Significance
The phenotypic analysis of mosaic animals has made possible the study of complex biological processes. We review the various tools available to Drosophila researchers today, many of which use the Gal4-upstream activating system and Flippase recognition target systems, as well as the use of fluorescence proteins. Emphasis is on the contributions that our group has made over the years to the development of these methods.
Keywords: clonal analysis, twin spot, Gal4-UAS, Flp-FRT
Abstract
Chimaeras, fanciful beasts that drew their force from being composed of parts of disparate animals, have stimulated our collective imagination for centuries. In modern terms, chimaeras are composite animals consisting of genetically distinct cell populations and are called “mosaics” if the different cell types have emerged from the same zygote. Phenotypic studies of chimeric animals formed from invertebrates, amphibians, birds, and mammals have provided many fundamental insights into biological processes, most notably in developmental biology. Many methods for generating both chimaeras and a range of markers for tracing their lineages have been developed over the years. Our laboratory has been intimately involved in the development of methods that facilitate the creation of genetic mosaics in Drosophila. Here, we review our contributions to the development of this field and discuss a number of approaches that will improve further the tool kit for generating mosaic animals.
In 1924, Spemann and Mangold (1) used grafting experiments with cells from distinct Salamander species to establish the organizing activity of the dorsal lip of the blastopore during amphibian gastrulation. Ever since, chimaeras have been central to the elucidation of many developmental mechanisms. Subsequent imaginative transplantation procedures in insects, amphibians, birds, and mammals (2) led to numerous insights on cell lineages, tissue induction, self recognition in the immune system, and sexual differentiation. Furthermore, in some cases, because the grafted mutant cells and/or tissues develop beyond the point that they typically reach in wholly mutant animals, phenotypic analyses can be extended to later stages; thus, chimaeras provide a powerful means of characterizing mutant phenotypes and the affected cell types.
In addition to cell and tissue transplantation experiments, advances in both genetics, to manipulate genomes, and in the use of fluorescent proteins, to detect marked cells, have facilitated the design and creation of mosaic animals, promoting flurries of cell lineage and mutant analysis studies. Because of the ease of performing genetics studies in this system, methods and approaches developed in Drosophila have led the field. Over the years, tools have become increasingly sophisticated, allowing both control of the genotype and detection of mutant cells in a precise spatial and temporal manner. These powerful tools have made possible the study of complex biological processes in vivo in exquisite detail. In particular, they have allowed the analysis of pleiotropic genes, i.e., genes that have multiple functions over time. These genes are particularly difficult to analyze because their roles at later time points may not be detectable because of earlier detrimental functions. Here, we describe the various tools available to Drosophila researchers today and emphasize, in the context of the evolution of the field, the contributions of our group.
Mitotic Recombination to Create Mosaics in the Soma and Germ Line
In 1936 Stern (3) first coined the term “twin spots” to refer to the two homozygous daughter cells generated by mitotic recombination (MR) in a heterozygous animal (Fig. 1). MR events are very rare in wild-type animals, but their frequency can be increased significantly by X-ray irradiation that generates DNA double-stranded breaks (Fig. 1A). Subsequent DNA repair enzyme activity is not always strand specific, leading to crossover exchange of genetic material between homologous chromosomes. Importantly, because the timing of irradiation can be controlled, MR events can be induced at different time points to generate clones of different sizes (the earlier the irradiation, the larger the clone size). Further, because the frequency of events depends upon the number of cells present, analysis of the resultant clone sizes can be used to estimate progenitor numbers and their division rates (e.g., see refs. 4 and 5).
Fig. 1.
