS-CoV. (A–C) Ad5-hDPP4–transduced BALB/c mice were inoculated with 1 × 105 pfu of MERS-CoV or DMEM and rested for 5 wk before infection with 1 × 104 pfu of SARS-CoV in 50 μL of DMEM. Control mice received 300 μL of immune serum from mice previously (5 wk) infected with a sublethal dose of SARS-CoV (500 pfu). Mortality (A) and weight (B) were monitored daily (n = 12 in all groups). (C) To obtain virus titers, lungs were homogenized at the indicated time points and titered on Vero E6 cells. Titers are expressed as pfu/g tissue (n = 4 mice per group per time point). Data are representative of two independent experiments. *, P values of <0.05 compared with DMEM group; Δ, P values of <0.05 compared with MERS-CoV–immunized group. (D–F) BALB/c mice were infected with 500 pfu of SARS-CoV or DMEM and rested for 5 wk before Ad5-hDPP4 transduction and infection with 1 × 105 pfu of MERS-CoV. Control mice received 300 μL of immune serum from mice previously (5 wk) infected with 1 × 105 pfu of MERS-CoV. Mortality (D) and weight (E) were monitored daily (n = 12 in all groups). (F) Virus titers in the lungs were measured at the indicated time points (n = 4 mice per group per time point). Data are representative of two independent experiments. *, P values of <0.05 compared with DMEM group; Δ, P values of <0.05 compared with SARS-CoV–immunized group.
Discussion
Here, we developed a novel platform strategy for sensitizing mice to MERS-CoV infection. We demonstrated that both innate, antibody, and T-cell responses are important for protection from MERS-CoV. Similar to infected patients, Ad5-hDPP4–transduced mice with normal immune systems developed mild disease whereas immunocompromised mice, like patients with underlying diseases, were more profoundly affected. MERS-CoV–infected mice were successfully used to evaluate an antiviral drug and a vaccine. Of note, poly I:C is inexpensive and has been approved for use in humans (26, 27). VRP-S induced a protective immune response (Fig. 3C); VRPs are excellent subunit vaccine candidates (28).
Although there are a few limitations of the Ad-hDPP4 transduction system (level of expression, tissue distribution), development of adenovirus vectors expressing the MERS-CoV receptor was efficient and rapid, resulting in the generation of an easily reproducible murine model for MERS-CoV within 2–3 wk. This short time course compares favorably with the much longer time (several months to years) required to develop transgenic or knock-in mice expressing the human receptor. Further, hDPP4 expression after AD5-hDDP4 intranasal inoculation is restricted to the lungs whereas transgenic expression may not be targeted to the correct organ (29, 30). Although hDPP4 expression may be more physiological in knock-in mice, the low level of native DPP4 expression in the mouse lung presents limitations (8). Another advantage of the Ad5-hDPP4 transduction strategy is that it can be used in genetically deficient mice, facilitating rapid identification of host genes and pathways that play protective or pathogenic roles in disease. Because Ad vectors are polytropic, this approach will have broad utility in rendering animals from multiple species susceptible to infection with emerging respiratory viruses, where speed of development is often critical to enable drug screening and vaccine validation.
Materials and Methods
Mice, Virus, and Cells.
