Significance
Gram-negative bacteria contain an unusual outer membrane that prevents the entry of most currently available antibiotics. This membrane contains a complex glycolipid, LPS, on the exterior. It is not understood how such a large molecule, which can contain hundreds of sugars and six fatty acyl chains, is transported across the cell envelope from its site of synthesis in the cytoplasmic membrane to the cell surface. Using a combination of genetics, biochemistry, and structural biology, we characterized residues in the protein that powers LPS transport to gain mechanistic insight into how ATP hydrolysis is coupled to the biological function of the transporter. These tools help us understand how to design antibiotics targeting this essential pathway.
Keywords: ABC transporter, antibiotic target, membrane biogenesis
Abstract
The cell surface of Gram-negative bacteria contains lipopolysaccharides (LPS), which provide a barrier against the entry of many antibiotics. LPS assembly involves a multiprotein LPS transport (Lpt) complex that spans from the cytoplasm to the outer membrane. In this complex, an unusual ATP-binding cassette transporter is thought to power the extraction of LPS from the outer leaflet of the cytoplasmic membrane and its transport across the cell envelope. We introduce changes into the nucleotide-binding domain, LptB, that inactivate transporter function in vivo. We characterize these residues using biochemical experiments combined with high-resolution crystal structures of LptB pre- and post-ATP hydrolysis and suggest a role for an active site residue in phosphate exit. We also identify a conserved residue that is not required for ATPase activity but is essential for interaction with the transmembrane components. Our studies establish the essentiality of ATP hydrolysis by LptB to power LPS transport in cells and suggest strategies to inhibit transporter function away from the LptB active site.
ATP-binding cassette (ABC) systems represent one of the largest protein superfamilies across all domains of life (1, 2). Many of these systems are transporters that share a common architecture of two transmembrane domains (TMDs) and two cytoplasmic nucleotide-binding domains (NBDs), which bind and hydrolyze ATP. They couple the energy of ATP binding and hydrolysis to the transport of a variety of substrates against a concentration gradient. Although eukaryotic ABC transporters involved in human diseases have received much attention (3), the canonical ABC systems that have been most extensively studied are Gram-negative bacterial importers (2).
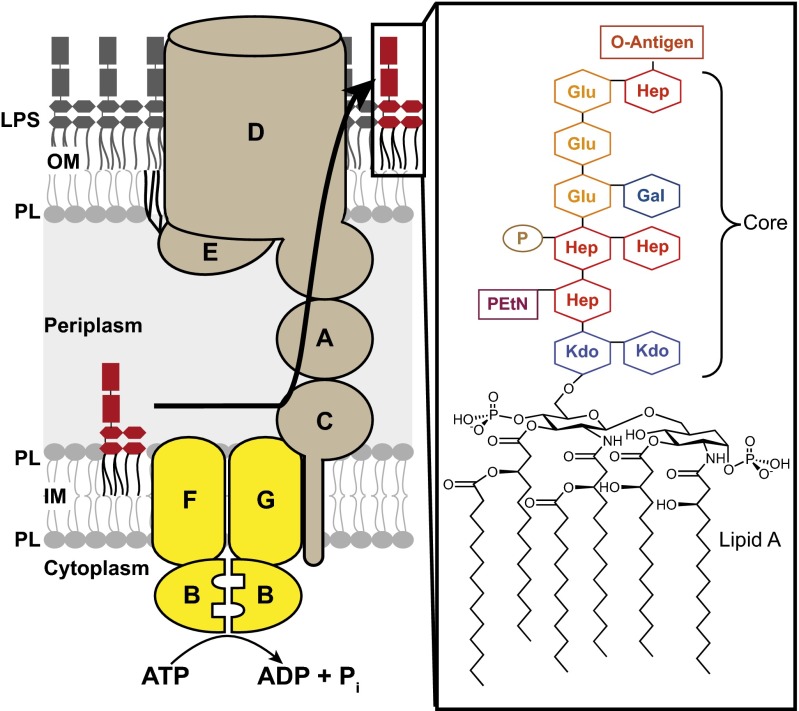
Gram-negative bacteria, such as Escherichia coli, have a unique double-membrane architecture that allows them to colonize harsh environments. The inner membrane (IM) contains phospholipids, whereas the outer membrane (OM) is an asymmetric lipid bilayer composed of phospholipids in the inner leaflet and LPS in the outer leaflet (4). These two membranes are separated by an aqueous periplasmic compartment. LPS is a complex glycolipid composed of hundreds of sugars attached to a core containing fatty acyl chains (Fig. 1). Millions of LPS molecules must be properly assembled during each division cycle (4) on the cell surface to establish a permeability barrier that prevents the entry of hydrophobic molecules, including antibiotics (5). Therefore, understanding how LPS is assembled at the OM could lead to the development of better strategies to target Gram-negative infections. In 1972, it was established that the biosynthesis of LPS is completed on the outer leaflet of the IM. Because LPS cannot move passively across the aqueous periplasm and through the OM, it was recognized that there must be machinery to transport LPS across the cell envelope (6, 7).