Mitotic recombination and generation of twin spots for clonal analyses. (A) X-ray irradiation causes chromosomal breaks and induces MR in the G2 phase of the cell cycle. (B and C) The underlying mechanisms of MR are the same in somatic (B) and germ-line (C) tissues, but the techniques for visualization of induced twin spots are different. MR causes an exchange of chromosomal arms distal to the site of crossing over. All genes downstream of the chromosomal breakpoint are homozygozed. In a heterozygote, the pattern of chromosomal segregation determines genotype. In G2-X segregation the recombined chromosomes migrate to different poles in a 1:3/2:4 configuration. Cytokinesis generates a mosaic fly with homozygous twin spots: one wild-type (+/+) and the other mutant (−/−); the rest of the cells are heterozygous (+/−). All twin spot genotypes shown in this and following figures arise from this type of segregation pattern. (B) In the soma, MR creates within the original heterozygous background multicellular homozygous clones of wild-type and mutant tissue of varying sizes, depending upon the number of further cell divisions. In early clonal analysis studies the homozygous mutant twin spot was identified by external phenotypes such as cuticle color or forked thoracic bristles; later, with the development of fluorescent protein markers, clones were identified within internal organs. (C) The DFS technique. Flies heterozygous and homozygous for DFS do not lay eggs; however, following MR in a heterozygote, double-positive germ cells will develop and produce normal eggs. If a mutation is positioned in trans to DFS, m/m homozygous clones will be generated in the germ line. OvoD1 ovaries degenerate very early, and wild-type eggs are lost. Thus, the only eggs that develop, if m does not interfere with germ-line development, will be homozygous mutants.
MR provides a powerful tool to mark all of the descendants of a single cell during the development of a multicellular organism (Fig. 1B). A number of cell-autonomous cuticle markers such as yellow, forked, and multiple wing hairs have been used extensively to detect twin spots and analyze the viability, cell autonomy, and phenotype of recessive mutations. Importantly, because daughter cells are equivalent in most proliferative tissues, comparison of the size of the wild-type marked twin provides a natural internal control to the mutant twin. Significantly, clonal analysis in imaginal discs of Minute mutations, which slow cell growth by affecting ribosome number, led to the fundamental discovery of compartments [e.g., anterior–posterior or dorsal–ventral (4)], now recognized as embodying a universal organizing principle in the patterning and differentiation of animal embryos and organs.
Apart from cell lineage analysis, such mosaic techniques also are useful to determine where the function of a particular gene is required within a tissue. For example, in the eye, the receptor tyrosine kinase Sevenless is required in photoreceptor 7, whereas its ligand, Bride of Sevenless, is required in photoreceptor 8 (6). Mosaic techniques also allow the determination of whether a gene is required cell-autonomously in the expressing cell or whether it acts nonautonomously. For example, in the Wingless (Wg) pathway, porcupine, which is required for Wg secretion, acts nonautonomously, whereas dishevelled and armadillo, which are required for transduction of the signal downstream of the Frizzled receptor, act cell autonomously (7).
The production of mosaics in the germ line is of special interest for the investigation of both oogenesis and the contribution of maternally synthesized gene products to early zygotic development. Perrimon and Gans (8) developed a simple and efficient method, the dominant female sterile (DFS) technique, for detecting germ-line clones based on the unique properties of the germ line-dependent ovoD1 DFS mutation (Fig. 1C). Although ovoD1/+ germ-line cells fail to produce eggs, double-positive germ-line clones induced in ovoD1/+ females by MR produce normal progeny. Thus, germ-line clones homozygous for a specific mutation (m) can be induced readily in transheterozygous animals for both ovoD1 and m. This approach allowed the characterization of the tissue specificity of female sterile mutations (8) as well as the analysis of the maternal effects of pleiotropic genes associated with zygotic lethality (9, 10). Importantly, because ovoD1 is located on the X chromosome, and other DFS mutations with similar features did not exist, our group transposed the ovoD1 mutation onto autosomal sites to extend the DFS technique to the autosomes, thus allowing the systematic analysis of the maternal effect of autosomal zygotic lethal mutations, among others (11).