Specific pathogen-free 6- to 12-wk-old and 18- to 22-mo-old C57BL/6 and BALB/c mice were purchased from the National Cancer Institute. RAG1−/−, μMT, TCR−/−, and IFNAR−/− mice were purchased from The Jackson Laboratory. MyD88−/− mice were obtained from Dr. S. Akira (Osaka University, Osaka, Japan) (31). MAVS−/− mice were obtained from Dr. S. Akira and developed as previously described (32). Mice were maintained in the animal care facility at the University of Iowa. All protocols were approved by the University of Iowa Institutional Animal Care and Use Committee. The EMC2012 strain of MERS-CoV (passage 8, designated MERS-CoV) was provided by Drs. Bart Haagmans and Ron Fouchier (Erasmus Medical Center). Mouse-adapted SARS-CoV (MA15) was a kind gift from Dr. Kanta Subbarao (National Institutes of Health, Bethesda, MD) (33). A human DPP4 ORF clone was purchase from Origene. Recombinant adenoviral vectors expressing-hDPP4 (Ad5-hDPP4) were prepared as previously described (15) by the University of Iowa Gene Transfer Vector Core at titers of 1010 ∼1011 pfu/ml in A195 buffer (29.5 mM histidine, 54 mM Tris⋅HCl, 10.8 mM MgCl2, 108 μM EDTA, 0.0216% Tween 80, 0.54% ethanol, 5% sucrose in PBS). African Green monkey kidney-derived Vero E6 cells and Vero 81 cells (CCL81; ATCC) were grown in DMEM (GIBCO) supplemented with 10% (vol/vol) FBS. MERS-CoV and SARS-CoV were passaged once on Vero 81 or Vero E6 cells, respectively, and titered on the same cell line, as described (34). MLE15 cells, a mouse type-II pneumocyte cell line, were cultured in 2% FBS RPMI 1640 supplemented with 5 μg/mL insulin, 5 μg/mL selenium, 10 μg/mL transferrin, 10 nM hydrocortisone, and 10 nM β-estradiol, all from Sigma and 2% FBS (35).
Chemicals, Cytokines, and Peptides.
Poly I:C (Sigma), IFN-γ (R&D Systems), and IFN-β (PBL) were purchased. MERS-CoV–specific peptides, predicted using online programs (RANKpep, SYFPEITHI, NetMHC 3.4) (36–38), were synthesized by BioSynthesis Inc.
Transduction and Infection of MLE15 Cells and Western Blot Analysis.
MLE15 cells were transduced with Ad5-hDPP4 or Ad5-Empty at multiplicity of infection (MOI) = 20 for 4 h at 37 °C. Extracts were prepared 48 h posttransduction. Identical amounts of protein were separated on a 4–20% SDS/PAGE gel and transferred to PVDF membranes. Membranes were stained with a mouse anti-human DPP4 antibody (clone 11D7; Origene), a rabbit anti-FLAG antibody (Sigma), or a mouse anti–α-tubulin (clone DM1A; Sigma). Proteins were detected using a SuperSignal West Pico Trial Kit (Thermo Scientific). For infection, MLE15 cells were transduced with AD5-hDPP4 or Ad5-Empty for 48 h before infection with MERS-CoV (MOI = 1) at 37 °C for 1 h. Supernatants were collected at the indicated time points and analyzed for infectious virus by plaque assay.
Transduction and Infection of Mice.
Mice were lightly anesthetized with isoflurane and transduced intranasally with 2.5 × 108 pfu of Ad5-hDPP4 or Ad5-Empty in 75 μL of DMEM. Five days posttransduction or at the indicated time points, mice were infected intranasally with MERS-CoV (1 × 105 pfu), or SARS-CoV (500 pfu, sublethal, or 1 × 104 pfu, 1 LD50 dose) in a total volume of 50 μL of DMEM. Mice were monitored daily for morbidity and mortality. All work with MERS-CoV and SARS-CoV was conducted in the University of Iowa Biosafety Level 3 (BSL3) Laboratory.
Virus Titers.
To obtain virus titers, lungs were removed into PBS and homogenized using a manual homogenizer. Virus was titered on Vero 81 cells or Vero E6 cells. Cells were fixed with 10% formaldehyde and stained with crystal violet 3 d.p.i.. Viral titers are expressed as pfu/g tissue for MERS-CoV and SARS-CoV.
Histology and Immunohistochemistry.