Fig. 1.
LPS is transported to the cell surface of E. coli by essential Lpt proteins. (Right) The E. coli LPS structure is composed of lipid A (endotoxin), a core region, and the O-antigen. (Left) Lpt proteins form a transenvelope complex that transports LPS from its site of synthesis (IM), across the aqueous periplasm, to the cell surface. LPS transport and assembly are believed to be powered by the ABC transporter LptB2FG in the IM (yellow), because there is no ATP in the periplasm. A, LptA; B, LptB; C, LptC; D, LptD; E, LptE; EtN, ethanolamine; F, LptF; G, LptG; Gal, d-galactose; Glu, d-glucose; Hep, l-glycero-d-mannoheptose; Kdo, 3-deoxy-d-manno-oct-2-ulosonic acid; P, phosphate; PL, phospholipids.
In the past decade, all of the components essential for the transport and assembly of LPS have been identified (8–14). These LPS transport (Lpt) proteins comprise a complex that spans all compartments in the cell, from the cytoplasm to the OM (15, 16) (Fig. 1). E. coli has seven different Lpt proteins that are essential for LPS transport and cell viability (17, 18). Three of these Lpt proteins form an ABC system composed of a heteromeric TMD complex (LptF and LptG) and a homodimeric NBD complex (LptB). LptB, both alone and in a complex with LptF and LptG, has ATPase activity in vitro (19–21). In addition, the LptB2FG complex is closely associated with the bitopic IM protein LptC, which binds LPS (20–23) and is part of the transenvelope bridge (15, 16). Based on these findings, the current model is that LptB2FG extracts LPS from the outer leaflet of the IM and is the sole energy input responsible for the entire process of transport and assembly of LPS on the cell surface against a concentration gradient (22). Coupling this ABC transporter with the Lpt periplasmic bridge and OM translocon enables cytoplasmic ATP to drive periplasmic transit. Although its heteromeric architecture with separate NBDs and TMDs resembles that of bacterial importers, LptB2FG has to perform a unique function that places it in a class distinct from traditional importers and exporters; LptB, the cytoplasmic ATPase, must power the extraction of a glycolipid from the periplasmic face of the IM.
To understand how the LptB2FG transporter works, we changed three residues implicated in important activities of LptB and showed that they are essential for in vivo function. One residue is the proposed essential active site glutamate, but the other two residues are not essential for the catalytic activity of purified LptB. We show that one of these residues affects the ATPase activity of the intact Lpt IM complex, whereas the other is an essential site for binding the TMDs. Based on high-resolution crystal structures of LptB pre- and post-ATP hydrolysis, we propose that the former residue facilitates phosphate exit from the active site. These studies highlight the importance of combining structural studies with genetics and biochemistry on full complexes, as well as individual components, to understand how this unique ABC transporter functions.
Results
Identification of LptB Residues Required for Cell Viability.
Using sequence homology to other NBDs, we identified conserved residues located in three different regions of LptB. E163 is at the end of the Walker B motif and is essential for catalysis in other ABC transporters. It is the proposed general base that deprotonates the nucleophilic water molecule that attacks the γ-phosphate of ATP (2, 24, 25). H195 is located in the conserved switch region adjacent to the active site of NBDs and has been implicated in catalytic activity, but its specific function is debated (2, 25–27). Lastly, F90 is located in the Q-loop, which is proposed to link the ATP-binding site to the structurally diverse region that interacts with the coupling helices of the TMDs of ABC transporters (2, 25, 28, 29). LptB has been shown to be required for LPS transport and, consequently, cell viability in E. coli (11, 12). Therefore, we mutated lptB to determine the importance of each of these residues for LPS biogenesis. Because defects in LPS assembly result in phenotypes that range from increased OM permeability to cell death (10, 17, 30), we assessed both susceptibility to OM-impermeant antibiotics and cell viability.