The Flippase Recognition Target System: Design and Early Applications
Although X-ray irradiation provides a relatively efficient way to generate MR, the low frequency of events, random targeting of the crossovers along the chromosome, and considerable mortality of treated animals were impediments to the technique. Golic and Lindquist (12) solved these issues by designing an easy, efficient, and neutral way to induce clones based on the Flippase (Flp)-Flp recognition target (FRT) system from the 2-μM plasmid of Saccharomyces cerevisiae (Fig. 2A). They showed that the recombinase Flp could drive recombination at the specific target site of the recombinase, FRT. These recombination events can occur either in cis on the same chromosome arm (12), or in trans across different chromosome arms (13). Key elements of this strategy are its controllability and high inducibility: By putting the recombinase under the control of the heat shock promoter hsp70, recombination is induced by a short temperature pulse that is harmless to the fly.
Fig. 2.
The Flp-FRT method and the PMML strategy. (A) The yeast recombinase Flippase is under the control of the hsp70 promoter in hs-Flp. Heat shock induces Flp expression, which activates recombination specifically at target FRT sites previously inserted into the genome. (B) In the PMML strategy, FRT sites are differentially flanked by a 5′-tubulin promoter on one homologous chromosome and a 3′-Lac-Z on the other. (C) Heat-shock induces MR, creating a mosaic animal with one unmarked twin spot and one twin spot in which a functional fused tubulin promoter-LacZ cds leads to expression of β-galactosidase.
The introduction of the Flp-FRT system allowed the improvement of previous MR strategies and the design of new applications for creating mosaics. In particular, to facilitate the production of germ-line mosaics, we developed the “Flp-DFS technique” (14, 15), allowing efficient generation of germ-line clones by MR on all chromosomal arms. Further, we took the first step toward the development of a direct positive marking technique for clonal analysis, the positively marked mutant lineage (PMML) technique (Fig. 2 B and C) (16). In the initial version, we used MR to fuse the tubulin promoter region on one chromosome with the lacZ-coding region on the other to generate a β-galactoside protein as a marker for the cells that had received the functional tubulin promoter/lac-Z combination. This analysis showed that MR mediated by yeast Flp could be used to generate and mark clones directly in embryonic, larval, and adult internal tissues. In an alternative approach, Struhl and Basler (17) found a clever way to induce clones (although not twins) efficiently using the Flp-FRT system. In the “flp-out” technique, FRT sites are arranged as direct repeats at either end of a segment of DNA containing a transcriptional stop signal, the whole of which is flanked by a 5′ constitutive promoter and a 3′ coding region. Whenever Flp mediates site-specific recombination between the cis-acting FRTs, the intervening DNA containing the stop signal is excised (or “flpped-out”), and the promoter becomes free to drive transcription of the now-fused coding region and, ultimately, expression of a functional protein. Because the change is irreversible and heritable, the dividing cells form multicellular clones that can be identified by ectopic expression of either the Lac-Z marker gene or developmentally important proteins that subsequently can be marked with tagged antibodies. Finally, another important application was the design of Flp-FRT–based genetic screens in which mutant cells, induced in heterozygous animals by the Flp-FRT system, are identified by their lack of expression of a marker gene. Xu and Rubin (18) placed FRT sites at the base of all Drosophila chromosome arms as well as cell-autonomous markers that localize to either the nucleus or membrane, allowing the clonal analysis of almost all Drosophila genes during development and in dividing adult tissues.
