Significance
Investigations of complex environments rely on large volumes of sequence data to adequately sample the genetic diversity of a microbial community. The assembly of short-read data into longer, more interpretable sequence currently is not possible for much of the research community because it requires specialized computational facilities. We present approaches that make de novo assembly of complex metagenomes more accessible. These approaches scale data size with community richness and subdivide the data into tractable subsets representing individual species. We applied these methods toward the assembly of two large soil metagenomes to identify important metagenomic references and show that considerably more data are needed to study the terrestrial microbiome comprehensively.
Abstract
The large volumes of sequencing data required to sample deeply the microbial communities of complex environments pose new challenges to sequence analysis. De novo metagenomic assembly effectively reduces the total amount of data to be analyzed but requires substantial computational resources. We combine two preassembly filtering approaches—digital normalization and partitioning—to generate previously intractable large metagenome assemblies. Using a human-gut mock community dataset, we demonstrate that these methods result in assemblies nearly identical to assemblies from unprocessed data. We then assemble two large soil metagenomes totaling 398 billion bp (equivalent to 88,000 Escherichia coli genomes) from matched Iowa corn and native prairie soils. The resulting assembled contigs could be used to identify molecular interactions and reaction networks of known metabolic pathways using the Kyoto Encyclopedia of Genes and Genomes Orthology database. Nonetheless, more than 60% of predicted proteins in assemblies could not be annotated against known databases. Many of these unknown proteins were abundant in both corn and prairie soils, highlighting the benefits of assembly for the discovery and characterization of novelty in soil biodiversity. Moreover, 80% of the sequencing data could not be assembled because of low coverage, suggesting that considerably more sequencing data are needed to characterize the functional content of soil.
Complex microbial communities operate at the heart of many crucial terrestrial, aquatic, and host-associated processes, providing critical ecosystem functionality that underpins much of biology (1–7). DNA sequencing has begun to reveal the enormous biological diversity and heterogeneity within these systems, making them difficult to study in situ (2, 4, 5). With ultradeep sequencing, we now have unprecedented access to even the rare species in these environments. However, in complex environments such as soil [where an estimated 50 Tbp is required to sample a gram adequately (8)], converting these large volumes of sequencing data to biologically useful information remains a major challenge.
As the sizes of sequencing datasets grow at an exponential rate, significant computational resources for data storage and analysis are required. A single metagenomic project can readily generate as much or more data than is in global reference databases; for example, a human-gut metagenome sample containing 578 Gbp [ERA000116 (5)], produced more than twice the data in the National Center for Biotechnology Information (NCBI) RefSeq (Release 56) database. In its simplest form, these data (millions to billions of short reads) are error prone and contain only minimal signal for homology searches, limiting direct annotation approaches against reference databases (9). Furthermore, in systems where little of the microbial diversity has been characterized, these annotation approaches are challenged by a lack of reference genomes, and more than half of identified genes share little or no similarity to any experimentally studied genes (1, 5).
Consequently, investigators of environmental metagenomic datasets are confronted by overwhelming volumes of data for which they have neither the computational resources nor effective bioinformatics tools (because of short read lengths or a lack of reference genomes) to analyze efficiently. De novo assembly of sequence data offers several advantages for analyzing metagenomic datasets. It provides improved accuracy of sequences by removing most random sequencing errors and results in longer and more specific contigs than found in unassembled sequencing reads (10). Furthermore, assembly significantly reduces the total volume of data required for downstream analysis (e.g., gene annotation). Also, de novo assembly does not rely on the existence of reference genomes, thus allowing the discovery of novel genomic elements. The main challenge for metagenomic applications of de novo assembly is that current assembly tools do not scale to the high diversity and large volume of metagenomic data. Metagenomes from rumen, human gut, and permafrost soil sequencing could be assembled only by discarding low-abundance sequences before assembly (2, 4, 5). Although many metagenome-specific assemblers have been developed recently for the assembly of low-complexity communities, they cannot work with the volume of reads necessary to achieve high coverage for extremely diverse environmental metagenomes (10–12).