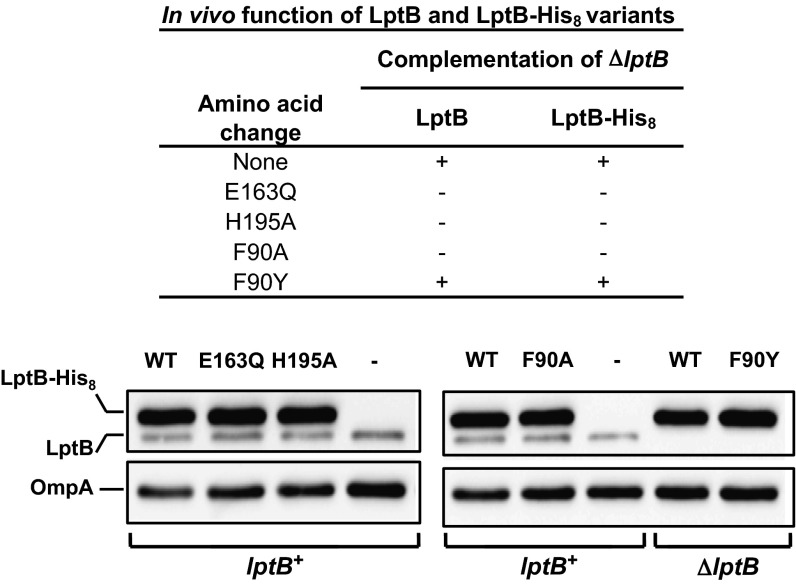
Plasmid-encoded LptB-His8 (used for purification, below) or untagged LptB (to assess the effect of the His8 tag) variants were classified as nonfunctional if they could not support cell viability in the absence of a wild-type lptB allele. To assess this type of complementation, we first constructed a strain in which the only source of wild-type LptB is encoded by pRC7KanLptB, an unstable, segregation-defective plasmid that is rapidly lost in a population, thereby causing cell death (details of strain construction are provided in SI Materials and Methods and Tables S1–S3). When a plasmid encoding a functional LptB or LptB-His8 variant was introduced into this strain, cells that lost pRC7KanLptB were viable; in contrast, when a plasmid encoding a nonfunctional LptB or LptB-His8 variant was introduced into this strain, cells that lost pRC7KanLptB died. With this approach, we found that unlike wild-type LptB-His8 and LptB, the E163Q, H195A, and F90A variants are all nonfunctional (Fig. 2). This loss of function is not a result of reduced protein levels (Fig. S1A). We also tested an F90Y variant because an aromatic residue is typically found at this position in other NBDs. We found that the LptB and LptB-His8 F90Y variants are functional in vivo.
Fig. 2.
Genetic studies indicate residues essential for LptB functionality. Functionality of untagged and His8-tagged LptB variants with alterations in the active site and Q-loop was determined by their ability to complement a chromosomal ΔlptB allele. Anti-LptB Western blots show that residue changes (shown above lanes) do not alter LptB-His8 levels. WT refers to wild-type LptB-His8. Chromosomal lptB alleles of strains are shown below LptB-His8 bands. Cross-reacting OmpA is shown as a loading control.
We also found that introduction of plasmid-encoded LptB-E163Q-His8 or LptB-H195A-His8 variants into the wild-type strain resulted in increased OM permeability (Fig. S1B). The simplest explanation for this dominant-negative effect is that the nonfunctional variants can substitute for wild-type LptB in Lpt IM complexes, thereby reducing the number of viable transporters and compromising LPS transport. In contrast, the merodiploid strain harboring the LptB-F90A-His8 variant did not display increased OM permeability (Fig. S1B), suggesting that it cannot replace wild-type LptB in the IM complex.
Residue F90 Is Essential for Proper Formation of the Lpt IM Complex.
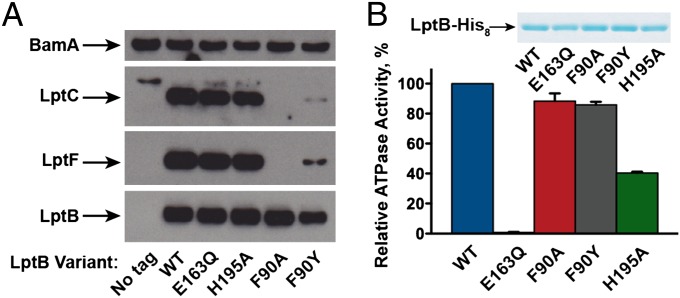
We next assessed whether the LptB variants produced in the merodiploid strains interact with other Lpt IM components. Wild-type LptB-His8 interacts with the Lpt IM complex in cells as it copurifies with endogenous LptF and LptC (Fig. 3A). A lack of LptG antiserum prevented us from monitoring LptG, but it is likely that both LptF and LptG are required for the formation of a stable complex with LptB and LptC (20). The E163Q and H195A variants also pull down these Lpt components, but the F90A variant does not. These results are consistent with our interpretation of the dominant-negative phenotypes exerted by the E163Q and H195A variants. They also show that the F90A variant cannot form a stable complex with the Lpt IM components, and thus does not confer a dominant phenotype in a merodiploid strain.
Fig. 3.
Biochemical studies indicate residues in LptB essential for catalysis and proper coupling with other Lpt components. (A) Western blots show affinity purifications of LptB variants expressed in merodipoloid strains harboring plasmids expressing LptB variants. All plasmids express LptB-His8, except for the untagged variant (labeled “No tag”). Levels of LptF and LptC that copurify with LptB are shown. Levels of BamA, an OM protein that nonspecifically interacts with the purification resin, are shown as a sample preparation and loading control. (B) LptB-His8 variants were overexpressed and purified, and their ATPase activity was measured. Data represent the average and SD of three experiments. WT, no amino acid change in LptB-His8.
