Significance
In New Zealand, nine species of moa (large, wingless ratite birds) went extinct shortly after Polynesian settlement. In this study, we characterize the gene pools of four moa species during the final 4,000 y of their existence and gain new insights into moa biology and their population sizes. Our analyses show that moa populations were large and viable prior to human arrival in New Zealand, and their demise therefore represents a striking example of human overexploitation of megafauna.
Abstract
The extinction of New Zealand's moa (Aves: Dinornithiformes) followed the arrival of humans in the late 13th century and was the final event of the prehistoric Late Quaternary megafauna extinctions. Determining the state of the moa populations in the pre-extinction period is fundamental to understanding the causes of the event. We sampled 281 moa individuals and combined radiocarbon dating with ancient DNA analyses to help resolve the extinction debate and gain insights into moa biology. The samples, which were predominantly from the last 4,000 years preceding the extinction, represent four sympatric moa species excavated from five adjacent fossil deposits. We characterized the moa assemblage using mitochondrial DNA and nuclear microsatellite markers developed specifically for moa. Although genetic diversity differed significantly among the four species, we found that the millennia preceding the extinction were characterized by a remarkable degree of genetic stability in all species, with no loss of heterozygosity and no shifts in allele frequencies over time. The extinction event itself was too rapid to be manifested in the moa gene pools. Contradicting previous claims of a decline in moa before Polynesian settlement in New Zealand, our findings indicate that the populations were large and stable before suddenly disappearing. This interpretation is supported by approximate Bayesian computation analyses. Our analyses consolidate the disappearance of moa as the most rapid, human-facilitated megafauna extinction documented to date.
The causes of Late Quaternary megafauna extinctions continue to be debated (e.g., refs. 1–3). Climate has been invoked as a major factor driving demographic shifts over evolutionary timescales, but it is undeniable that most recent megafauna extinctions occurred in the presence of humans. However, the role of humans in the extinction process differs among continents and the species studied (4), and it has proven difficult to find evidence for a direct causative link between anthropogenic activity and megafauna loss.
Ancient DNA (aDNA) research has contributed significantly to the extinction debate. DNA extracted from fossil material spanning thousands of years can yield insights into the demographic histories of extirpated populations and extinct species in the period leading up to their loss (5–7). However, most efforts have involved continental-scale data where the large geographic distances hamper fine-scale inferences, limiting our ability to determine the causative agents of discrete demographic events. In contrast, island extinctions offer analytical advantages not afforded by studies of widely distributed species. In island endemics, the absence of gene flow from mainland populations allows us to disregard the spatial dimension in the genetic analyses and focus on extinction dynamics through time.
New Zealand is central to the megafauna extinction debate. It was the last major landmass to be colonized by humans and harbored a diverse assemblage of avian megafauna (8–10). Among them were nine species of moa (11): large, wingless ratite birds ranging in size from the ∼12-kg North Island morph of Euryapteryx curtus to the ∼250-kg females of the two Dinornis species (8). Moa inhabited a variety of habitats across the New Zealand archipelago until their extinction shortly after the arrival of Polynesian settlers, estimated at approximately the late 13th century (8–10, 12). The abundance of well-preserved archaeological sites containing evidence of large-scale exploitation of moa (e.g., ref. 13) brings the controversy of the role of humans in the extinction event into sharp focus.
Early claims of environmental changes or poor adaptive abilities of moa as causes for the extinction (reviewed in ref. 8) have now been largely replaced by the view that direct or indirect human impacts—including hunting, fires, and the introduction of exotic species—were the primary drivers (14–18). Ecological modeling suggests that such human-mediated extinction could have happened within 100 y of Polynesian colonization (10). In contrast, it has been argued, based on limited mitochondrial DNA (mtDNA) data, that moa populations had already collapsed before human arrival, as a consequence of volcanic eruptions or diseases, suggesting that humans were just one of several additive factors responsible for the extinction (19).
To address this issue, we investigated the demographic trajectories of four sympatric moa species in the four millennia leading up to their extinction. We genotyped 281 individuals of Dinornis robustus (Dinornithidae), Euryapteryx curtus, Pachyornis elephantopus, and Emeus crassus (all Emeidae) using mtDNA and six nuclear microsatellite markers developed specifically for moa (20, 21). Moa were recovered from five fossil sites within a 10-km radius in North Canterbury on New Zealand’s South Island (Fig. 1). Through a series of genetic analyses, we have gained insights into moa paleobiology, population sizes, and reproductive success, and we have specifically addressed whether moa populations were in decline before Polynesian settlement of New Zealand.