Here, we apply two preassembly read-filtering strategies, digital normalization and partitioning, that together provide a general strategy for scaling and improving metagenome assembly for large, complex datasets (e.g., billions of reads). Digital normalization reduces the size of the dataset by setting aside reads from high-coverage regions and results in more uniform sequence coverage overall; digital normalization has been used previously for both genome and mRNA-seq assembly (13). We apply digital normalization to metagenomes to scale assembly by sample richness rather than by diversity. Further, we demonstrate that digital normalization combined with a partitioning approach to separate reads based on transitive connectivity (e.g., grouping reads with sequencing overlap) (14) can be applied to complex metagenomes. Because these approaches have yet to be applied to environmental metagenomes for which the true content is unknown, we first evaluated this strategy on a human-gut mock community (HGMC) dataset containing 21 known genomes and found that these methods result in assemblies that are nearly identical to assemblies from the unprocessed HGMC dataset. Moreover, we show that partitioning separates most reads into species-level bins, providing an alternative to abundance-based and k-mer approaches to species clustering. We next used these approaches to assemble two previously intractable metagenomes from matched soils, 100-y-cultivated Iowa agricultural corn soil and native Iowa prairie soil. We use the resulting assemblies to evaluate our ability both to sample and to characterize small, 3- to 6-g soil samples and their associated functional diversity. Even with 300 Gbp of data, we are unable to achieve deep coverage of the majority of organisms in the sample, highlighting the need for more extensive sequencing.
Results
Normalization Results in Similar Assemblies with Minimal Loss of Information.
The HGMC dataset contains sequences from mixed DNA from isolates at varying abundances ranging from fourfold to 2,000-fold sequencing coverage using the Illumina sequencing platform (Table S1). We evaluated our ability to describe the original HGMC genomes and to estimate the abundances of these genomes from our filtered assembly as compared with the unfiltered, original assembly (Fig. 1, Assembly I and Assembly II).
Fig. 1.
Summary of approaches for large-scale assembly of complex metagenomes presented in this study. Unprocessed (I), normalized (II), and partitioned assemblies (III) were evaluated and compared with the HGMC metagenome. These approaches were used toward the assembly of metagenomes.
After sequencing, the mock metagenome (unassembled) encompassed a total of 93% of the genomic content of the reference genomes (Fig. S1). After digital normalization, reads were removed based on their coverage within the dataset (Materials and Methods), resulting in a total of 5.9 million reads (40% of the total reads) from the original HGMC dataset (Table 1) with coverage of 91% of the reference genomes (recovery per genome in Fig. S1). The resulting assembly of filtered HGMC reads (normalized) was compared with the assembly of all original reads, evaluating the recovery of reference genomes and the length distribution of assembled contigs for each reference. Using the Velvet assembler (15), we recovered 43% and 44% of the reference genomes in the original and filtered assemblies, respectively. The assembly of the original dataset contained 29,063 contigs and 38 million bp; the filtered assembly contained 30,082 contigs and 35 million bp (Table 3). Comparable recoveries of references between original and filtered datasets also were obtained with other assemblers [SOAPdenovo (16) and Meta-IDBA (17)]. Overall, the unfiltered and filtered assemblies were very similar, sharing 95% of genomic content (Table S2), and the distributions of contig lengths in unfiltered and filtered assemblies also were comparable. For the large majority of genomes, the filtered assembly recovered similar fractions of each reference. In genomes with lower coverage, such as NC_003112.2 and NC_006085.1, improved assemblies from normalization were observed. In genomes with high sequencing coverage, such as the plasmids NC_005008.1, NC_005007.1, and NC_005003.1, the unfiltered assembly recovered significantly more of the original sequence (Table S1).
Table 1.
Total number of reads in unfiltered, normalized, and partitioned datasets
| Dataset | Unfiltered reads (Mbp) | Normalized reads (Mbp) | Partitioned reads (Mbp) |
| HGMC | 14,494,884 (1,136) | 8,656,520 (636) | 8,560,124 (631) |
| HGMC spike | 14,992,845 (1,137) | 8,189,928 (612) | 8,094,475 (607) |
| HGMC multispike | 17,010,607 (1,339) | 9,037,142 (702) | 8,930,840 (697) |
| Iowa corn | 1,810,630,781 (140,750) | 1,406,361,241 (91,043) | 1,040,396,940 (77,603) |
| Iowa prairie | 3,303,375,485 (256,610) | 2,241,951,533 (144,962) | 1,696,187,797 (125,105) |
Table 3.
Assembly summary statistics for unfiltered, normalized, and normalized + partitioned datasets
| Dataset | No. contigs | Unfiltered length (Mbp) | Maximum contig (bp) | No. contigs | Normalized filtered length (Mbp) | Maximum contig (bp) | No. contigs | Partitioned length (Mbp) | Maximum contig (bp) | Assembler |
| HGMC | 29,063 | 38 | 146,795 | 30,082 | 35 | 90,497 | 30,115 | 35 | 90,497 | V |
| HGMC | 24,300 | 36 | 86,445 | — | — | — | 27,475 | 36 | 96,041 | M |
| HGMC | 36,689 | 37 | 32,736 | — | — | — | 29,295 | 37 | 58,598 | S |
| Iowa corn | — | — | — | — | — | — | 1,862,962 | 912 | 20,234 | V |
| Iowa corn | — | — | — | — | — | — | 1,334,841 | 623 | 15,013 | M |
| Iowa corn | — | — | — | — | — | — | 1,542,436 | 675 | 15,075 | S |
| Iowa prairie | — | — | — | — | — | — | 3,120,263 | 1,510 | 9,397 | V |
| Iowa prairie | — | — | — | — | — | — | 2,102,163 | 998 | 7,206 | M |
| Iowa prairie | — | — | — | — | — | — | 2,599,767 | 1,145 | 5,423 | S |
M, MetaIDBA assembler; S, SOAPdenovo assembler; V, Velvet assembler. Assemblies of Iowa corn and prairie metagenomes could not be completed on unfiltered or normalized-only datasets.