The Binary Gal4-Upstream Activating System to Control Gene Expression Spatially and Temporally
To complement loss-of-function studies, we adapted the yeast Gal4-upstream activating system (UAS) transcriptional activation system to drive targeted gene expression in the fly (Fig. 3) (19). In yeast, induction of the GAL structural genes by galactose is dependent on the transcriptional activator Gal4 that operates through a UAS. Gal4 selectively activates any gene-coding sequence (cds) that has been cloned downstream of UAS, conferring upon it the tissue, time, and position specificity of its own promoter/enhancer. As shown in Fig. 3B, if UAS-GFP and UAS-gene X previously have been inserted into the background, Gal4 drives coexpression of GFP and gene X to mark clones specifically for phenotypic screening. An important genetic aspect of the system (Fig. 3A) is that the physical separation of the Gal4 transactivator from its target UAS-cds construct into two distinct transgenic fly lines assures that there is minimal leakiness of expression: The target gene, which is present in one fly line, is silent in the absence of its activator; the activator protein is present in the other line but has no gene to activate. Consequently, in the absence of any significant baseline expression, there is no selective pressure, so the lines can be maintained indefinitely as laboratory stocks. The method also offers a significant technical advantage in that construction of a single Gal4 “driver” line exhibiting interesting patterns of expression can be used to drive the expression of any number of cds, each represented as an individual fly line. Thousands of such target UAS-cds lines as well as Gal4 driver lines are available to the scientific community (http://flystocks.bio.indiana.edu/). In addition, two approaches that allow control of Gal4 activity are available. One is the repression of Gal4 activity by the use of a natural repressor of Gal4 known as Gal80, which later was mutated to a more labile, temperature-sensitive derivative (20) (Fig. 3C). GeneSwitch Gal4, whereby the expression of UAS effector lines is controlled by a chimeric Gal4 protein that becomes active in the presence of the steroid RU486 (21), is the second.
Fig. 3.
The Gal4 binary expression system expands levels of control. (A) The genetic components of the Gal4-UAS system are present in two distinct transgenic fly lines so that leaky expression and potential genetic selection are avoided. The driver and target UAS fly lines are crossed, and the full complement of genetic elements comes together exclusively in the progeny genome. (B) In the first phase of binary expression, the Gal4 promoter determines the time and cell specificity of transcription. After translation, Gal4 protein binds to UAS sequences placed upstream of any and all cds, causing their coordinate expression, so a single UAS-marker reveals the specific expression pattern of all UAS-linked sequences. The box above the colorless cell shows UAS-target sequences inserted into the background. (C) A tertiary level of control is possible through the use of a specific Gal4 repressor, Gal80TS, which is temperature sensitive. At lower temperatures, the repressor is active, and there is no Gal4 transcription; at higher temperatures, the repressor cannot function, and the active repressor pool becomes depleted. Gal4 transcription progressively resumes, and binary expression is reactivated.
Gal4-UAS and Flp-FRT—a Powerful Combination
Combining the Gal4-UAS and Flp-FRT systems opened the door to the development of a flurry of imaginative methods for creating mosaics (22) and illuminated how the use of cross-species components considerably enhances genetic manipulation in existing models. Importantly, the use of GFP from the jellyfish Aequoria victoria (23) and of other fluorescent proteins allowed the easy visualization of labeled cells in mosaics.
In particular, we developed a directed mosaic system by using the Gal4 system to control the expression of Flp in a spatial and temporal fashion (24), thereby allowing mosaic screens to be restricted to specific tissues. Lee and Luo (25) developed the mosaic analysis with a repressible cell marker (MARCM) system that uses Gal80. With MARCM, MR is used to segregate away both copies of a constitutively active form of the repressor gene to one twin cell, thus relieving the Gal4 repression in the other. In the latter twin cell, lack of turnover eventually depletes it of its repressor Gal80 pool, allowing reactivation of the binary system; if a UAS-GFP construct has been placed in the genetic background, this twin is uniquely marked with green fluorescence. The system can be used to generate labeled MARCM clones that are homozygous for a specific mutation. The temperature-sensitive form of the repressor makes possible an additional level of control, allowing inducible inhibition of its function by a rise in temperature. Finally, an interesting method, the split Gal4 system (26) permits Gal4 activation of UAS constructs only at the intersection of two expression domains. One construct expresses a Gal4 DNA-binding domain–leucine zipper motif fusion protein, and the other expresses a leucine zipper motif–transcriptional activation domain fusion protein. Gal4 activation of UAS constructs will occur only in cells where both constructs are expressed and where the two fusion proteins can dimerize via their leucine zipper motifs.