The His-tagged F90Y variant pulls down a lesser amount of IM Lpt components compared with the wild-type, E163Q, and H195A variants (Fig. 3A), showing that it does not associate as well with the IM components as the wild-type variant. This defective interaction has functional consequences because a haploid strain producing LptB-F90Y-His8 shows increased OM permeability compared with the wild-type strain (Fig. S1C). Taken together, these results implicate F90 in the association of LptB with LptF/G/C. They also show that the essential role of E163 and H195 in LptB function is unrelated to Lpt complex formation.
Nonfunctional LptB Variants Show Variable Catalytic Activity.
We next tested the ATPase activity of the different LptB variants to gain more insight into the nature of their functional defects. We overexpressed and purified all variants as stable proteins. As expected (19), LptB-E163Q-His8 is catalytically inactive (Fig. 3B). Because this variant forms a stable IM complex, its inability to support cell viability can be attributed to this defective catalytic activity. Therefore, ATP hydrolysis by LptB is required for LPS transport in the cell. We found that LptB-F90A-His8 and LptB-F90Y-His8 show wild-type levels of ATPase activity. Even though these substitutions do not affect protein folding or catalytic activity, both affect LptB function in vivo. Finally, the LptB-H195A-His8 variant shows a reduction of ∼60% in ATPase activity even though it does not support cell viability. This activity is somewhat surprising because changing this conserved histidine in other ABC transporter systems results in nearly complete loss of ATPase activity (26, 31–34).
Crystal Structures of LptB Bound to ADP and ATP.
To understand the roles of F90 and H195 better, we obtained crystal structures of LptB before and after ATP hydrolysis. We overexpressed and purified functional LptB-His8 and obtained crystals with both the native protein and a selenomethionine derivative following incubation with ATP/MgCl2 (Fig. S2A and Tables S4 and S5). There was unambiguous density for ADP-Mg2+, indicating ATP hydrolysis had occurred during crystallization (Fig. S2B). This 1.55-Å structure will hereafter be referred to as LptB-ADP.
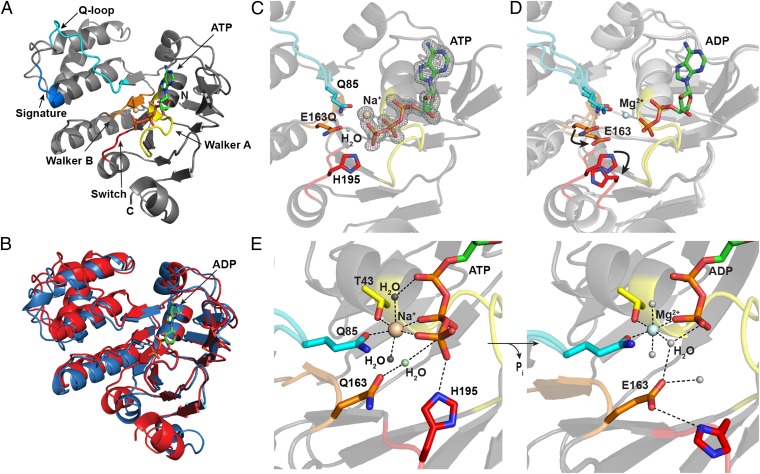
To obtain a prehydrolysis LptB/ATP complex, we purified the catalytically inactive variant LptB-E163Q (Fig. 3B) and obtained crystals after preincubation with ATP. The resulting 1.65-Å LptB-E163Q-ATP structure will hereafter be referred to as LptB-ATP (Fig. 4A and Fig. S2C). Examination of the active site demonstrated clear electron density for intact ATP (Fig. 4C). Additionally, unlike the LptB-ADP structure, the prehydrolysis form crystallized as a canonical nucleotide-sandwich dimer (Fig. S2D). Secondary structure matching alignments (35) of the LptB-ATP sandwich dimer with those of ATP-bound E. coli MalK, the NBD of the maltose/maltodextrin importer [Protein Data Bank (PDB) ID code 1q12], and Methanocaldococcus jannaschii MJ0796 (a homolog of LolD, the NBD of the lipoprotein exporter, PDB ID code 1l2t) have rmsd values less than 1.75 Å. In all cases, ATP is sandwiched between the Walker A motif of one subunit and the signature motif of the opposing unit (Fig. S2E).
Fig. 4.