To understand the representation of genes and genomes in the metagenome, we evaluated our ability to estimate genome abundance in the HGMC metagenome and both unfiltered and filtered assemblies. Abundance was estimated through the alignment of unassembled reads to both the known reference genomes or assembled contigs (experimentally hypothesized references). Sequencing coverage was determined as the median base-pair coverage of all aligned reads. For assembled contigs with a coverage greater than 5, the majority of reads that could be aligned to contigs also were mapped to reference genomes (Fig. S2). Below this threshold, reads were mapped to reference genomes but were less likely to be associated with assembled contigs. When the unfiltered and filtered assemblies were compared, the estimated abundance of the HGMC genomes from the filtered assembly were significantly closer to abundances predicted from reference genomes (n = 28,652; P = 0.032; see SI Materials and Methods).
Partitioning Separates Most Reads by Species.
To subdivide large metagenomic datasets, we next partitioned the normalized dataset based on De Bruijn graph connectivity. This approach should separate disconnected sequences from distinct species and allow the assembly of each partition independently (Fig. 1, Assembly III). Notably, conserved regions shared by multiple genomes (e.g., 16S rRNA genes) may be connected within a single partition; we examined these partitions through the HGMC dataset. Overall, it was partitioned into 85,818 disconnected partitions containing a total of 9 million reads. Among these, only 2,359 (2.7%) of the partitions contained reads originating from more than one genome, indicating that partitioning separated the large majority of reads from distinct genomes. Partitioning had minimal effects on the assembly of a mock metagenome; the HGMC assemblies of the unpartitioned and partitioned dataset were very similar, sharing 99% identical genomic content.
The number of partitions in the mock metagenome depends largely on the sequencing coverage of its content. In general, reference genomes with high sequence coverage were associated with fewer partitions; a total of 112 partitions contained reads from high-abundance reference genomes (coverage above 25), whereas 2,771 partitions were associated with lower-abundance genomes (coverage below 25). This result is consistent with previous observations that low coverage in sequences causes “breaks” in connectivity within the assembly graph (18, 19).
To evaluate partitioning and its separation of species further, we introduced spiked, simulated reads from several Escherichia coli genomes into the HGMC dataset. First, simulated reads from a single genome (E. coli strain E24377A, NC_009801.1 with a 2% substitution error and 10× coverage) were added to the HGMC dataset, and the resulting dataset, HGMC.Ecoli1, was normalized by coverage, partitioned, and assembled. Similar amounts of data reduction were observed after digital normalization and partitioning (Table 1). Among the resulting 81,154 partitioned sets of reads in the HGMC.Ecoli1 dataset, only 2,580 partitions (3.2%) contained reads from multiple genomes. In total, 424 partitions contained reads from the spiked E. coli genome (201 partitions contained only spiked reads), and, when assembled, the contigs aligned to 99.5% of the E. coli strain E24377A genome (4,957,067 of 4,979,619 bp).
Next, we introduced five closely related E. coli strains [97.3–98.7% average nucleotide identity (20)] into the original HGMC dataset. This dataset, referred to as HGMC.Ecoli5, was normalized, partitioned, and assembled, resulting in 81,425 partitions. Among these, 1,154 partitions (1.4%) contained reads associated with multiple genomes. Among the partitions that contained reads associated with a single genome, 658 partitions contained reads originating from one of the spiked E. coli strains. In partitions containing reads from more than one genome, 224 partitions contained reads from a spiked E. coli strain and one other reference genome (either from another spiked strain or from the HGMC dataset). Independently assembling the partitions containing reads originating from the spiked E. coli strains resulted in 6,076 contigs, all but three originating from a spiked E. coli genome. The remaining three contigs were more than 99% similar to HGMC reference genomes (NC_000915.1, NC_003112.2, and NC_009614.1). The contigs associated with the five E. coli strains aligned to more than 98% of each of the five genomes. Many of these contigs contained similarities to reads originating from multiple genomes found in the HGMC, and more than half of the contigs (3,075) could be aligned to reads that originated from more than one spiked genome.