Gal4 and Transgenic RNAi: A Perfect Marriage
Following the initial discovery by Fire et al. (27) that dsRNAs provide an effective knock-down approach, many groups explored the possibility of using the Gal4-UAS system to target RNAi reagents to specific cells and tissues (28). RNAi reagents consist of either long dsRNAs that are processed into siRNAs or small hairpin microRNA-based (shRNA) RNAi constructs that generate a single siRNA. Importantly, RNAi constructs are most effective when targeted to integration sites in the genome that are optimal for transgene expression through use of the phiC31-mediated site-specific integration approach (29). Our group in particular characterized a number of genomic sites that provide high levels of gene expression (30), developed a number of vectors that provide maximum expression (31, 32), and produced a large-scale resource of transgenic shRNA lines (33). Large-scale screens in the soma (34) and the germ line (35, 36) now are performed easily simply by crossing a Gal4 line of interest with UAS-RNAi lines, which now number in the tens of thousands (28). The progeny from each individual cross then can be screened for specific RNAi-induced phenotypes (Fig. 4 A and B). Importantly, we showed that shRNAs are more efficient than long dsRNAs for RNAi, particularly in the female germ line (33). This last feature allowed large screens to identify genes expressed during oogenesis that are required for germ-line or embryonic development (Fig. 4C) (36).
Fig. 4.
Somatic and germ-line RNAi screens. (A and B) Experimental protocol for large-scale in vivo screens in which RNAi induces systematic knockdown of gene expression. (A) RNAi screens are initiated by systematically crossing virgin females from a single Gal4 driver line with males from individual UAS target lines, each carrying a specific UAS-RNAi (either dsRNA or shRNA) directed against a known gene. The promoter of Gal4 specifies the time and tissue specificity of Gal4 expression in the driver line, but transactivation of target UAS-RNAi can occur only when both elements are united in the progeny genome. Gal4 > RNAi, Gal4 activation of dsRNA or shRNA transcription. (B) Systematic Gal4-driven expression of RNAis induces knockdown of target gene expression to allow systematic phenotypic characterization of the progeny of each cross. (C) Examples of embryonic phenotypes detected from germ-line screens using shRNAs (35, 36). Adapted from ref. 35.
Recent Tools for Clonal Analysis and Other Binary Systems
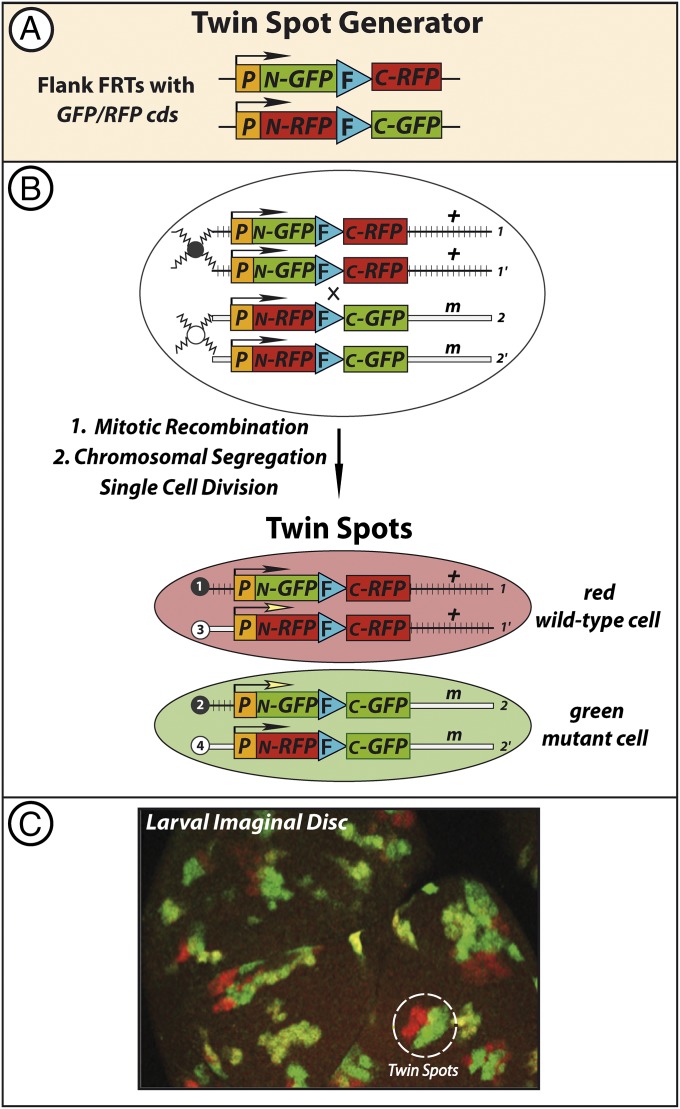