Conformational changes upon ATP hydrolysis show how reorganization of the active site causes changes in the region of LptB believed to interact with LptF/G. (A) Cartoon rendering of LptB-ATP (prehydrolysis), with conserved ATPase and ABC motifs indicated (Walker A, yellow; Walker B, orange; signature motif, blue; Q-loop, cyan; switch region, red). We assigned the active site metal of the LptB-ATP structure to a Na+ ion. The distances between the metal and the coordinating water molecules correspond more to those characteristic of Na+ (∼2.4 Å) than to those characteristic of Mg2+ (≤2.1 Å), which were observed in the LptB-ADP structure. In addition, LptB-ATP crystallized in magnesium-free buffer in a condition containing sodium chloride. (B) Structural overlay of the LptB-ATP (red) and LptB-ADP (blue) structures shows conformational changes upon ATP hydrolysis. The product (ADP) is shown in the active site. (C) Close-up view of the ATP-bound active site, with the side chains of conserved residues in the Walker B, switch, and Q-loop regions shown. The Fo-Fc omit map is contoured at 3σ and shows clear electron density for a γ-phosphate. (D) Structural overlay, as in B, of the pre- and posthydrolysis structures, with arrows indicating movement of side chains upon hydrolysis. Coloring of conserved regions is the same as in A. (E) Rearrangement of active site residues upon ATP hydrolysis to stabilize ADP is shown. The octahedral coordination sphere of the active site cation is shown, and the putative hydrolytic water is indicated in the ATP-bound active site.
LptB possesses an overall fold resembling that of NBD structures (Fig. 4A). It contains the canonical L-shaped architecture (36) composed of a RecA-like α/β-ATPase domain and a structurally diverse α-helical domain. The RecA-like domain contains the Walker A and Walker B motifs present in many nucleoside triphosphate-binding proteins (37) (Fig. 4A). This domain also furnishes Mg2+- and nucleotide-binding motifs specific to ABC proteins, namely, the Q-loop, which links the more highly conserved α/β-ATPase domain to the α-helical domain, and the switch region, which contains the conserved H195. As observed in other NBD structures, the LSSG(E/Q) signature motif is found in the helical domain (25, 38) (Fig. 4A). Additionally, both LptB structures reveal an interface where we predict LptB interacts with TMDs LptF/G, based on comparisons with the structures of other NBDs and those of full ABC transporters (Fig. S2F). This interface contains grooves (Fig. 5) that could accommodate coupling helices of TMDs, which are involved in communication between the NBDs and TMDs in other ABC transporters (39, 40).
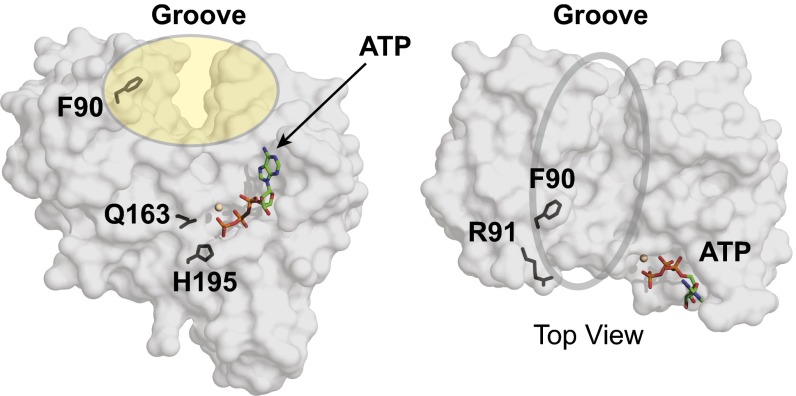
Fig. 5.
Structural observations of LptB implicate a key binding site for TMDs. Surface renderings of a monomer of LptB-ATP (frontal facing and top views) show residues Q163 and H195 near ATP, residue F90 facing the interior of the groove, and residue R91 facing away from the groove.
ATP Hydrolysis Induces Conformational Changes.
Comparison of the pre- and posthydrolysis complexes of LptB demonstrates considerable movement in a number of regions associated with binding and/or hydrolysis of the γ-phosphate (Fig. 4B). Conformational changes in the switch region, containing H195, and in the region surrounding the Walker B motif, containing E163, are apparently driven by reorganization of the active site following ATP hydrolysis. This reorganization is coupled to changes in the signature motif and the Q-loop, which contains F90 (Fig. 4 A and B).
A closer view of the LptB-ATP active site reveals that key residues that orient and stabilize the γ-phosphate must reorganize during the catalytic cycle to maintain contact with ADP following phosphate-bond cleavage (Fig. 4 C and D). Residue 163 (Q163 in LptB-ATP and E163 in the native LptB-ADP structure), located at the end of the Walker B motif, and H195, located in the switch region, make contacts with a bridging water molecule and the γ-phosphate, respectively, in the LptB-ATP structure (Fig. 4E). Glutamine-85, which is at the start of the Q-loop (giving it its name), forms part of the octahedral coordination sphere surrounding the metal ion associated with the nucleotide (Fig. 4E).
E163 is essential for catalysis (Fig. 3B). A closer look at the carbonyl oxygen in the side chain of residue 163 reveals that it is ∼2.2 Å closer to the nucleotide in the posthydrolysis structure based on a structural alignment of the Walker A motifs (36-GPNGAGKT-43). Through a bridging water molecule, this glutamate contacts the β-phosphate of the nucleotide (Fig. 4E). In LptB-ATP, Q163 is oriented slightly farther from the nucleotide because it is separated by both a bridging water molecule and the γ-phosphate (Fig. 4E). Based on structural alignments with E. coli MalK bound to transition-state mimics (ADP-vanadate and ADP-aluminum fluoride) (41), and by comparison with the ATP-bound MJ0796 structure (42), we believe this water is well positioned to be the nucleophilic water in the hydrolysis reaction. Consistent with this hypothesis, this water molecule is not present in the LptB-ADP structure (Fig. 4E).