For comparison, the HGMC.Ecoli5 dataset also was assembled without using any filtering approaches (e.g., no digital normalization or partitioning). In comparing the unfiltered and filtered HGMC.Ecoli5 assemblies, we found that the fractions of contigs associated with multiple genomes were similar. The assembly of the unfiltered dataset resulted in a greater proportion of contigs (66% or 4,702 contigs vs. 51% or 3,075 normalized/partitioned contigs) associated with multiple genomes.
Assembly of Two Soil Metagenomes.
We next applied digital normalization and partitioning approaches to the de novo assembly of two soil metagenomes. Unfiltered Iowa corn and prairie datasets (containing 1.8 billion and 3.3 billion reads, respectively) could not be assembled by Velvet in 500 GB of RAM. A 75-million-reads subset of the Iowa corn dataset alone required 110 GB of memory, suggesting that assembly of the 3.3-billion-read dataset might need as much as 4 TB of RAM. Applying both normalization and partitioning approaches reduced the Iowa corn and prairie datasets to 1.4 billion and 2.2 billion reads, respectively, and after partitioning a total of 1.0 billion and 1.7 billion reads remained, respectively. These prefiltering approaches required 300 GB of RAM or less (Table 2). Notably, the large majority of k-mers in the soil metagenomes are relatively low abundance (Fig. 2), and consequently digital normalization did not remove as many reads in the soil metagenomes as in the mock dataset (Table 1).
Table 2.
Computational resources (memory and time) required
| Filter I: normalization, GB(h) | Filter II: partitioning, Gb(h) | |
| HGMC | 4(<2) | 4(<2) |
| HGMC spike | 4(<2) | 4(<2) |
| HGMC multispike | 4(<2) | 4(<2) |
| Iowa corn | 188(83) | 234(120) |
| Iowa prairie | 258(178) | 287(310) |
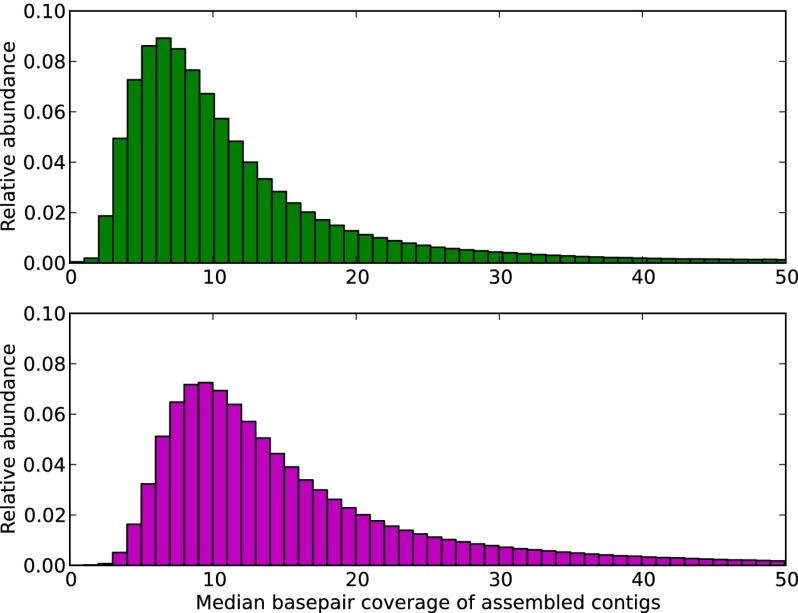
Fig. 2.
Coverage (median base pair recovered) distribution of assembled contigs from the Iowa corn soil (Upper) and Iowa prairie soil (Lower) metagenomes.
Based on the HGMC dataset, we estimated that above a sequencing depth of five, the large majority of sequences that could be aligned to reference genomes are also assembled into contigs greater than or equal to 300 bp (Fig. S2). Given the greater diversity expected in the soil metagenomes, we normalized these datasets to a sequencing depth of 10 (i.e., setting aside redundant reads within dataset above this coverage). After partitioning the filtered datasets, we identified a total 31,537,798 and 55,993,006 partitions (containing more than five reads) in the corn and prairie datasets, respectively. For assembly, we grouped partitions together into files containing a minimum of 10 million reads. Data reduction and partitioning were completed in less than 300 GB of RAM; once partitioned, each group of reads could be assembled in less than 14 GB and 4 h, readily enabling the evaluation of multiple assemblers and assembly parameters with practical computational resources.