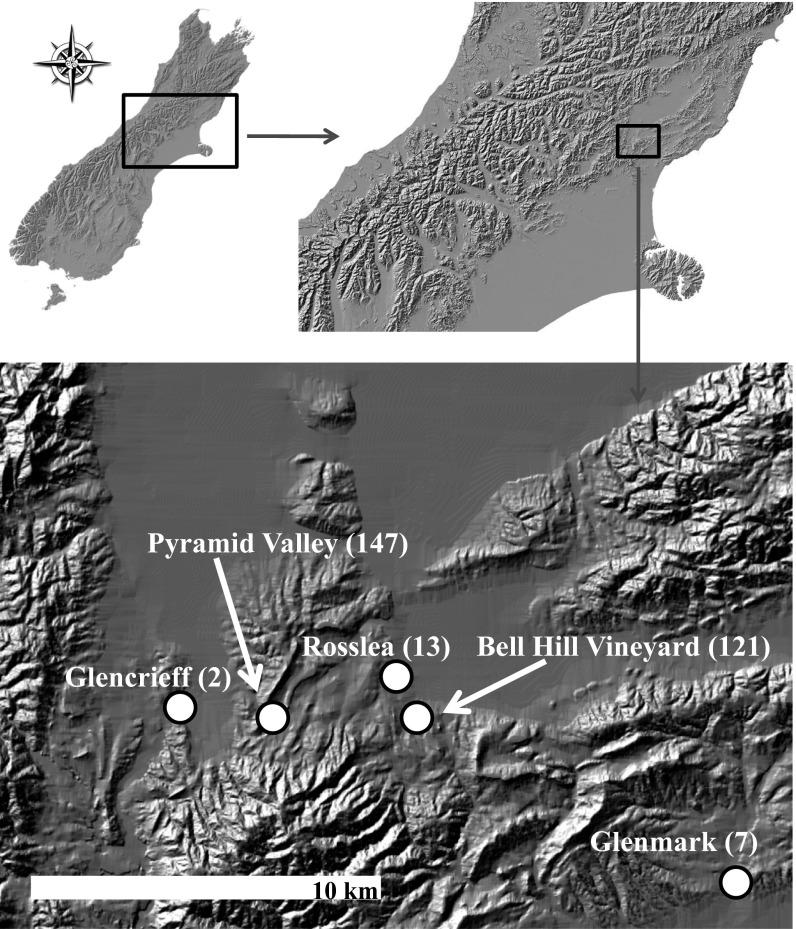
Fig. 1.
Map of the study sites in North Canterbury, South Island, New Zealand. All sites represent well-described moa fossil assemblages, with geographical coordinates as follows: Pyramid Valley (42°58′22.0″S, 172°35′49.0″E) (42), Bell Hill Vineyard (42°58′19.36″S, 172°39′56.15″E) (37), Rosslea (42°57′53.83″S, 172°39′22.39″E) (44), Glenmark (43°00′00.0″S, 172°46′50.0″E) (43), and Glencrieff (42°58′07.45″S, 172°34′01.84″E) (57). Sample sizes are in parentheses.
Results
Summary Statistics.
The 217 radiocarbon-dated individuals covered a calibrated chronology from 12,966 B.P. to 602 B.P. (Table 1), although only 13 samples were from >4,000 y B.P. (see individual sample ages in Dataset S1). We successfully amplified mtDNA sequences from 281 of 290 samples [GenBank accession nos. KJ533749–KJ533834 (D. robustus), KJ533835–KJ533912 (E. crassus), KJ533913–KJ533991 (E. curtus), KJ533992–KJ534022 (P. elephantopus)] and retrieved reliable (20) microsatellite genotypes from 188 of the 217 radiocarbon-dated individuals.
Table 1.
Time span of calibrated radiocarbon ages for sites and species
| Site/taxon | Sampled | Dated | Age |
| Site | |||
| Pyramid Valley | 147 | 127 | 4136–602 B.P. |
| Bell Hill Vineyard | 121 | 74 | 2876–851 B.P. |
| Rosslea | 13 | 8 | 7839–1482 B.P. |
| Glenmark | 7 | 6 | 5152–782 B.P. |
| Glencrieff | 2 | 2 | 12966–4837 B.P. |
| Total | 290 | 217 | 12966–602 B.P. |
| Taxon | |||
| D. robustus | 89 | 88 | 12966–602 B.P. |
| P. elephantopus | 32 | 32 | 4067–853 B.P. |
| E. curtus | 84 | 40 | 5812–945 B.P. |
| E. crassus | 85 | 57 | 5791–874 B.P. |
| Total | 290 | 217 | 12966–602 B.P. |
Of 24 taxon–locus combinations, only two significant deviations from Hardy–Weinberg proportions were observed (α = 0.05) and only one after Bonferroni correction (Table S1). No tests for linkage disequilibrium among loci were significant (P > 0.07) except for one instance in P. elephantopus (Moa_MS2/Moa_MA1; P = 0.0005). The linkage could have resulted from a 106-bp homozygous profile in the Moa_MS2 locus, which was accompanied by a 90-bp homozygous profile in the Moa_MA1 locus in five individuals. This finding was probably an effect of the heterochronous data. Using established criteria (20), we did not find evidence of scoring bias from null alleles, allelic dropout, or stuttering in any of the datasets.