Comparison of the pre- and posthydrolysis structures also shows that H195 in the switch region undergoes a major conformational change (Fig. 4D). The side chain of H195 makes direct contact with the γ-phosphate in the ATP-bound structure. However, its Cα shifts by ∼4 Å in the ADP-bound structure. Not only does the switch region move, but the side chain of H195 flips. One important unanswered question is how inorganic phosphate (Pi) exits the active sites of NBDs. There is no clear electron density for a Pi group in the posthydrolysis crystal structure (Fig. S2B), which is consistent with claims that Pi leaves the active site before ADP is released (42–44). Based on an electrostatic potential surface of LptB, the H195 side chain flips to face a negatively charged part of the protein (Fig. S3). It is possible that the γ-phosphate remains bound to H195 and that the movement of the switch region forces out the Pi by electrostatic repulsion in this negatively charged area. It is also possible that the dramatic movement of the switch region observed during ATP hydrolysis plays a critical role in communicating changes in the active site to changes in the TMDs.
Because H195 is essential in vivo and the LptB-ATP structure shows that the imidazole side chain directly interacts with the γ-phosphate of ATP, we suspected that the ATPase activity observed for the isolated NBD (Fig. 3B) reported inaccurately on the activity of the full complex (21). Therefore, we overexpressed and purified LptB2FGC in detergent and measured the ATPase activity of the wild-type complex and complexes containing LptB-E163Q and LptB-H195A (Fig. S4A). The complex containing LptB-E163Q is catalytically inactive, and the complex containing LptB-H195A has ∼10% of the ATPase activity of the wild-type complex. These results confirm our hypothesis that although H195 is not the sole catalytic residue, its positioning is important for some step of the catalytic cycle, as suggested by the crystal structures.
Groove Region of LptB Is Essential for Interaction with IM Partners.
Taken together, ATP hydrolysis initiates global movement in LptB that couples changes in regions surrounding the active site (the switch and the Walker B domains) to changes in the structurally diverse helical domain (the signature motif and the Q-loop). These changes near the helical domain result in a shift in the groove located adjacent to the Q-loop (Fig. S4B and Fig. 5). This coupling of ATP hydrolysis with movement in the groove region could be important for communicating with the TMDs, thereby affecting LPS transport. Therefore, we were interested in the positioning of F90 in the Q-loop, because we have interpreted in vivo and biochemical results described above to mean that F90 is important for binding to putative coupling helices of the TMDs of this ABC system.
The side chain of F90 in the Q-loop faces the interior of the groove opening in both the ATP- and ADP-bound struc-tures (Fig. 5), suggesting this residue might be functionally important for interacting with the TMDs during part or all of the catalytic cycle. Indeed, this role for residue F90 would explain why the lptB-F90A allele is nonfunctional. To test whether positions with side chains facing away from the groove interior are as important as residues pointing into the groove, we assessed the importance of LptB-R91A, which has a side chain facing away from the interior of the groove in both crystallographic snapshots (Fig. 5). We found that the LptB-R91A variant is functionally similar to wild-type LptB (Fig. S1 A and C).
Taken together, these results support the hypothesis that residues in the Q-loop of LptB that face the interior of the groove are important for assembly of the complex, for its function, or for both. The high conservation of F90 in LptB orthologs (Fig. S4C), combined with the fact that the lptB-F90Y allele confers mild OM permeability defects in haploid strains (Fig. S1C), suggests that a conservative change from a phenylalanine to a tyrosine is tolerated, but not optimal, as LptB evolved to have a phenylalanine at this position. These experiments lead us to conclude that F90 forms a critical part of the binding site for LptF/G.
Discussion
We describe the crystal structures of a catalytically inactive variant of LptB bound to ATP and wild-type LptB bound to ADP. By combining biochemistry with a genetic analysis of LptB variants, we have identified residues in LptB that are important for ATP hydrolysis (E163), assembly with LptF/G (F90), and the function of the complex (H195). Structural studies reveal that residues involved in binding ATP and catalyzing γ-phosphate hydrolysis reorganize to stabilize the product, ADP. Our crystallographic snapshots suggest that movement of residues around the LptB active site is coupled to movement of regions at the interface with LptF/G that might be essential for LPS transport. In addition, using a catalytically inactive LptB-E163Q variant, we have demonstrated that ATP hydrolysis by LptB is essential for cell viability; in contrast, ATP hydrolysis is not required for the formation of the Lpt IM complex.