The final assembly of the corn and prairie soil metagenomes resulted in a total of 1.9 million and 3.1 million contigs (minimum length of 300 bp), respectively, and a total assembly length of 912 million bp and 1.5 billion bp, respectively. To estimate abundance of assembled contigs and evaluate incorporation of reads, all quality-trimmed reads (including filtered reads) were aligned to assembled contigs. Overall, for the Iowa corn assembly, 8% of single reads and 10% of paired-end reads mapped to the assembly. Among paired-end reads, 95.5% of the reads aligned concordantly. In the Iowa prairie assembly, 10% of the single reads and 11% of the paired-end reads aligned to the assembled contigs, and 95.4% of the paired ends aligned concordantly (Table 4). Based on the alignment of sequencing reads to assembled contigs, we estimated the distribution of sequencing coverage in the resulting assemblies (Fig. 2). Overall, the coverage of each metagenome was low; 48% and 31% of total contigs in Iowa corn and prairie assemblies, respectively, had a read coverage less than 10.
Table 4.
Unassembled single-end (SE) and paired-end (PE) reads mapped to Iowa corn and prairie Velvet assemblies
| Type of read | Iowa corn assembly | Iowa prairie assembly |
| Total unfiltered reads | 1,810,630,781 | 3,303,375,485 |
| Total unfiltered SE READS | 141,517,075 | 358,817,057 |
| SE aligned one time | 11,368,837 | 32,539,726 |
| SE aligned more than one time | 562,637 | 1,437,284 |
| SE aligned, % | 8.43 | 9.47 |
| Total unfiltered PE reads | 834,556,853 | 1,472,279,214 |
| PE aligned one time | 54,731,320 | 110,353,902 |
| PE aligned more than one time | 1,993,902 | 3,133,710 |
| PE aligned disconcordantly, % | 0.47 | 0.63% |
| PE aligned, % | 9.68 | 11.20 |
Because the resulting assemblies are consensus representatives of the unassembled datasets, we also investigated the degree of variation (i.e., polymorphism) present among aligned reads to assembled contigs (SI Materials and Methods). For both the Iowa corn and prairie metagenomes, more than 99.9% of contigs contained base calls that were supported by a 95% consensus from mapped reads over 90% of their lengths, demonstrating an unexpectedly low polymorphism rate.
We annotated assembled contigs (greater than 300 bp) through the MG-RAST pipeline. This annotation resulted in 2,089,779 and 3,460,496 predicted protein coding regions in the corn and prairie metagenomes, respectively. The large majority of these regions, 61.8% in corn and 70.0% in prairie, had less than 60% similarity (over a minimum length of 15 aa) with any gene in the MG-RAST database M5NR (release 52). In total, 613,213 (29.3%) and 777,454 (22.5%) protein coding regions were assigned to an existing function. Many contigs were greater than 1 kbp, including 85,581 contigs in the corn metagenome (maximum length = 20,234) and 11,728 contigs in the prairie genome (maximum length = 2,579), and the distribution of lengths among assembled contigs was similar between sequences which could be assigned a function and those that could not (e.g., unknown sequences) (Figs. S3 and S4).
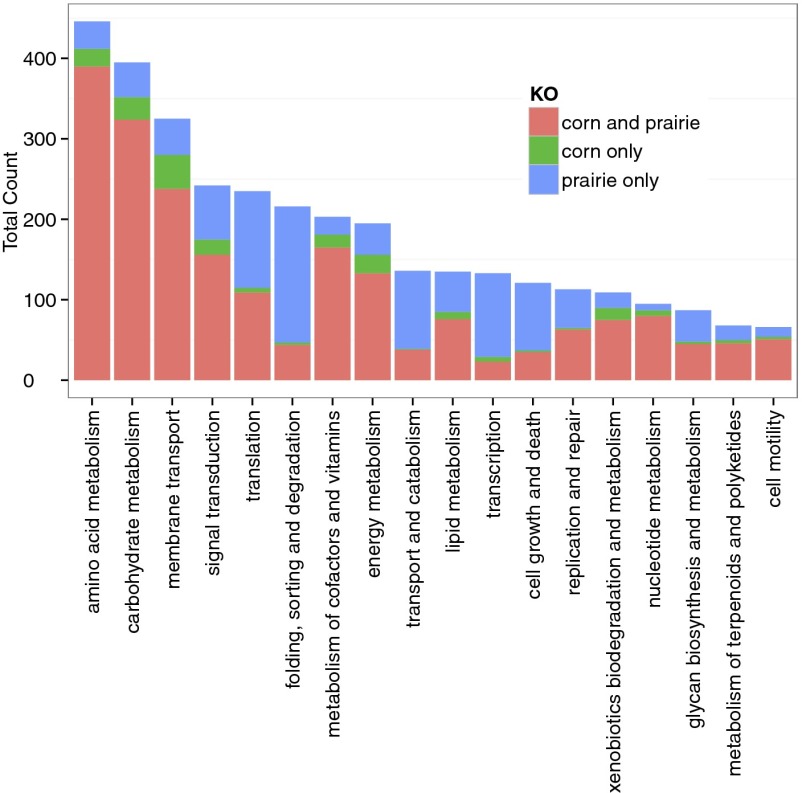
Annotations of the assembled corn and prairie soil metagenomes also were identified against the MG-RAST Kyoto Encyclopedia of Genes and Genomes Orthology (KEGG KO) database (Release 56). In total, 143,666 corn metagenome sequences and 164,318 prairie metagenome sequences matched sequences within the KO database with a minimum identity of 60%, a minimum length of 30 aa, and E-value <1e-10. The assembled contigs had significantly longer alignments to KEGG proteins than did unassembled reads (89 ± 39 aa vs. 32 ± 1 aa) (Fig. S5). Among these, a total of 3,553 unique KO identifiers were identified (2,201 shared between corn and prairie metagenomes, 223 in corn alone, and 1,129 in prairie alone) and were found to represent broad metabolic functions (Fig. 3 and Fig. S6) involved in metabolism, genetic and environmental information processing, and cellular processes.