Genetically engineered systems continue to evolve and provide powerful new tools and approaches for cell lineage analyses. The Gal4 technique for real-time and clonal expression (G-TRACE) labels cells after Gal4-dependent excision of an FRT cassette that was placed to stop the expression of a reporter 1 before flp-out, whereas Gal4 expression is revealed with an alternate reporter 2 (37). Twin-spot MARCM labels the homozygous daughter cells, derived from MR, because of the differential loss of either repressor 1 or repressor 2 (38). In addition, our group developed the twin-spot generator (TSG) that labels cells after interchromosomal recombination (Fig. 5) (39). TSG generates positively labeled clones without the use of a repressor, thus minimizing the lag time between clone induction and appearance of label. In TSG the FRT site is flanked with complementary, hybrid sequences encoding GFP and RFP (Fig. 5A). After MR, full-length GFP and RFP are reconstructed by the chromosomal exchange (Fig. 5B). The FRT site is located within an intron so that when the corresponding primary RNAs undergo splicing all traces of the interrupting intron are erased, and a normal GFP or RFP functional protein generates differential fluorescence signals in the twin daughter cells (Fig. 5C). The system can be used to generate marked mutant clones (e.g., to evaluate their growth rates compared with the twin wild-type control).
Fig. 5.
TSG differential labeling of twin spots. (A) TSG is a Flp-FRT–based MR strategy, as in Fig. 2, except that the FRT site, located in an intron, is flanked with complementary GFP and RFP sequences to create TSG hybrid cassettes (Materials and Methods). (B) Heat shock-induced MR generates full-length but interrupted cds for GFP on one recombined chromosome and for RFP on the other. Splicing removes the FRT site along with the rest of the intron to reconstruct continuous full-length GFP or RFP cds. In a heterozygote, placing the wild-type allele distal to the 3′ RFP and the mutant allele distal to the 3′ GFP enables direct identification of both the red wild-type control and green mutant twins. (C) TSG provides a direct readout of toxicity. In control experiments in which both twin spots are homozygous wild type, statistical analysis showed equal numbers of cells present in the red and green twin spots, as expected and confirming that ectopic expression of GFP or RFP had no apparent toxic effects (compare with Fig. 6C). Adapted from ref. 39.
Success with the Gal4/UAS/Gal80 repressible binary expression system together with the need to manipulate or mark different cell populations motivated the development of additional binary systems to control gene expression. Two systems have been developed recently based on LexA from bacteria and QF from Neurospora (40, 41). LexA binds to specific sequences called lexA operator (lexO), and a number of tools allowing the use of LexA together with the Gal4 system were introduced to allow various mosaic manipulations. The Q-system consists of three components: the QF transcription factor, the QS suppressor, and a QUAS effector. QUAS effectors show robust expression in the presence of QF, an expression that can be suppressed by ubiquitous expression of QS. Importantly, temporal control of the Q-system is possible by feeding flies quinic acid, an inhibitor of the QS suppressor.