With respect to the mode of catalysis, our observations are consistent with the prevailing general base catalysis model, in which a carboxylate side chain at the end of the Walker B motif (in residue E163) deprotonates a water molecule so that it can serve as a nucleophile in the hydrolysis reaction. The locations of the general base and potential nucleophile are also consistent with structures of the transition state of the E. coli maltose importer (41). The pre- and postcatalysis structures reveal that ATP hydrolysis induces conformational changes in LptB. The most dramatic conformational change in our structural snapshots is in the switch region. Not only does the switch loop move, but the imidazole side chain of the conserved H195 flips away from the active site toward a negatively charged channel. This movement, combined with the observation that H195 coordinates the γ-phosphate in the prehydrolysis complex, leads us to speculate that this residue helps move the γ-phosphate from the active site into the acidic channel, from which it is ejected by electrostatic repulsion (Fig. S3). This shift of the conserved histidine near a negatively charged surface is also observed in a structure of the NBD MJ0796 (42). Such a role is consistent with the low-level ATPase activity observed for LptB-H195A in complex with the other Lpt IM components.
Our crystal structures reveal a groove in LptB that undergoes movement upon ATP hydrolysis. We hypothesize this groove is involved in the interaction with its TMD partners, LptF/G, and that its movement is essential for connecting ATP hydrolysis by LptB with LPS extraction by LptF/G. Our genetic and biochemical studies implicate groove residue F90 as an important binding site for LptF/G. The crystal structures suggest that this residue is critical, because its aromatic side chain faces the inside of the groove that likely interacts with the coupling helices of the TMDs. Structural alignments with other NBDs, both in isolation and in complex with their TMDs, reveal that, despite being part of a structurally diverse region, there is often an aromatic residue at the same location as F90 in LptB. For example, F90 in LptB aligns with F429 in Salmonella typhimurium MsbA (PDB ID code 3b60), with Y87 in MalK (PDB ID code 2r6g), and with Y94 in Sulfolobus solfataricus GlcV (PDB ID code 1oxx). In the structure of Sav1866 bound to a nonhydrolyzable ATP analogue (PDB ID code 2onj), the guanidinium group of R206 in the TMD is in close enough proximity to make a π-cation interaction with the aromatic ring of F427 in the NBD, which aligns with F90 in LptB (Fig. S4D), suggesting that this residue is critical for interaction with the TMDs. For LptB, we have established the importance of F90 by genetic analysis and affinity purifications: The F90A variant fails to complement and results in destabilization of the ABC transporter. The aromatic character of this residue is critical for mediating the interaction of LptB with LptF/G, because the F90Y substitution only results in a partial loss of function in vivo, suggesting that the interaction is specific. Further studies are underway to examine how this key interaction with the TMDs might be important for linking ATP hydrolysis to the extraction of LPS from the opposite side of the membrane via the conformational changes in the groove region.
We have done a comprehensive analysis of selected residues in the ATPase component of the Lpt IM complex that combines genetic studies of essentiality, biochemical studies of catalytic activity and protein–protein interaction, and crystallographic analysis of pre- and posthydrolysis complexes. Our results highlight the importance of combining multiple techniques to understand the roles of individual residues in the process of transporting LPS from the IM to the cell surface. Neither catalytic activity nor cell viability alone can illuminate the functions of certain residues. Clearly, interfering with catalytic activity and coupling with other Lpt components are viable strategies for the development of new antibiotics targeting LPS biogenesis.
Materials and Methods
In Vivo Experiments.
Strains, growth conditions, and construction of plasmids and a ΔlptB allele are described in SI Materials and Methods and Tables S1–S3. Functionality of mutant lptB alleles was assessed by two complementary methods also described in SI Materials and Methods. When necessary, the chromosomal ΔlptB allele was transferred by P1 transduction (45).
OM permeability tests were performed using disk diffusion assays (46). LptB protein levels were monitored by immunoblotting using anti-LptB antisera raised in a rabbit using the LptB peptide DDLSAEQREDRANE as the immunogen (ProSci, Inc.).
Affinity Purifications and ATPase Assays.
Modifications to affinity purifications (15) are explained in SI Materials and Methods. ATPase assays were conducted as described with 5 mM ATP/MgCl2 for both LptB and LptB2FGC variants (21).
Protein Overexpression and Purification.
LptB-His8 and the catalytically inactive E163Q variant were overexpressed in E. coli and purified by nickel-affinity chromatography and gel filtration chromatography as described (19), with notable changes described in SI Materials and Methods. Overexpression of a selenomethionine-containing derivative of LptB (SeMet-LptB-His8) is described in SI Materials and Methods.
LptB2FGC complexes (wild-type, E163Q, and H195A variants) containing His6-LptB were overexpressed in E. coli and purified as described in SI Materials and Methods.
Protein Crystallization, Data Collection, and Structure Determination.