Fig. 3.
Distribution of most abundant KEGG Orthology groups identified in corn and prairie soil metagenomes.
The shared presence of contigs without functional annotations in both the corn and prairie datasets also was evaluated. Assembled contigs that shared no homology to known sequences in the M5NR database were used as references for the complementing soil metagenome (e.g., corn assembly reference for prairie unassembled reads). Among these, a total of 34,436 contigs (31,058 and 3,416 corn and prairie contigs, respectively) were found to be shared between the two soil metagenomes (SI Materials and Methods).
Discussion
Coverage-Based Filtering Approaches Reduce Datasets Without Information Loss.
Our described approach for scalable metagenomic assembly was effective in reducing the HGMC dataset size without significant loss of information. Although the diversity and sequencing depth represented by the HGMC dataset are extremely low as compared with most environmental metagenomes, it represents a simplified, unevenly sampled model for a metagenomic dataset that allows the evaluation of novel approaches through the availability of source genomes. Our approaches normalized the abundance of reads in the dataset to a specific sequencing coverage while reducing the dataset volume and removing errors introduced by extraneous reads. Furthermore, the partitioning approach subdivides large datasets into biologically relevant subsets that can be assembled separately and by any assembler.
Based on our evaluations of a mock metagenome, we observed that the specific effects of filtering by digital normalization vary, depending on the conservation of genomic regions and abundance of genomes. In general, filtered (normalized and partitioned) assemblies were similar to or improved upon the assembly of the unprocessed dataset, suggesting that removing and subsetting these data do not result in substantial loss of information. For low-coverage genomes, the removal of erroneous sequences during normalization resulted in improved assemblies. The assembly of highly abundant genomes sharing conserved regions [such as the plasmids of the Staphylococcus epidermidis (NC_005008.1, NC_005007.1, and NC_005003.1)] was negatively affected by normalization. The greater number of reads representing these sequences within the unfiltered mock metagenome likely enabled assemblers to extend the assembly of these sequences more effectively, and this advantage ultimately was observed as an increased recovery of these genomes in this assembly as compared with the normalized assembly. This result identifies a shortcoming of our approach for metagenomic assembly and, indeed, of most short-read assembly approaches, related to repetitive regions and/or polymorphisms. Although data reduction may cause some information loss, we exchanged this disadvantage for the ability to assemble previously intractable datasets. Our evaluation of the mock metagenome suggests that this information loss is minimal overall and that our approach results in a comparable assembly whose abundance estimations are slightly improved.
Partitioning Separates Metagenomes into Tractable Subsets Representative of Species.
Metagenomes contain many distinct genomes that are largely disconnected from each other but that often share sequences as the result of sequence conservation or lateral transfer. Our prefiltering normalization approach removes both common multigenome elements and most artificial connectivity stemming from the sequencing process. The removal of these sequences does not significantly alter the recovery of HGMC reference genomes through de novo assembly, in which the resulting assemblies of unfiltered, normalized, and partitioned datasets were nearly identical. Further, for the mock metagenome, the large majority of partitions contained reads from a single reference genome, supporting our previous hypothesis that most connected subgraphs contain reads from distinct genomes (14). When an E. coli genome of 10× sequencing coverage was spiked into this dataset, it was divided into 424 partitions, likely because of the presence of introduced sequencing errors. Although fewer than half of these partitions (n = 201) contained reads unique to the original reference genome, the combined assembly of each partition could recover nearly all of the original reference. When five similar E. coli genomes were mixed with the mock metagenome, we observed more partitions (n = 658) containing E. coli sequences, one-third of which contained only E. coli sequences. When these partitions were assembled, the large majority of the genomic content of these strains was recovered, albeit largely in chimeric contigs. This particular result is not unique to our approach, however, because the comparable unfiltered assembly dataset resulted in a slightly higher fraction of assembled contigs associated with multiple references. This observation suggests that partitioning is an effective method for subdividing a metagenomic dataset, even one with highly similar strains, for assembly. Furthermore, these much-reduced subsets of sequences could be targeted for more sensitive assembly approaches for highly variable regions such as overlap-layout-consensus approaches or abundance binning approaches (21).