Animals were anesthetized and transcardially perfused with PBS followed by zinc formalin. Lungs were removed, fixed in zinc formalin, and paraffin-embedded. Sections were stained with hematoxylin/eosin for histological analysis. A MERS-CoV–specific antibody was developed by immunizing rabbits with a peptide encompassing residues 244–257 of the N protein (AAAKNKMRHKRTST) per company protocol (BioGenes). For hDPP4 staining, high-pH microwave antigen retrieval was performed on sections using Antigen Unmasking Solution (Vector Laboratories) and a Mouse-on-Mouse (M.O.M.) kit (BMK-2202; Vector Labs) to reduce endogenous mouse IgG staining. Sections were then blocked using goat serum (MERS-CoV) or M.O.M.-blocking protein (DDP4) and incubated overnight with rabbit anti-hDPP4 antibody (TA500733, 1:800; Origene) or rabbit anti-N antibody (1:600). Sections were incubated with a biotinylated goat anti-rabbit secondary antibody (Vector Laboratories). Binding of the secondary antibody was detected with an alkaline phosphatase–biotin–avidin mixture. Sections were then visualized with Vector Blue (Vector Laboratories) and counterstained with Nuclear Fast Red.
Preparation of Cells from Lungs.
Mice were killed at the indicated time points. Lungs were removed, cut into small pieces, and digested in HBSS buffer containing 2% FCS, 25 mM Hepes, 1 mg/mL Collagenase D (Roche), and 0.1 mg/mL DNase (Roche) for 30 min at room temperature. Tissues were dispersed using a 70-μM cell strainer, and single-cell suspensions were prepared. Live cells were enumerated by 0.2% trypan blue exclusion.
In Vivo Cytotoxicity Assay.
In vivo cytotoxicity assays were performed on day 7–8 after MERS-CoV infection, as previously described (34, 39). Briefly, splenocytes from CD45.1 congenic naive mice were stained with either 2 μM or 100 nM carboxyfluorescein succinimidyl ester (CFSE) (Molecular Probes) and then pulsed with the indicated peptides (3 μM each) at 37 °C for 1 h. Then, 5 × 105 cells from each group were mixed together (1 × 106 cells in total) and transferred intranasally into mice. At 12 h after transfer, total lung cells were isolated. Target cells were identified on the basis of CD45.1 staining and were distinguished from each other by differential CFSE staining. After gating on CD45.1+ cells, the percentage lysis was calculated as previously described (34).
Flow Cytometry.
The following monoclonal antibodies were used: rat anti-mouse CD8α (53-6.7), rat anti-mouse CD45.1 (A20), rat anti-mouse NKG2D (CX5), all from BD Bioscience; rat anti-mouse IFN-γ (XMG1.2) from eBioscience; and mouse anti-hDPP4 (BA5b) from Biolegend. For surface staining, 106 cells were blocked with 1 μg of anti-CD16/32 antibody and 1% rat serum and stained with the indicated antibodies at 4 °C. For intracellular cytokine staining (ICS), 1 × 106 cells per well were cultured in 96-well dishes at 37 °C for 5–6 h in the presence of 1 μM peptide and brefeldin A (BD Biosciences). Cells were then labeled for cell-surface markers, fixed/permeabilized with Cytofix/Cytoperm Solution (BD Biosciences), and labeled with anti–IFN-γ antibody. All flow-cytometry data were acquired on a BD FACSCalibur or BD FACSVerse and were analyzed using FlowJo software (Tree Star, Inc.).
Venezuelan Equine Encephalitis Replicon Particles.
Venezuelan equine encephalitis replicon particles (VRPs) expressing the MERS-CoV spike glycoprotein were constructed as previously described (6, 40). Briefly, a VRP construct expressing MERS-CoV S glycoprotein was generated using overlap PCR by fusing an amplicon containing the S gene in frame with an amplicon containing sequences from the Venezuelan equine encephalitis (VEE) replicon. The primers for VEE replicon have been described previously (1), and the primers used for generating the S gene amplicon are available upon request. Ligated DNA was digested with ApaI and PacI and inserted into the pVR21 plasmid. VRPs were packaged using helper RNAs encoding structural proteins, as described before (2). A hemagglutinin (HA) tag was added to the C terminus of S protein for titering in BHK21 cells as described previously (41, 42).
Statistical Analysis.