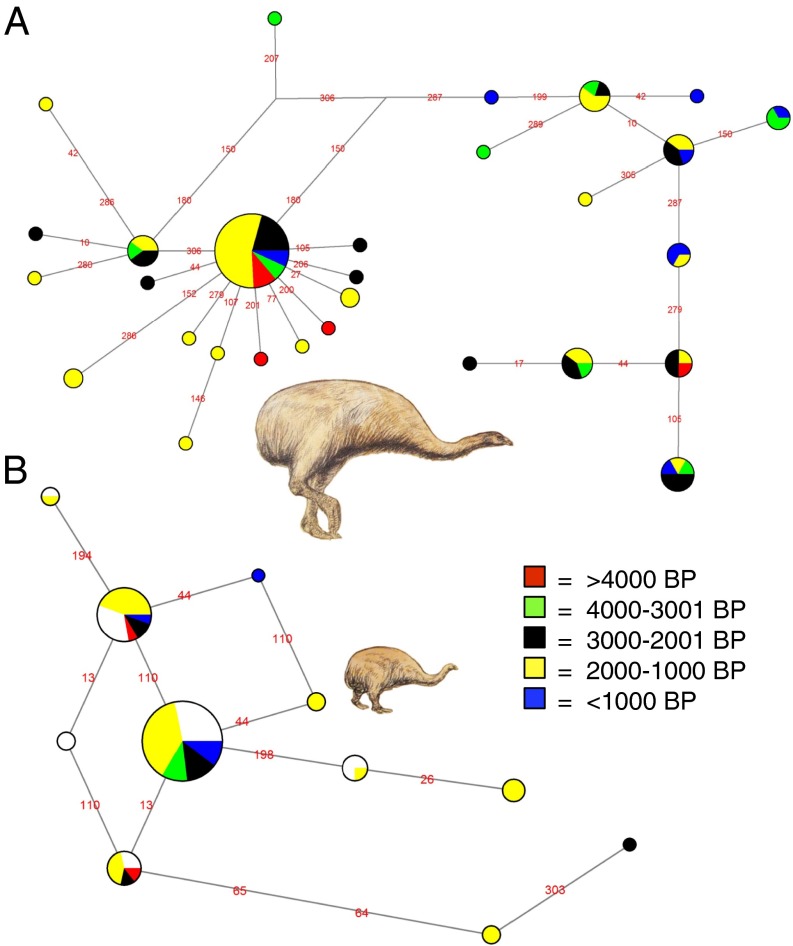
The four species differed in their levels of genetic diversity in both the mtDNA and microsatellite data (Figs. 2 and 3 and Table 2). Differences are visualized in the mtDNA haplotype networks (Fig. 2), where the E. crassus gene pool is largely dominated by two haplotypes separated by a single mutation. Haplotype networks for P. elephantopus and E. curtus are provided in Fig. S1.
Fig. 2.
mtDNA haplotype networks of D. robustus (n = 87) (A) and E. crassus (n = 81) (B), based on ∼340 bp of mtDNA. The color composition of each haplotype is defined by the individual radiocarbon ages of the fossils. See Table 2 for summary statistics.
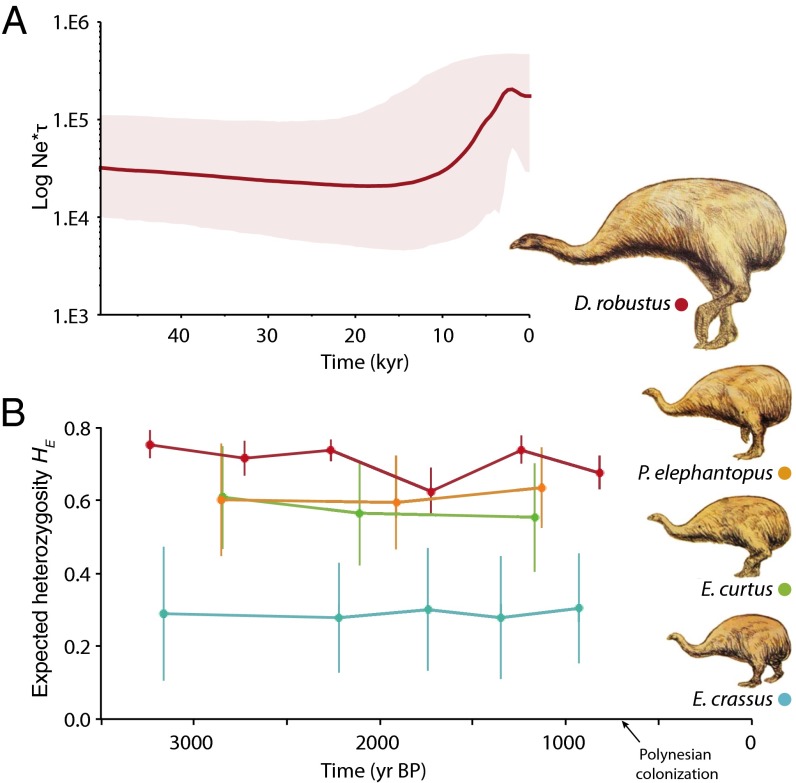
Fig. 3.
Demographic history and genetic diversity. (A) Bayesian skyline plot for D. robustus (n = 87), where the y axis depicts the effective female population size multiplied by generation time. Year zero corresponds to the age of the youngest sample at 602 B.P. (B) Expected heterozygosity (HE) for six microsatellite loci, measured across time in the four moa species (n = 188). Data points represent the mean age and mean HE (with SE) of the moa individuals in 1,000-y time bins.
Table 2.
Genetic diversity
| mtDNA |
Microsatellites |
|||||||||||
| Taxon | n | Length, bp | h | S | π | k | n | NA | NE | HO | HE | FIS |
| D. robustus | 87 | 341 | 29 | 24 | 0.010 | 3.44 | 74 | 9.5 | 3.8 | 0.687 | 0.721 | 0.040 |
| P. elephantopus | 31 | 337 | 13 | 15 | 0.008 | 2.55 | 30 | 7.2 | 3.9 | 0.561 | 0.601 | 0.041 |
| E. curtus | 82 | 338 | 16 | 15 | 0.007 | 2.29 | 29 | 8.7 | 4.6 | 0.600 | 0.570 | −0.079 |
| E. crassus | 81 | 337 | 11 | 9 | 0.004 | 1.19 | 55 | 4.7 | 2.4 | 0.305 | 0.288 | −0.048 |
Intraspecific summary statistics for the four species. n, number of analyzed individuals for mtDNA and nuclear microsatellite loci; length of analyzed DNA fragment excluding primers; h, observed number of haplotypes; S, number of segregating sites; π, nucleotide diversity; k, average number of nucleotide differences between two sequences; NA, average number of observed alleles per locus; NE, average number of effective alleles per locus; HO, observed heterozygosity; HE, expected heterozygosity; FIS, fixation index.