The most valuable result of partitioning is that it subdivides our datasets into sets of reads that can be assembled (or analyzed) with minimal computational resources. For the HGMC dataset, this gain was small, reducing unfiltered assembly at 12 GB RAM and 4 h to less than 2 GB RAM and 1 h. However, for the soil metagenomes, previously impossible assemblies could be completed in less than a day and in under 14 GB RAM of memory, enabling the use of multiple assembly parameters (e.g., k-length; see SI Materials and Methods) and multiple assemblers (Velvet, SOAPdenovo, and Meta-IDBA; Table 3).
Benefits of Soil Assembly.
This study represents the largest published soil metagenomic sequencing and assembly effort to date. Despite a significant sequencing effort (141 Gbp and 257 Gbp for Iowa corn and prairie soil, respectively), our resulting assemblies show that these metagenomes are largely characterized by low-coverage sequences (Fig. 2). Based on our evaluation of the mock community, the assembly of low-coverage sequences using our approaches results in minimal loss of information (Fig. S1). As improved sampling of soils is accompanied by increased sequencing coverage, these approaches will further enable the analysis of larger volumes of soil metagenomes in the future.
In our soil assemblies, we identified millions of putative genes, with hundreds of thousands of functions, even though only 10% of sequences were sufficiently sampled for assembly. The resulting corn and prairie soil metagenome assemblies resulted in a total length of 912 million bp and 1.5 billion bp, respectively, equivalent to ∼500 E. coli genomes’ worth of DNA. The contigs agreed well with the raw sequencing data, as evidenced by evaluation of paired-end concordance (Table 4), which was even slightly greater than the fraction of unpaired read alignments. Further, these contigs contained very low degrees of variation when unassembled reads were aligned, suggesting that they do not originate from polymorphic species within the soil. Combined, these results support the quality of these metagenomic assemblies and their representation of soil diversity.
The overall representation of assembled soil contigs was low (average coverage of 10×) (Fig. 2), demonstrating the high diversity even in these localized soils and emphasizing the need to increase sampling of these metagenomes considerably for them to represent its microbial diversity. As these datasets become increasingly available, our approaches enabling assembly offer a number of advantages for metagenomic analysis. First, the assembly resulted in significant data compression, reducing the volume of our data to be annotated (including sequencing errors) from 397 Gbp (unassembled) to 2.4 Gbp (assembled) and thus allowing more efficient and effective annotation and analysis of the resulting sequences. Furthermore, the length of the assembled sequences is significantly greater than their unassembled counterparts. In the soil metagenomes, more than 97,000 contigs were longer than 1,000 bp, allowing the possible identification of multiple genes and operon structure. Notably, nine sequences were assembled into contigs longer than 10 kbp (corn metagenome), and the most abundant sequences (17,507 bp and 16,126 bp) were related to sequences of phage origin (Pseudomonas phage PaP2) (Table S3).
The longer lengths of assembled sequences relative to unassembled metagenomes allowed both greater numbers and improved identification (lengths of alignment) of metabolic pathways within the framework of the KEGG Orthology database (e.g., 79,477 KO in the corn assembly vs. 68,037 in unassembled corn metagenomes) (Fig. S5), representing a broad catalog of the majority of known metabolic pathways in corn and prairie soils (Fig. S6). We identified unique metabolic contributions of the prairie microbial communities relative to that of the corn, especially those involved in cellular processes (e.g., cell growth and death and transport and catabolism) and genetic information processing (e.g., folding, sorting, and degradation; translation; and transcription) (Fig. 3). This result may reflect the varying management history of these two soils. Unlike the prairie soils, which have never been tilled, the corn soils have been cultivated for more than 100 y and have had annual additions of animal manure that potentially could enrich specific metabolic pathways with decreased diversity.
A key challenge facing future soil investigations is the lack of culturable representatives from soil and consequently the poor availability of reference genomes. This problem is highlighted by our observation that more than half of the assembled contigs were not similar to any sequence in the MG-RAST m5nr databases, suggesting that soil holds considerable unexplored taxonomic and functional novelty. These “unknown” sequences are broadly distributed in both length and abundance (Figs. S3 and S4) and represent the potential of gene and organism discovery. These sequences highlight the value of using de novo assemblies as reference datasets that are more representative of site-specific genes than are the publicly available references (where the average homology of assembled sequences against the SEED database was 68% over an average of 70 bp). For example, we identified 17 Mbp of unknown sequences in 34,436 contigs that were shared at relatively high abundance (C >10) by the corn and prairie soil metagenomes. These broadly present, novel sequences are targets for further investigations of proteins about which nothing is known. As increasing numbers of metagenomes become available, the co-occurrence of these assembled sequences with known genes and genomes will enable further characterization.