A Student t test was used to analyze differences in mean values between groups. All results are expressed as means ± SEs of the means (SEM). P values of <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Bart Haagmans and Ron Fouchier (Erasmus Medical Center) for providing MERS-CoV (isolate HCoV-EMC/2012) and Dr. David Meyerholz for analysis of lung sections. We thank Beverly Davidson for helpful discussions. We acknowledge the excellent technical support of the University of Iowa Gene Transfer Vector Core. This research was supported in part by National Institutes of Health Grants RO1AI091322 (to S.P.), PO106099 (to S.P., P.B.M., T.G., and L.E.), AI074973 and AI083019 (to M.J.G.), and AI057157 (to R.S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323279111/-/DCSupplemental.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10(Suppl 12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assiri A, et al. KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assiri A, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. Lancet Infect Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Engl J Med. 2013;368(16):1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scobey T, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E, et al. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS ONE. 2013;8(7):e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman CM, Matthews KL, Goicochea L, Frieman MB. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol. 2014;95(Pt 2):408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietzsch J, et al. A mouse model for HIV-1 entry. Proc Natl Acad Sci USA. 2012;109(39):15859–15864. doi: 10.1073/pnas.1213409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474(7350):208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crystal RG, et al. Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis. Nat Genet. 1994;8(1):42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- 13.Knowles MR, et al. A controlled study of adenoviral-vector-mediated gene transfer in the nasal epithelium of patients with cystic fibrosis. N Engl J Med. 1995;333(13):823–831. doi: 10.1056/NEJM199509283331302. [DOI] [PubMed] [Google Scholar]
- 14.Nabel GJ. Genetic, cellular and immune approaches to disease therapy: past and future. Nat Med. 2004;10(2):135–141. doi: 10.1038/nm990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson RD, Haskell RE, Xia H, Roessler BJ, Davidson BL. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7(12):1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, Muruve DA. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 2003;10(11):935–940. doi: 10.1038/sj.gt.3302036. [DOI] [PubMed] [Google Scholar]
- 17.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kindler E, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4(1):e00611–e00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zielecki F, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87(9):5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan RW, et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87(12):6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Totura AL, Baric RS. SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falzarano D, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19(10):1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 24.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):e00473. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J, Zhao J, Legge K, Perlman S. Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121(12):4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada H, et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with alpha-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J Clin Oncol. 2011;29(3):330–336. doi: 10.1200/JCO.2010.30.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butowski N, et al. A phase II clinical trial of poly-ICLC with radiation for adult patients with newly diagnosed supratentorial glioblastoma: A North American Brain Tumor Consortium (NABTC01-05) J Neurooncol. 2009;91(2):175–182. doi: 10.1007/s11060-008-9693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis NL, et al. Alphavirus replicon particles as candidate HIV vaccines. IUBMB Life. 2002;53(4-5):209–211. doi: 10.1080/15216540212657. [DOI] [PubMed] [Google Scholar]
- 29.McCray PB, Jr, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81(2):813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng CT, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81(3):1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adachi O, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9(1):143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 32.Suthar MS, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6(2):e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Zhao J, Van Rooijen N, Perlman S. Evasion by stealth: Inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5(10):e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wikenheiser KA, et al. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90(23):11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundegaard C, et al. NetMHC-3.0: Accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(Web Server issue):W509-12. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reche PA, Glutting JP, Zhang H, Reinherz EL. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56(6):405–419. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 38.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 39.Barber DL, Wherry EJ, Ahmed R. Cutting edge: Rapid in vivo killing by memory CD8 T cells. J Immunol. 2003;171(1):27–31. doi: 10.4049/jimmunol.171.1.27. [DOI] [PubMed] [Google Scholar]
- 40.Deming D, et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3(12):e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pushko P, et al. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: Expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239(2):389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 42.Heise MT, Simpson DA, Johnston RE. A single amino acid change in nsP1 attenuates neurovirulence of the Sindbis-group alphavirus S.A.AR86. J Virol. 2000;74(9):4207–4213. doi: 10.1128/jvi.74.9.4207-4213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.