The microsatellite markers were developed specifically for D. robustus (20, 21), and we therefore cannot eliminate ascertainment bias as being responsible for the lower levels of genetic diversity observed in the other species. However, the congruence between the relative diversity levels of the mtDNA and microsatellite data supports the validity of our data (Table 2). Also, ascertainment cannot explain the much lower genetic diversity in E. crassus in comparison with that in the other emeids (E. curtus and P. elephantopus); these three species are phylogenetically equidistant from D. robustus (11).
Genetic Structuring.
We found strong genetic differentiation among the four moa species (Fig. S2). Only one of the 188 individuals with microsatellite profiles was assigned to the wrong species. Morphologically, this tibiotarsus (AV8470 from Pyramid Valley) could only be a female D. robustus, and the individual exhibited a rare combination of alleles for this species. We found no temporal genetic structuring within species, even when manipulating datasets to comprise only the 10 oldest and 10 youngest individuals for each species (Fig. S2). This finding was supported by a lack of isolation by time using the Mantel test (P values ranging from 0.24 to 0.48) and small insignificant FST values across time bins. In D. robustus, <2.8% of the genetic variation was explained by temporal differentiation, and no pairwise FST values were significant (Table S2).
Demographic History.
Bayesian Skyline plots were generated using the software BEAST (22) and the plots from the three emeid species (P. elephantopus, E. curtus, and E. crassus) displayed flat lines with large highest posterior densities (HPDs), likely reflecting a lack of power in the data to model a reliable genealogy, as has been reported for other aDNA megafauna data (4). We found support for a modest population expansion in D. robustus before human settlement of New Zealand. Despite the relatively large HPDs in the skyline plot, we found an increase in population size in ∼13,000 B.P. (Fig. 3 and Table S3), although we were unable to reject the constant population size model as an alternative fit for the data. This increase was supported by the free model of demographic change, which always yielded a posterior distribution with positive values for growth rate. Also, an expand model proved superior to a decline model, which consistently yielded -infinity likelihood values (Table S3).
The results from BEAST were supported by approximate Bayesian computation (ABC) analyses, which encompassed both mtDNA and microsatellite data. Under the free-model approach, the modal value for the demographic change parameter (Nanc/Ncur) was 0.9 (90% HPD interval 0.26–25.84), indicating that the population was unlikely to have changed dramatically in the period 31,700–100 y before the youngest sample (Table 3). The time of onset of demographic change had a wide posterior distribution (Table 3), as expected if the population size were constant or only changed slightly. The effective population size at the time of human arrival was estimated to be 9,200 individuals, but also with wide HPDs (Table 3).
Table 3.
ABC analysis
| Model | Ncur | Nanc/Ncur | T | µ | p | Marginal density |
| Priors | ||||||
| Free model | 500–50,000 | 0.01–100 | 100–31,700 | 10−3–10−5 | 0.4–0.9 | 0.31 |
| Model selection: decline | 500–50,000 | 1–100 | 100–31,700 | 10−3–10−5 | 0.4–0.9 | 0.24 |
| Model selection: expand | 500–50,000 | 0.01–1 | 100–31,700 | 10−3–10−5 | 0.4–0.9 | 1.26 |
| Posteriors | ||||||
| Free model | 9,200 [900–45,600] | 0.90 [0.26–25.84] | 1,100 [100–15,900] | 5.1e-5 [2.4e-5–1.1e-4] | 0.82 [0.75–0.89] | |
| Model selection: expand | 10,200 [3800–37,900] | 0.47 [0.04–1.00] | 1,900 [1100–19,500] | 4.8e-5 [2.4e-5–9.8e-5] | 0.81 [0.72–0.89] |
ABC analysis of D. robustus mtDNA and microsatellite data combined. Priors were incorporated on a log scale with a uniform distribution, but here they are shown on a natural scale for clarity. Ncur is “current” population size, representing effective population size at the last sampling point (602 B.P.). Nanc/Ncur is the size of the “ancient” effective population size at the onset of demographic change relative to Ncur. Numbers are diploid individuals rounded to the nearest 100, converted from haploids as used in ABCtoolbox. T is the time at the onset of the demographic change measured in years relative to the youngest sampling point, rounded to the nearest 100, and converted from generations, as used in ABCtoolbox (generation time assumed to be 10 y for D. robustus). µ is the microsatellite mutation rate per generation, and p is the shape parameter of the geometric distribution of the General Stepwise Model. Marginal densities were compared for the two scenarios in the model selection approach, and the Bayes factor support for the expand model was 5.25 (1.26/0.24), indicating substantial support (Jeffreys; ref. 58). Posterior parameter estimates represented by mode values, and 90% HPDs are shown for the two models with highest marginal density (free and expand models).
In the model selection approach, we calculated a Bayes factor of 5.25 in favor of the expand model over the decline model. We observed good posterior coverage and high information content of the summary statistics, making us confident that the ABC analyses were informative with regard to model choice and parameter estimation (Table S4).
Despite allowing the effective population size of the mtDNA (Nemt) to remain independent of the autosomal (microsatellite) effective size (Nems) so as to assess whether there was support for an unequal reproductive contribution of the two sexes, Nemt remained close to the theoretically expected value of Nems/4 (posterior mode of Nemt = 3,200; posterior mode of Nems = 15,800). Hence, we fixed Nemt to a quarter of Nems in all reported simulations.
Discussion
No Precolonization Decline in Moa.
Our genetic data have provided a unique source of information regarding community-level megafauna population dynamics preceding the extinction event. We applied a range of methods to analyze mtDNA and microsatellite data and failed to detect any evidence of moa decline, suggesting that the populations were large and viable throughout the Holocene until their sudden loss.