Conclusions
We have presented two strategies that readily enable the assembly of very large environmental metagenomes by compressing and subdividing the data before assembly. The strategies are generic and broadly applicable to any metagenome. We demonstrate their effectiveness by first evaluating them on the assembly of a mock community metagenome and then applying them to two previously intractable soil metagenomes. Digital normalization scales the data size with community richness rather than diversity and is particularly effective for mixed-abundance communities. After digital normalization, partitioning enables the extraction of read subsets that belong to individual species. These read partitions are small enough that a variety of genomic-based analysis techniques can easily be applied to them individually, as evidenced by the application of multiple assemblers for our soil metagenomes with considerably reduced computational resources. By acting as prefilters, digital normalization and partitioning let downstream assemblers focus on improving their performance on low-coverage or high-variability data without a strong consideration for computational resources. This ability should enable significant improvement of metagenome assembly techniques going forward and provide the critical references that will enable future investigations of soils and other complex environments. Importantly, our assembly results also demonstrate that 300 Gbp of read data are insufficient to cover even a small, localized soil sample deeply, confirming that considerably more data are needed to study the content of soil metagenomes comprehensively.
Materials and Methods
Assemblies of the HGMC and soil metagenomes using various software were performed on (i) quality-filtered unassembled sequences and (ii) the same sequences filtered by digital normalization [HGMC coverage threshold (C) = 20, soil coverage threshold (C) = 10], removal of high-coverage sequences (C >50), and partitioning disconnected sets of reads. Coverage of assembled sequences or reference genomes was estimated through consensus alignment of raw sequences, and assembled contigs were compared with one another or reference genomes through BLASTn alignment (see SI Materials and Methods for specific thresholds). Annotation of assembled metagenomes and quality-filtered unassembled reads was performed through the MG-RAST and the M5NR (version 1) database and are available publicly (see SI Materials and Methods for accession numbers).
Supplementary Material
Acknowledgments
We thank Krystle Chavarria and Regina Lamendella for help in extracting DNA from Great Prairie soil samples; Fan Yang for helpful comments on this paper; Eddy Rubin and Tijana Glavina del Rio at the Department of Energy Joint Genome Institute (DOE JGI); and John Johnson and Eric McDonald at the Michigan State University High Performance Computing Center. This project was supported by Agriculture and Food Research Initiative Competitive Grant 2010-65205-20361 from the US Department of Agriculture and by National Institute of Food and Agriculture and National Science Foundation Grant IOS-0923812 (both to C.T.B.). A.C.H. was supported by National Science Foundation Postdoctoral Fellowship Award 0905961 and the Great Lakes Bioenergy Research Center (Department of Energy BER DE-FC02-07ER64494). The work conducted by the DOE JGI is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences discussed in this paper have been deposited in the MG-RAST database (http://metagenomics.anl.gov/). The unassembled reads can be found as accession nos. 4539514.3–4539528.3 (Iowa corn) and 4539571.3–4539594.3 (Iowa prairie). The assembled metagenomes can be found as accession nos. 4504979.3 (Iowa corn) and 4504798.3 (Iowa prairie).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402564111/-/DCSupplemental.
References
- 1.Arumugam M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 3.Iverson V, et al. Untangling genomes from metagenomes: Revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 4.Mackelprang R, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480(7377):368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 7.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 8.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309(5739):1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 9.Wommack KE, Bhavsar J, Ravel J. Metagenomics: Read length matters. Appl Environ Microbiol. 2008;74(5):1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loman NJ, et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA. 2013;309(14):1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 11.Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. Ray Meta: Scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13(12):R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholz MB, Lo C-C, Chain PSG. Next generation sequencing and bioinformatic bottlenecks: The current state of metagenomic data analysis. Curr Opin Biotechnol. 2012;23(1):9–15. doi: 10.1016/j.copbio.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 13. Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH (2012) A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802. Accessed February 6, 2014.
- 14.Pell J, et al. Scaling metagenome sequence assembly with probabilistic de Bruijn graphs. Proc Natl Acad Sci USA. 2012;109(33):13272–13277. doi: 10.1073/pnas.1121464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Y, Leung HCM, Yiu SM, Chin FYL. Meta-IDBA: A de novo assembler for metagenomic data. Bioinformatics. 2011;27(13):i94–i101. doi: 10.1093/bioinformatics/btr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18(2):324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci USA. 2001;98(17):9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 21.Sharon I, et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23(1):111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.