A detailed description of the structure determination is provided in SI Materials and Methods. Briefly, all protein crystals were grown by vapor diffusion in hanging drops. Experimental phasing was obtained by multiple isomorphous replacement with anomalous scattering using a selenomethionine derivative crystal and a tantalum derivative crystal. The X-ray diffraction data were collected at the National Synchrotron Light Source beamline X29 at Brookhaven National Laboratory, except for the selenomethionine dataset, which was collected at 24-ID-E of the Advanced Photon Light Source at Argonne National Laboratory.
Supplementary Material
Acknowledgments
We thank Rebecca M. Davis for strain construction and Dr. Seong-Joo Lee for collecting the selenomethionine dataset. This work was supported by a National Science Foundation Graduate Research Fellowship (to D.J.S.); the Harvard Blavatnik Biomedical Accelerator Fund; National Institutes of Health Grants AI081059 (to D.K.), GM066174 (to D.K.), and GM094263 (to S.W.); and startup funds from The Ohio State University (to N.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4P31, 4P32, and 4P33).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323516111/-/DCSupplemental.
References
- 1.Dassa E, Bouige P. The ABC of ABCS: A phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152(3-4):211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 2.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11(7):1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborn MJ, Gander JE, Parisi E. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J Biol Chem. 1972;247(12):3973–3986. [PubMed] [Google Scholar]
- 7.Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247(12):3962–3972. [PubMed] [Google Scholar]
- 8.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101(25):9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45(5):1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105(14):5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandeo P, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189(1):244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperandeo P, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190(13):4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperandeo P, Pozzi C, Dehò G, Polissi A. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol. 2006;157(6):547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103(31):11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chng SS, Gronenberg LS, Kahne D. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49(22):4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freinkman E, Okuda S, Ruiz N, Kahne D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry. 2012;51(24):4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: The long road of discovery. Nat Rev Microbiol. 2009;7(9):677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sperandeo P, Dehò G, Polissi A. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta. 2009;1791(7):594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Gronenberg LS, Kahne D. Development of an activity assay for discovery of inhibitors of lipopolysaccharide transport. J Am Chem Soc. 2010;132(8):2518–2519. doi: 10.1021/ja910361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita S, Tokuda H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 2009;583(13):2160–2164. doi: 10.1016/j.febslet.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 21.Sherman DJ, Okuda S, Denny WA, Kahne D. Validation of inhibitors of an ABC transporter required to transport lipopolysaccharide to the cell surface in Escherichia coli. Bioorg Med Chem. 2013;21(16):4846–4851. doi: 10.1016/j.bmc.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338(6111):1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran AX, Dong C, Whitfield C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem. 2010;285(43):33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277(24):21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oswald C, Holland IB, Schmitt L. The motor domains of ABC-transporters. What can structures tell us? Naunyn Schmiedebergs Arch Pharmacol. 2006;372(6):385–399. doi: 10.1007/s00210-005-0031-4. [DOI] [PubMed] [Google Scholar]
- 26.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24(11):1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst R, et al. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc Natl Acad Sci USA. 2008;105(13):5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karpowich N, et al. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure. 2001;9(7):571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt L, Benabdelhak H, Blight MA, Holland IB, Stubbs MT. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: Identification of a variable region within ABC helical domains. J Mol Biol. 2003;330(2):333–342. doi: 10.1016/s0022-2836(03)00592-8. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol. 2006;1(6):385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 31.Davidson AL, Sharma S. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J Bacteriol. 1997;179(17):5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido K, Ames GF. One intact ATP-binding subunit is sufficient to support ATP hydrolysis and translocation in an ABC transporter, the histidine permease. J Biol Chem. 1999;274(38):26727–26735. doi: 10.1074/jbc.274.38.26727. [DOI] [PubMed] [Google Scholar]
- 33.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22(1):1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 34.Shyamala V, Baichwal V, Beall E, Ames GF. Structure-function analysis of the histidine permease and comparison with cystic fibrosis mutations. J Biol Chem. 1991;266(28):18714–18719. [PubMed] [Google Scholar]
- 35.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 36.Hung LW, et al. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396(6712):703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 37.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101(7):789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 39.Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446(7132):213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 40.Mourez M, Hofnung M, Dassa E. Subunit interactions in ABC transporters: A conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 1997;16(11):3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldham ML, Chen J. Snapshots of the maltose transporter during ATP hydrolysis. Proc Natl Acad Sci USA. 2011;108(37):15152–15156. doi: 10.1073/pnas.1108858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith PC, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10(1):139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janas E, et al. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J Biol Chem. 2003;278(29):26862–26869. doi: 10.1074/jbc.M301227200. [DOI] [PubMed] [Google Scholar]
- 44.Zaitseva J, et al. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 2006;25(14):3432–3443. doi: 10.1038/sj.emboj.7601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 46.Ruiz N, Falcone B, Kahne D, Silhavy TJ , eds (2005) Chemical conditionality: A genetic strategy to probe organelle assembly. Cell 121(2):307–317. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.