Temporal sampling is required to directly observe changes in genetic variability over time. To date, relatively few aDNA studies have included microsatellite data; those that did have shown that microsatellite analyses can be a powerful tool to detect temporal loss of genetic diversity and changes in allele frequencies (23–26). We observed no loss of genetic diversity in any of the four moa species (Fig. 3). Moreover, we observed a star-like haplotype network in D. robustus, which suggests an increase in population size (27, 28), although the signal may also reflect an artifact of analyzing sequence data sampled across different points in time (29, 30). However, our detailed demographic analyses of D. robustus, using BEAST and ABC, take temporal sampling into account and support a scenario of a slowly increasing population size during the Holocene.
All moa were sampled from within a 10-km radius, and we can therefore disregard geographic structuring patterns and focus exclusively on genetic patterns through time. Our observation of Hardy–Weinberg proportions and no linkage disequilibrium in the moa populations could be interpreted as random mating. However, these samples span several millennia, and, rather, the result reflects that the microsatellite allele frequencies remained approximately constant during the entire second half of the Holocene. This analysis is also supported by the absence of temporal genetic structuring and lack of isolation by time. Our results reflect very low levels of genetic drift, in agreement with a recent mtDNA analysis of the moa genus Pachyornis, which also failed to detect significant demographic shifts during this period (31).
Despite comprehensive genetic analyses of four moa species, we found no genetic signatures of a hypothesized Holocene prehuman decline. The previous study describing this population collapse (19) may have been compromised by some questionable assumptions (discussed in ref. 32). Rather, our results indicate that the D. robustus population increased slowly at the onset of the Holocene, which seems reasonable from an ecological perspective. Moa were primarily forest and shrubland dwellers, with some entering herbfields and the subalpine zone (8, 33), and pollen records from the South Island reflect an establishment of postglacial shrubland in 14,000–10,000 y B.P., followed by podocarp forest expansion during approximately 13,600–7,500 y B.P. (34, 35).
Population Size of D. robustus.
Our data suggest that D. robustus had an effective population size (Ne) of ∼9,200 individuals when Polyneisans reached New Zealand. Although the posterior distributions are wide, this is the modal value from the free-model ABC analysis (Table 3), which encompasses both mtDNA and nuclear microsatellite data and is without any directional restrictions on simulated growth rate. The conversion of effective population size to census population size (Nc) is problematic, especially for extinct taxa with limited biological information, and no Ne:Nc ratios have been published for extant ratites. However, our best estimate of Ne:Nc ratio for D. robustus is 0.4 (SI Materials and Methods). With an Ne of 9,200, we therefore estimate a census size of ∼23,000 D. robustus individuals, which is the same order of magnitude as the 14,100 individuals estimated from ecological modeling (SI Materials and Methods and Datasets S2 and S3). Such a large population size of a heavy and slowly maturing species (36) suggests a larger, panmictic South Island population, rather than a local enclave isolated in North Canterbury. This finding suggests that D. robustus individuals could disperse over long distances and sustain population connectivity in a landscape fragmented by rivers, glaciers, and mountain ranges. It seems reasonable to infer that for flightless birds, larger body size translates into higher dispersal rates, resulting in larger effective population sizes and higher levels of genetic diversity. This hypothesis could also explain why the smallest species of the region (E. crassus) had the lowest observed genetic diversity (Table 2).
The fossil record suggests that moa had skewed sex ratios with an excess of females (8, 37). If the sexes did not contribute evenly to the effective population size, we would expect a deviation from a 1:4 ratio between mtDNA and microsatellite (autosomal) effective population sizes. Indeed, we observe a 1:4 ratio in the ABC analysis of D. robustus, suggesting that putative skewed sex ratios did not cause differential reproductive success between the sexes. Our result could indicate mating competition among females and accordance with the observation of pronounced reverse sexual size-dimorphism (38, 39) and hypotheses of female territoriality (37) in moa.
Genetic Diversity in the Moa Community.
We found differing levels of genetic diversity in the four moa species, although each remained constant through time (Fig. 3 and Table 2). In the four millennia preceding extinction, we found only half the genetic diversity in E. crassus compared to the three other moa species, suggesting either a smaller population size or a previous demographic bottleneck. Unlike E. curtus and P. elephantopus, E. crassus was not present in the wetter western and northwestern South Island during the Pleistocene (40); the more limited distribution likely resulted in smaller population sizes. If a bottleneck was the cause of lower levels of genetic diversity in E. crassus, it could reflect population isolation in a habitat refugium during the Otiran glaciation, during ∼74,000–17,900 y B.P. (41), followed by recolonization of the eastern lowlands, including North Canterbury, when suitable habitat increased at the onset of the Holocene (e.g., ref. 11). Despite markedly lower genetic diversity, E. crassus appears to have been thriving in the Eastern forests, where late Holocene fossils have been found in great abundance (42, 43).
Conclusion
This study has highlighted the paleobiological insights that can be gained from ancient population genetics beyond the traditional analyses of mtDNA. We profiled 281 individual moa of four species in the 4,000 y preceding their extinction by combining radiocarbon dating, mtDNA sequencing, and nuclear microsatellite genotyping. We observed differing levels of genetic diversity between species in the Holocene moa community. For D. robustus, we found an equal reproductive output between sexes and estimated the population size and demography leading up to the extinction event. Interestingly, the moa extinction process did not leave any genetic traces in our data, very likely because it was too short for increased genetic drift to have an effect on the gene pools. Our results do not support a collapse in any of the moa populations in the millennia preceding Polynesian settlement of New Zealand (19). Rather, our detailed analysis of D. robustus indicated that this moa species increased in numbers during the Holocene. When humans arrived in New Zealand they encountered a large and perhaps still increasing D. robustus population with an estimated effective size of 9,200 individuals. From the archaeological record, we know that moa were hunted intensively and that D. robustus disappeared along with eight other moa species within just one or two centuries following human arrival (10). Together, these findings point strongly toward human contact as the only factor responsible for the extinction.