Future Directions: Clonal Screens in Mosaic animals
Building on TSG, we currently are extending the method to control gene expression in one or both daughter cells by developing mosaic analysis protocols that progressively integrate the Gal4 and Q binary expression systems. The underlying principle is the same as in TSG: We induce MR via the Flp-FRT system in hybrid cassettes to reconstruct full-length cds, but in this approach the TSG cassettes are composed of complementary sequences from either Gal4/RFP or Gal4/Q. In the Gal4/RFP version, one twin expresses RFP directly, and the other expresses Gal4, which drives expression of a UAS-GFP cds inserted on a different chromosome (Fig. 6 A and B, Left). In a proof-of-principle experiment with only the UAS-GFP marker sequence present, we tested a hybrid Gal4 sequence (Gal4-VP16). Gal4-VP16 is a potent transcriptional activator that contains the Gal4-specific DNA-binding domain and the phage λ VP-16 transcription activator domain (42). As shown in Fig. 6C, both red and green fluorescing clones were readily detectable, demonstrating that the Gal4 binary system is functional in TSG; however, we also observed that the twin spots were not of equal size: The red clones were larger and in several instances showed no corresponding green twin spot. We attribute this difference to the inherent toxicity of the Gal4-VP16 protein when expressed at high levels. These results demonstrate that TSG renders differences in cell viability (via clone size) directly visible and quantifiable.
Fig. 6.
Extension of the TSG method to control differential gene expression in each twin spot cell. (A) As in Fig. 5, except that we replaced the GFP sequences with those of Gal4 to create TSG Gal-RFP hybrid cassettes (Left) and additionally replaced the RFP sequences with those of the Neurospora crassa QF transactivator to create TSG Gal-QF hybrid cassettes (Right). (B) Boxes at the top of the panel show the UAS target sequences that were inserted into one or the other of the TSG parental lines before TSG fly crosses. MR proceeds as in Fig. 1, except that both the RNAi-induced mutation and green marker fluorescence are generated indirectly by Gal4 activation of the UAS-RNAi and UAS-GFP sequences, respectively (Left), and the red marker fluorescence of the internal wild-type control twin is generated indirectly by QF activation of the QUAS-mTomato, a strong red-fluorescing marker (Right). (C) Proof-of-principle and toxicity assay. Only UAS-GFP is present in the background. Both red and green clones are visible, demonstrating that the Gal4-UAS binary expression system works in TSG. However, Gal4-VP16 expression affects clone size, presumably because of its toxicity.
In future applications of TSG, we anticipate that a combination of the Gal4 and Q transcription systems with an MR-dependent induction of differentially marked clones will provide a maximally flexible system for the control of gene expression in each of the twin clones (Fig. 6 A and B, Right), particularly because we can establish baseline toxicity in the same TSG assay. This system will have a number of applications, such as identifying reciprocal signaling events between normal and mutant cell populations during developmental and pathological processes.
Materials and Methods
The TSG strategy and methods used for Gal-RFP (Fig. 6) are described in Griffin et al. (39). Specifically, for TSG plasmid construction/integration strategy, see supplementary figure 1 in ref. 39. For G2-Z segregation, see supplementary figure 4 in ref. 39. For hybrid partner 5′ and 3′ GFP and RFP nucleotide sequences, see supplementary table 1 in ref. 39. For primers for GFP/RFP hybrid construction, see supplementary table 2 in ref. 39. For clone cell counts for figure 5 in ref. 39, see supplementary table 5 in ref. 39. The Gal4-VP16 sequence is identical to GAL4 fusion protein [Flexi Vector pFN11A (BIND)] GenBank ID: ABG01697.1 from amino acids 1–146 and, after a single valine residue linker, to amino acids 411–490 of transactivating tegument protein VP16 (Human herpesvirus 1) GenBank ID: ADD60084.1. To make hybrid Gal4/RFP and RFP/Gal4 hybrid cassettes, the Gal4-VP16 sequence was split at nucleotide 329.