Materials and Methods
Sampling, Extraction, Identification, and Age.
A total of 290 moa fossils from five adjacent Holocene fossil deposits (Fig. 1) were sampled according to established protocols (21, 44). To avoid including more than one sample from each individual, only left tibiotarsi were sampled, except for one right femur (Canterbury Museum, catalogue no. AV 41188) that was not associated with any of the tibiotarsi (based on consideration of fossil site, bone size, and preservation). DNA extractions from 200-mg aliquots of bone powder were performed in a dedicated aDNA facility (Murdoch University) by using a silica-column-based method (21). Genetic sex identifications were taken from previous work (37, 44), as were 158 of the 217 calibrated radiocarbon ages (45); the experimental procedures are detailed in these references. The dates presented in this study are median calibrated ages (years B.P.) using the SHCAL04 curve in OxCal (Version 4.1; Oxford Radiocarbon Accelerator Unit). An overview of the material is presented in Table 1, and individual data are in Dataset S1.
PCR and Authentication.
Two primer sets (185F/294R and 262F/441R) were used to amplify a 337- to 341-bp fragment (excluding primers) of the moa mtDNA control region. This region has proven informative for assessing intra- and interspecific genetic differentiation in moa (11); PCR conditions are described in previous studies (37, 38). PCR products were sequenced in both directions, and samples with DNA sequences that continually yielded ambiguous base calls (9 of 290 samples) were excluded from further analyses, yielding a total of 281 samples. Samples with mutations, appearing less than three times in the overall sequence alignments, were sequenced and observed at least twice from independent PCRs before being accepted. Information on the six microsatellite loci and their PCR conditions are provided in previous work (20, 21). To overcome the challenges introduced by allelic dropout, we followed the strict guidelines set out in ref. 20. We restricted the microsatellite profiling attempts to the 217 radiocarbon-dated individuals.
Summary Statistics.
The mtDNA sequences were aligned in Geneious (Version 4.8.3; ref. 46) and imported into DNASP (Version 5.10; ref. 47) to calculate genetic diversity. Microsatellite genetic diversity was calculated in GenAlEx (Version 6.4; ref. 48). Deviations from Hardy–Weinberg proportions were quantified as FIS (49), and significance was tested with the exact test in GENEPOP (Version 4.0.10; ref. 50). Deviations from linkage equilibrium were also tested with GENEPOP. We used MICRO-CHECKER (Version 2.2.3; ref. 51) to investigate the presence of null alleles and allelic dropout.
Genetic Structure.
To visualize the genetic structure and diversity of the mtDNA sequences in each of the four species, we generated median-joining haplotype networks with NETWORK (Version 4.5; Fluxus-engineering). We used STRUCTURE (Version 2.3.3; ref. 52) on the microsatellite data to perform several analyses to detect genetic structuring within and among species (SI Materials and Methods). The fixation index (FST) was used to quantify intraspecific genetic differentiation based on the microsatellite data. Because all individuals were sampled within a radius of 10 km, geographic differentiation was not relevant to our study, and we instead investigated genetic differentiation across time. Using the age of each sample, we constructed temporal groups by pooling individuals into 1,000 calendar years. FST values between all pairs of groups were calculated (49), and the significance of the differentiation was assessed by a permutation test using FSTAT (Version 2.9.3; ref. 53). A Mantel test is commonly applied to assess genetic isolation by distance, but it should be equally suitable for detection of genetic ”isolation by time.” Based on microsatellite data, matrices of temporal and genetic distances between all pairs of individuals (within each species separately) were generated and tested for correlations by using the Mantel test in GenAlEx (Version 6; ref. 48). The temporal distances were recorded as the number of calendar years that separated two individuals. We assessed the P value of each correlation with 1,000 randomizations.
Demographic History.
We used two overall methods to estimate demographic history: BEAST and ABC. We analyzed the 217 radiocarbon-dated mtDNA sequences in BEAST (Version 1.6.1; ref. 22). JMODELTEST (Version 0.1.1; ref. 54) and the Akaike Information Criterion (55) were used to estimate the most likely substitution model, favoring a HKY+I+G model for all four species. Prior values on parameters associated with this substitution model were estimated in JMODELTEST and incorporated in BEAST by using wide prior distributions. We analyzed each species dataset using the Bayesian skyline model, which allows population sizes to fluctuate freely through time as governed by the data. D. robustus represented the largest of the datasets (most radiocarbon dates; Table 1) and is the species for which a Holocene decline has been claimed (19). We therefore analyzed this species in more detail and tested an additional four demographic models (Table S3). These models were a constant population size model and three different single-change-point models, assuming a constant population size replaced by an exponential change in population size. These three models were (i) a free model, allowing both exponential growth and decline; (ii) an expand model allowing only growth, as expected when more favorable moa habitat became available after the end of the most recent glaciation in New Zealand, 74,000–17.900 y B.P. (41); and (iii) a decline model, allowing only population decline, following the scenario suggested previously (19). Parameter details are provided in SI Materials and Methods and Table S3. BEAST output files were analyzed in TRACER (Version 1.5; ref. 56) after removing the first 10% of the trees as burn-in. The different models were compared with marginal likelihoods after 1,000 bootstraps.
We used ABC simulations to further elucidate the demographic history of D. robustus. Comparable with the BEAST analyses, we used two different approaches: the free model approach, under which the population size in the most recent phase of the demographic history was allowed to change by a factor of 100 (decline or expansion), and the model selection approach, in which we compared two discrete scenarios of historical population dynamics with declining or expanding population size (Table 3) and compared their respective fit to our observed data using 19 summary statistics (Table S5). See SI Materials and Methods for details on the ABC analyses.
Supplementary Material
Acknowledgments
We thank the Museum of New Zealand, Te Papa Tongarewa (A. J. D. Tennyson), Canterbury Museum (P. Scofield), and the American Museum of Natural History (C. Mehling). We also thank Eske Willerslev, Malene Møhl, James Haile, and Ross Barnett for assistance with the sequencing and sampling; Jayne Houston, Emma McLay, Helen Hunt, and Frances Brigg for technical assistance; Daniel Wegmann, Simon Ho, Christian Anderson, Alexei Drummond, and Che Si Wu for advice on the analyses; and Marcel Giesen (Bell Hill Vineyard) and the Hodgen family (Pyramid Valley) for their support of our research. This work was supported by Marsden Fund of the Royal Society of New Zealand Contracts 06-PAL-001-EEB (to Palaecol Research Ltd.; R.N.H.) and 09-UOO-164 (to Otago University; C.J.). M.E.A. is supported by European Research Council Marie Curie Actions Grant Agreement 300554. M.B. is supported by Australian Research Council Future Fellowship FT0991741.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KJ533749–KJ533834 (D. robustus), KJ533835–KJ533912 (E. crassus), KJ533913–KJ533991 (E. curtus), and KJ533992–KJ534022 (P. elephantopus)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314972111/-/DCSupplemental.
References
- 1.Alroy J. A multispecies overkill simulation of the end-Pleistocene megafaunal mass extinction. Science. 2001;292(5523):1893–1896. doi: 10.1126/science.1059342. [DOI] [PubMed] [Google Scholar]
- 2.Koch PL, Barnosky AD. Late quaternary extinctions: State of the debate. Annu Rev Ecol Evol Syst. 2006;37:215–250. [Google Scholar]
- 3.Stuart AJ, Kosintsev PA, Higham TFG, Lister AM. Pleistocene to Holocene extinction dynamics in giant deer and woolly mammoth. Nature. 2004;431(7009):684–689. doi: 10.1038/nature02890. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzen ED, et al. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 2011;479(7373):359–364. doi: 10.1038/nature10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro B, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306(5701):1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 6.Campos PF, et al. Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. Proc Natl Acad Sci USA. 2010;107(12):5675–5680. doi: 10.1073/pnas.0907189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palkopoulou E, et al. Holarctic genetic structure and range dynamics in the woolly mammoth. Proc Biol Sci. 2013;280(1770):20131910. doi: 10.1098/rspb.2013.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worthy TH, Holdaway RN. The Lost World of the Moa. Christchurch, New Zealand: Canterbury Univ Press; 2002. p. 718. [Google Scholar]
- 9.Anderson A. Mechanics of overkill in the extinction of New-Zealand moas. J Archaeol Sci. 1989;16(2):137–151. [Google Scholar]
- 10.Holdaway RN, Jacomb C. Rapid extinction of the moas (Aves: Dinornithiformes): Model, test, and implications. Science. 2000;287(5461):2250–2254. doi: 10.1126/science.287.5461.2250. [DOI] [PubMed] [Google Scholar]
- 11.Bunce M, et al. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc Natl Acad Sci USA. 2009;106(49):20646–20651. doi: 10.1073/pnas.0906660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higham T, Anderson A, Jacomb C. Dating the first New Zealanders: The chronology of Wairau Bar. Antiquity. 1999;73(280):420–427. [Google Scholar]
- 13.Oskam CL, et al. Molecular and morphological analyses of avian eggshell excavated from a late thirteenth century earth oven. J Archaeol Sci. 2011;38(10):2589–2595. [Google Scholar]
- 14.Anderson A. Prodigious Birds. Cambridge, UK: Cambridge Univ Press; 1989. [Google Scholar]
- 15.Holdaway RN. New Zealand's pre-human avifauna and its vulnerability. N Z J Ecol. 1989;12:11–25. [Google Scholar]
- 16.Holdaway RN. Arrival of rats in New Zealand. Nature. 1996;384(6606):225–226. [Google Scholar]
- 17.McWethy DB, Whitlock C, Wilmshurst JM, McGlone MS, Li X. Rapid deforestation of South Island, New Zealand, by early Polynesian fires. Holocene. 2009;19(6):883–897. [Google Scholar]
- 18.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc Natl Acad Sci USA. 2008;105(22):7676–7680. doi: 10.1073/pnas.0801507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gemmell NJ, Schwartz MK, Robertson BC. Moa were many. Proc Biol Sci. 2004;271(Suppl 6):S430–S432. doi: 10.1098/rsbl.2004.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allentoft ME, et al. Profiling the dead: Generating microsatellite data from fossil bones of extinct megafauna—protocols, problems, and prospects. PLoS ONE. 2011;6(1):e16670. doi: 10.1371/journal.pone.0016670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allentoft ME, et al. Identification of microsatellites from an extinct moa species using high-throughput (454) sequence data. Biotechniques. 2009;46(3):195–200. doi: 10.2144/000113086. [DOI] [PubMed] [Google Scholar]
- 22.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7(214):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitehouse AM, Harley EH. Post-bottleneck genetic diversity of elephant populations in South Africa, revealed using microsatellite analysis. Mol Ecol. 2001;10(9):2139–2149. doi: 10.1046/j.0962-1083.2001.01356.x. [DOI] [PubMed] [Google Scholar]