Acknowledgments
We thank M. Dolega and S. Porte for advice on graphics, D. Yan for help in generating Fig. 6C, and C. Villalta for technical help. N.P. thanks all past and current members of the N.P. laboratory who contributed to the development of the methods described in this review. R.G. is funded by the Biomics Laboratory (Director X. Gidrol), Unité Mixte Centre National de la Recherche Scientifique FR3425, Institut National de la Santé et de la Recherche Médicale U1038, and Commissariat à l’Energie Atomique. N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Spemann H, Mangold H. Über induktion von Embryonalagen durch Implantation Artfremder Organisatoren. Roux'. Arch Entwicklungsmech Org. 1924;100:599–638. [Google Scholar]
- 2.LeDouarin N, McLaren A. Chimeras in Developmental Biology. London: Academic; 1984. [Google Scholar]
- 3.Stern C. Somatic Crossing over and Segregation in Drosophila Melanogaster. Genetics. 1936;21(6):625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Bellido A, Ripoll P, Morata G. Developmental compartmentalization in the dorsal mesothoracic disc of Drosophila. Dev Biol. 1976;48(1):132–147. doi: 10.1016/0012-1606(76)90052-x. [DOI] [PubMed] [Google Scholar]
- 5.Wieschaus E, Szabad J. The development and function of the female germ line in Drosophila melanogaster: A cell lineage study. Dev Biol. 1979;68(1):29–46. doi: 10.1016/0012-1606(79)90241-0. [DOI] [PubMed] [Google Scholar]
- 6.Cagan RL, Krämer H, Hart AC, Zipursky SL. The bride of sevenless and sevenless interaction: Internalization of a transmembrane ligand. Cell. 1992;69(3):393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- 7.Siegfried E, Wilder EL, Perrimon N. Components of wingless signalling in Drosophila. Nature. 1994;367(6458):76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- 8.Perrimon N, Gans M. Clonal analysis of the tissue specificity of recessive female-sterile mutations of Drosophila melanogaster using a dominant female-sterile mutation Fs(1)K1237. Dev Biol. 1983;100(2):365–373. doi: 10.1016/0012-1606(83)90231-2. [DOI] [PubMed] [Google Scholar]
- 9.Perrimon N, Engstrom L, Mahowald AP. The effects of zygotic lethal mutations on female germ-line functions in Drosophila. Dev Biol. 1984;105(2):404–414. doi: 10.1016/0012-1606(84)90297-5. [DOI] [PubMed] [Google Scholar]
- 10.Perrimon N, Engstrom L, Mahowald AP. Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics. 1989;121(2):333–352. doi: 10.1093/genetics/121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119(4):1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 12.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59(3):499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 13.Golic KG. Site-specific recombination between homologous chromosomes in Drosophila. Science. 1991;252(5008):958–961. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 14.Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131(3):643–653. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144(4):1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison DA, Perrimon N. Simple and efficient generation of marked clones in Drosophila. Curr Biol. 1993;3(7):424–433. doi: 10.1016/0960-9822(93)90349-s. [DOI] [PubMed] [Google Scholar]
- 17.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72(4):527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 18.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117(4):1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 19.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 20.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 21.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible Gal4. Proc Natl Acad Sci. 2001;98(22):12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy JB. GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis. 2002;34(1-2):1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 23.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 24.Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125(12):2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- 25.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24(5):251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 26.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52(3):425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 28.Perrimon N, Ni JQ, Perkins L. 2010. In vivo RNAi: Today and Tomorrow. Cold Spring Harb Perspect Biol 2(8):a003640.
- 29.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni JQ, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni JQ, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182(4):1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni JQ, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8(5):405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 35.Staller MV, et al. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193(1):51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan D, et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev Cell. 2014;28(4):459–473. doi: 10.1016/j.devcel.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans CJ, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6(8):603–5. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat Neurosci. 2009;12(7):947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffin R, et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods. 2009;6(8):600–602. doi: 10.1038/nmeth.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagi R, Mayer F, Basler K. Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc Natl Acad Sci USA. 2010;107(37):16166–16171. doi: 10.1073/pnas.1005957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: A repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141(3):536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]