- 24.Nyström V, et al. Microsatellite genotyping reveals end-Pleistocene decline in mammoth autosomal genetic variation. Mol Ecol. 2012;21(14):3391–3402. doi: 10.1111/j.1365-294X.2012.05525.x. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd LD, et al. Microevolution and mega-icebergs in the Antarctic. Proc Natl Acad Sci USA. 2005;102(46):16717–16722. doi: 10.1073/pnas.0502281102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper GL, Maclean N, Goulson D. Analysis of museum specimens suggests extreme genetic drift in the adonis blue butterfly (Polyommatus bellargus) Biol J Linn Soc Lond. 2006;88(3):447–452. [Google Scholar]
- 27.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9(3):552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 28.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129(2):555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debruyne R, Poinar HN. Time dependency of molecular rates in ancient DNA data sets, a sampling artifact? Syst Biol. 2009;58(3):348–360. doi: 10.1093/sysbio/syp028. [DOI] [PubMed] [Google Scholar]
- 30.Depaulis F, Orlando L, Hänni C. Using classical population genetics tools with heterochroneous data: Time matters! PLoS ONE. 2009;4(5):e5541. doi: 10.1371/journal.pone.0005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlence NJ, et al. The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quaternary Science Reviews. 2012;50:141–153. [Google Scholar]
- 32.Allentoft ME, Rawlence NJ. Moa's Ark or volant ghosts of Gondwana? Insights from nineteen years of ancient DNA research on the extinct moa (Aves: Dinornithiformes) of New Zealand. Ann Anat. 2012;194(1):36–51. doi: 10.1016/j.aanat.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Wood JR, et al. Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes) Proc Natl Acad Sci USA. 2013;110(42):16910–16915. doi: 10.1073/pnas.1307700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moar NT. Late quaternary vegetation. In: Winterbourn MJ, Knox GA, Burrows CJ, Marsden ID, editors. The Natural History of North Canterbury. Christchurch, New Zealand: Canterbury Univ Press; 2008. pp. 169–192. [Google Scholar]
- 35.McGlone MS, Turney CSM, Wilmshurst JM. Late-glacial and Holocene vegetation and climatic history of the Cass basin, central south island, New Zealand. Quat Res. 2004;62(3):267–279. [Google Scholar]
- 36.Turvey ST, Green OR, Holdaway RN. Cortical growth marks reveal extended juvenile development in New Zealand moa. Nature. 2005;435(7044):940–943. doi: 10.1038/nature03635. [DOI] [PubMed] [Google Scholar]
- 37.Allentoft ME, Bunce M, Scofield RP, Hale ML, Holdaway RN. Highly skewed sex ratios and biased fossil deposition of moa: Ancient DNA provides new insight on New Zealand's extinct megafauna. Quat Sci Rev. 2010;29(5-6):753–762. [Google Scholar]
- 38.Bunce M, et al. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature. 2003;425(6954):172–175. doi: 10.1038/nature01871. [DOI] [PubMed] [Google Scholar]
- 39.Huynen L, Millar CD, Scofield RP, Lambert DM. Nuclear DNA sequences detect species limits in ancient moa. Nature. 2003;425(6954):175–178. doi: 10.1038/nature01838. [DOI] [PubMed] [Google Scholar]
- 40.Worthy TH, Holdaway RN. Quaternary fossil faunas from caves in the Punakaiki area, West Coast, South Island, New Zealand. J R Soc N Z. 1993;23(3):147–254. [Google Scholar]
- 41.Lorrey AM, et al. Palaeocirculation across New Zealand during the last glacial maximum at similar to 21 ka. Quat Sci Rev. 2012;36:189–213. [Google Scholar]
- 42.Holdaway RN, Worthy TH. A reappraisal of the late quaternary fossil vertebrates of Pyramid Valley Swamp, North Canterbury, New Zealand. NZ J Zool. 1997;24(1):69–121. [Google Scholar]
- 43.Worthy TH, Holdaway RN. Quaternary fossil faunas, overlapping taphonomies, and palaeofaunal reconstruction in north Canterbury, South Island, New Zealand. J R Soc N Z. 1996;26(3):275–361. [Google Scholar]
- 44.Allentoft ME, et al. A molecular characterisation of a newly discovered megafaunal fossil site in North Canterbury, South Island, New Zealand. J R Soc N Z. 2012;42(4):241–256. [Google Scholar]
- 45.Allentoft ME, et al. The half-life of DNA in bone: Measuring decay kinetics in 158 dated fossils. Proc Biol Sci. 2012;279(1748):4724–4733. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, et al. 2010. Geneious (Biomatters, Auckland), Version 5.1.
- 47.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 48.Peakall R, Smouse PE. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6(1):288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 50.Rousset F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8(1):103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 51.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- 52.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices, Version 2.9.3. Available at www.unil.ch/izea/softwares/fstat. Accessed August 7, 2013.
- 54.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 55.Akaike H. New look at statistical model identification. Transactions on Automatic Control. 1974;19(6):716–723. [Google Scholar]
- 56.Rambaut A, Drummond A. 2007. Tracer. Available at http://beast.bio.ed.ac.uk/Tracer. Accessed August 7, 2013.
- 57.Rawlence NJ, et al. New palaeontological data from the excavation of the Late Glacial Glencrieff miring bone deposit, North Canterbury, South Island, New Zealand. J R Soc N Z. 2011;41(3):217–236. [Google Scholar]
- 58. Jeffreys H (1961) The Theory of Probability (Oxford Univ Press, Oxford), 3rd Ed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.